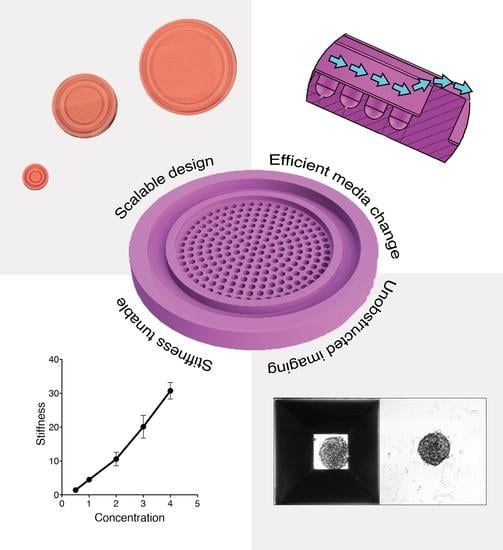
An Improved Scalable Hydrogel Dish for Spheroid Culture
Abstract
:1. Introduction
2. Materials and Methods
2.1. 3D Printing and Silicone Casting
2.2. Hydrogel Casting
2.3. Young’s Moduli Derivation
2.4. Media Change Simulation
2.5. Media Formulations
2.6. Cell and Spheroid Culture
2.7. Effects of Agarose Stiffness in Spheroid Formation
2.8. Confocal Microscopy
2.9. Multispectral Imaging Flow Cytometry
2.10. TGFβ-Induced Epithelial-to-Mesenchymal Transition
2.11. Porcine and Murine Primary Cell Isolation and 3D Culture
2.12. Image and Statistical Analyses
3. Results
3.1. Scalable Mold Design
3.2. Effect of Hydrogel Stiffness in Spheroid Formation
3.3. Comparing Spheroid Formation in Our Agarose Dish Compared to the Aggrewell™800 Plates
3.4. Inhibition of EMT in the Spheroid Migration Assay
3.5. Scaffold-Free Primary-Derived Spheroid Formation
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Witt Hamer, P.C.; Van Tilborg, A.A.; Eijk, P.P.; Sminia, P.; Troost, D.; Van Noorden, C.J.; Ylstra, B.; Leenstra, S. The genomic profile of human malignant glioma is altered early in primary cell culture and preserved in spheroids. Oncogene 2008, 27, 2091–2096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, J.; Zhang, X.; Xia, X.; Han, M.; Li, F.; Li, C.; Li, Y.; Gao, D. Organoid technology for tissue engineering. J. Mol. Cell Biol. 2020, 12, 569–579. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, T.; Sato, T. Advancing Intestinal Organoid Technology toward Regenerative Medicine. Cell Mol. Gastroenterol. Hepatol. 2018, 5, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Sachs, N.; Papaspyropoulos, A.; Zomer-van Ommen, D.D.; Heo, I.; Bottinger, L.; Klay, D.; Weeber, F.; Huelsz-Prince, G.; Iakobachvili, N.; Amatngalim, G.D.; et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 2019, 38, e100300. [Google Scholar] [CrossRef]
- Bresciani, G.; Hofland, L.J.; Dogan, F.; Giamas, G.; Gagliano, T.; Zatelli, M.C. Evaluation of Spheroid 3D Culture Methods to Study a Pancreatic Neuroendocrine Neoplasm Cell Line. Front. Endocrinol. 2019, 10, 682. [Google Scholar] [CrossRef] [PubMed]
- Panek, M.; Grabacka, M.; Pierzchalska, M. The formation of intestinal organoids in a hanging drop culture. Cytotechnology 2018, 70, 1085–1095. [Google Scholar] [CrossRef] [Green Version]
- Vinci, M.; Gowan, S.; Boxall, F.; Patterson, L.; Zimmermann, M.; Court, W.; Lomas, C.; Mendiola, M.; Hardisson, D.; Eccles, S.A. Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol. 2012, 10, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Folkman, J.; Moscona, A. Role of cell shape in growth control. Nature 1978, 273, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, J.; Seidel, C.; Ebner, R.; Kunz-Schughart, L.A. Spheroid-based drug screen: Considerations and practical approach. Nat. Protoc. 2009, 4, 309–324. [Google Scholar] [CrossRef]
