Natural Product Gene Clusters in the Filamentous Nostocales Cyanobacterium HT-58-2
Abstract
:1. Introduction
2. Materials and Methods
3. Results
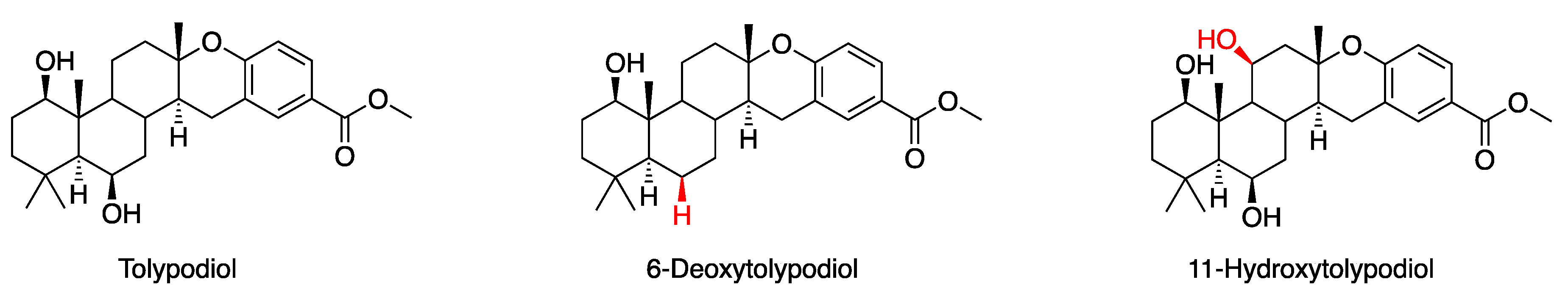
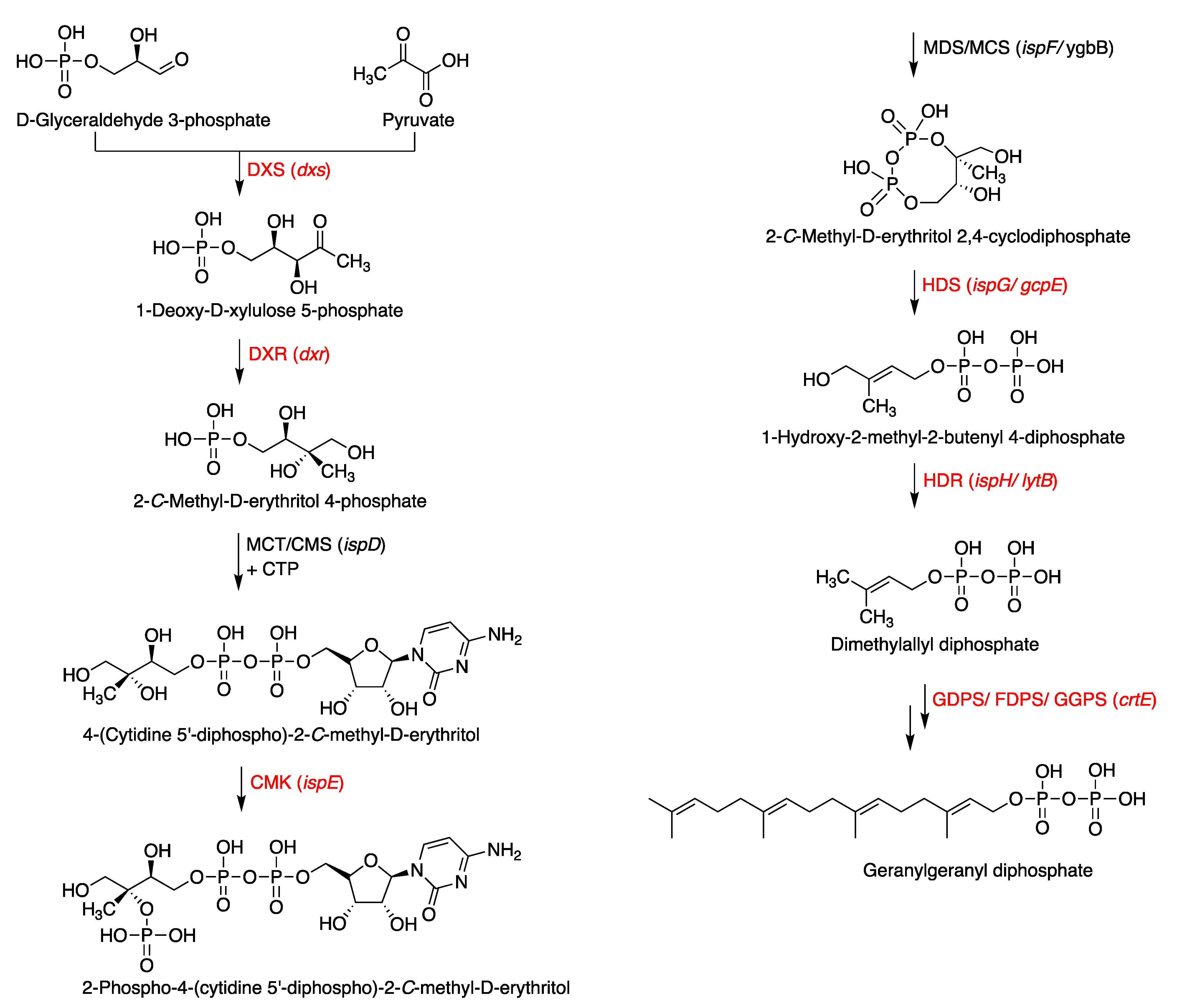