- Gao, W.; Wu, D.; Wang, Y.; Wang, Z.; Zou, C.; Dai, Y.; Ng, C.F.; Teoh, J.Y.C.; Chan, F.L. Development of a novel and economical agar-based non-adherent three-dimensional culture method for enrichment of cancer stem-like cells. Stem Cell Res. Ther. 2018, 9, 243. [Google Scholar] [CrossRef]
- Zanoni, M.; Piccinini, F.; Arienti, C.; Zamagni, A.; Santi, S.; Polico, R.; Bevilacqua, A.; Tesei, A. 3D tumor spheroid models for in vitro therapeutic screening: A systematic approach to enhance the biological relevance of data obtained. Sci. Rep. 2016, 6, 19103. [Google Scholar] [CrossRef]
- Chen, M.; Vial, M.L.; St John, J.A. 3D-tips: User-friendly mesh barrier pipette tips for 3D-spheroid culture. J. Biol. Eng. 2019, 13, 80. [Google Scholar] [CrossRef]
- Leung, B.M.; Lesher-Perez, S.C.; Matsuoka, T.; Moraes, C.; Takayama, S. Media additives to promote spheroid circularity and compactness in hanging drop platform. Biomater. Sci. 2015, 3, 336–344. [Google Scholar] [CrossRef] [Green Version]
- Drakhlis, L.; Biswanath, S.; Farr, C.M.; Lupanow, V.; Teske, J.; Ritzenhoff, K.; Franke, A.; Manstein, F.; Bolesani, E.; Kempf, H.; et al. Human heart-forming organoids recapitulate early heart and foregut development. Nat. Biotechnol. 2021. [Google Scholar] [CrossRef]
- Li, W.; Germain, R.N.; Gerner, M.Y. High-dimensional cell-level analysis of tissues with Ce3D multiplex volume imaging. Nat. Protoc. 2019, 14, 1708–1733. [Google Scholar] [CrossRef] [PubMed]
- Warr, C.; Valdoz, J.C.; Bickham, B.P.; Knight, C.J.; Franks, N.A.; Chartrand, N.; Van Ry, P.M.; Christensen, K.A.; Nordin, G.P.; Cook, A.D. Biocompatible PEGDA Resin for 3D Printing. ACS Appl. BioMater. 2020, 3, 2239–2244. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibish, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Napolitano, A.P.; Dean, D.M.; Man, A.J.; Youssef, J.; Ho, D.N.; Rago, A.P.; Lech, M.P.; Morgan, J.R. Scaffold-free three-dimensional cell culture utilizing micromolded nonadhesive hydrogels. Biotechniques 2007, 43, 494–500. [Google Scholar] [CrossRef]
- Maritan, S.M.; Lian, E.Y.; Mulligan, L.M. An Efficient and Flexible Cell Aggregation Method for 3D Spheroid Production. J. Vis. Exp. 2017. [Google Scholar] [CrossRef] [PubMed]
- Saleh, F.; Harb, A.; Soudani, N.; Zaraket, H. A three-dimensional A549 cell culture model to study respiratory syncytial virus infections. J. Infect. Public Health 2020, 13, 1142–1147. [Google Scholar] [CrossRef]
- George, T.C.; Basiji, D.A.; Hall, B.E.; Lynch, D.H.; Ortyn, W.E.; Perry, D.J.; Seo, M.J.; Zimmerman, C.A.; Morrissey, P.J. Distinguishing modes of cell death using the ImageStream multispectral imaging flow cytometer. Cytometry A 2004, 59, 237–245. [Google Scholar] [CrossRef]
- Koopman, G.; Reutelingsperger, C.P.; Kuijten, G.A.; Keehnen, R.M.; Pals, S.T.; van Oers, M.H. Annexin V for flow cytometric detection of phosphatidylserine expression on B cells undergoing apoptosis. Blood 1994, 84, 1415–1420. [Google Scholar] [CrossRef] [Green Version]
- Dongre, A.; Weinberg, R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef]
- Skrypek, N.; Goossens, S.; De Smedt, E.; Vandamme, N.; Berx, G. Epithelial-to-Mesenchymal Transition: Epigenetic Reprogramming Driving Cellular Plasticity. Trends Genet. 2017, 33, 943–959. [Google Scholar] [CrossRef]
- Vinci, M.; Box, C.; Zimmermann, M.; Eccles, S.A. Tumor spheroid-based migration assays for evaluation of therapeutic agents. Methods Mol. Biol. 2013, 986, 253–266. [Google Scholar]
- Grygielko, E.T.; Martin, W.M.; Tweed, C.; Thornton, P.; Harling, J.; Brooks, D.P.; Laping, N.J. Inhibition of gene markers of fibrosis with a novel inhibitor of transforming growth factor-beta type I receptor kinase in puromycin-induced nephritis. J. Pharmacol. Exp. Ther. 2005, 313, 943–951. [Google Scholar] [CrossRef] [Green Version]
- Avizienyte, E.; Fincham, V.J.; Brunton, V.G.; Frame, M.C. Src SH3/2 domain-mediated peripheral accumulation of Src and phospho-myosin is linked to deregulation of E-cadherin and the epithelial-mesenchymal transition. Mol. Biol. Cell 2004, 15, 2794–2803. [Google Scholar] [CrossRef] [Green Version]
- Smyrek, I.; Mathew, B.; Fischer, S.C.; Lissek, S.M.; Becker, S.; Stelzer, E.H.K. E-cadherin, actin, microtubules and FAK dominate different spheroid formation phases and important elements of tissue integrity. Biol. Open 2019, 8. [Google Scholar] [CrossRef] [Green Version]
- Fatehullah, A.; Tan, S.H.; Barker, N. Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 2016, 18, 246–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Koo, B.K.; Knoblich, J.A. Human organoids: Model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020, 21, 571–584. [Google Scholar] [CrossRef]
- Garreta, E.; Prado, P.; Tarantino, C.; Oria, R.; Fanlo, L.; Marti, E.; Zalvidea, D.; Trepat, X.; Roca-Cusachs, P. Gavaldà-Navarro, A.; et al. Fine tuning the extracellular environment accelerates the derivation of kidney organoids from human pluripotent stem cells. Nat. Mater. 2019, 18, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.; Tan, W.J.; Pek, M.M.X.; Tan, M.H.; Kurisawa, M. Mechanically and chemically defined hydrogel matrices for patient-derived colorectal tumor organoid culture. Biomaterials 2019, 219, 119400. [Google Scholar] [CrossRef] [PubMed]
- Costa, E.C.; Moreira, A.F.; de Melo-Diogo, D.; Gaspar, V.M.; Carvalho, M.P.; Correia, I.J. 3D tumor spheroids: An overview on the tools and techniques used for their analysis. Biotechnol. Adv. 2016, 34, 1427–1441. [Google Scholar] [CrossRef]
- Gyorfi, A.H.; Matei, A.E.; Distler, J.H.W. Targeting TGF-beta signaling for the treatment of fibrosis. Matrix Biol. 2018, 68–69, 8–27. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, I.E.; Eickelberg, O. The impact of TGF-beta on lung fibrosis: From targeting to biomarkers. Proc. Am. Thorac. Soc. 2012, 9, 111–116. [Google Scholar] [CrossRef]
- Higashiyama, H.; Yoshimoto, D.; Kaise, T.; Matsubara, S.; Fujiwara, M.; Kikkawa, H.; Asano, S.; Kinoshita, M. Inhibition of activin receptor-like kinase 5 attenuates bleomycin-induced pulmonary fibrosis. Exp. Mol. Pathol. 2007, 83, 39–46. [Google Scholar] [CrossRef]
- Smoktunowicz, N.; Alexander, R.E.; Franklin, L.; Williams, A.E.; Holman, B.; Mercer, P.F.; Jarai, G.; Scotton, C.J.; Chambers, R.C. The anti-fibrotic effect of inhibition of TGFbeta-ALK5 signalling in experimental pulmonary fibrosis in mice is attenuated in the presence of concurrent gamma-herpesvirus infection. Dis. Model. Mech. 2015, 8, 1129–1139. [Google Scholar]
- Guerra, F.S.; Oliveira, R.G.; Fraga, C.A.M.; Mermelstein, C.D.S.; Fernandes, P.D. ROCK inhibition with Fasudil induces beta-catenin nuclear translocation and inhibits cell migration of MDA-MB 231 human breast cancer cells. Sci. Rep. 2017, 7, 13723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knipe, R.S.; Probst, C.K.; Lagares, D.; Franklin, A.; Spinney, J.J.; Brazee, P.L.; Grasbereger, P.; Zhang, L.; Black, K.E.; Sakai, N.; et al. The Rho Kinase Isoforms ROCK1 and ROCK2 Each Contribute to the Development of Experimental Pulmonary Fibrosis. Am. J. Respir. Cell Mol. Biol. 2018, 58, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Huang, X.; Hecker, L.; Kurundkar, D.; Kurundkar, A.; Liu, H.; Jin, T.H.; Sesai, L.; Bernard, K.; Thannickal, V.J. Inhibition of mechanosensitive signaling in myofibroblasts ameliorates experimental pulmonary fibrosis. J. Clin. Investig. 2013, 123, 1096–1108. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Mun, H.; Sung, C.O.; Cho, E.J.; Jeon, H.J.; Chun, S.M.; Jung, D.J.; Shin, T.H.; Jeong, G.S.; Kim, D.K.; et al. Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat. Commun. 2019, 10, 3991. [Google Scholar] [CrossRef] [PubMed]
- Driehuis, E.; Kretzschmar, K.; Clevers, H. Establishment of patient-derived cancer organoids for drug-screening applications. Nat. Protoc. 2020, 15, 3380–3409. [Google Scholar] [CrossRef]
- Boretto, M.; Maenhoudt, N.; Luo, X.; Hennes, A.; Boeckx, B.; Bui, B.; Heremans, R.; Perneel, L.; Kobayashi, H.; Zundert, I.V.; et al. Patient-derived organoids from endometrial disease capture clinical heterogeneity and are amenable to drug screening. Nat. Cell Biol. 2019, 21, 1041–1051. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valdoz, J.C.; Jacobs, D.J.; Cribbs, C.G.; Johnson, B.C.; Hemeyer, B.M.; Dodson, E.L.; Saunooke, J.A.; Franks, N.A.; Poulson, P.D.; Garfield, S.R.; et al. An Improved Scalable Hydrogel Dish for Spheroid Culture. Life 2021, 11, 517. https://doi.org/10.3390/life11060517
Valdoz JC, Jacobs DJ, Cribbs CG, Johnson BC, Hemeyer BM, Dodson EL, Saunooke JA, Franks NA, Poulson PD, Garfield SR, et al. An Improved Scalable Hydrogel Dish for Spheroid Culture. Life. 2021; 11(6):517. https://doi.org/10.3390/life11060517
Chicago/Turabian StyleValdoz, Jonard Corpuz, Dallin J. Jacobs, Collin G. Cribbs, Benjamin C. Johnson, Brandon M. Hemeyer, Ethan L. Dodson, Jordan A. Saunooke, Nicholas A. Franks, Peter Daniel Poulson, Seth R. Garfield, and et al. 2021. "An Improved Scalable Hydrogel Dish for Spheroid Culture" Life 11, no. 6: 517. https://doi.org/10.3390/life11060517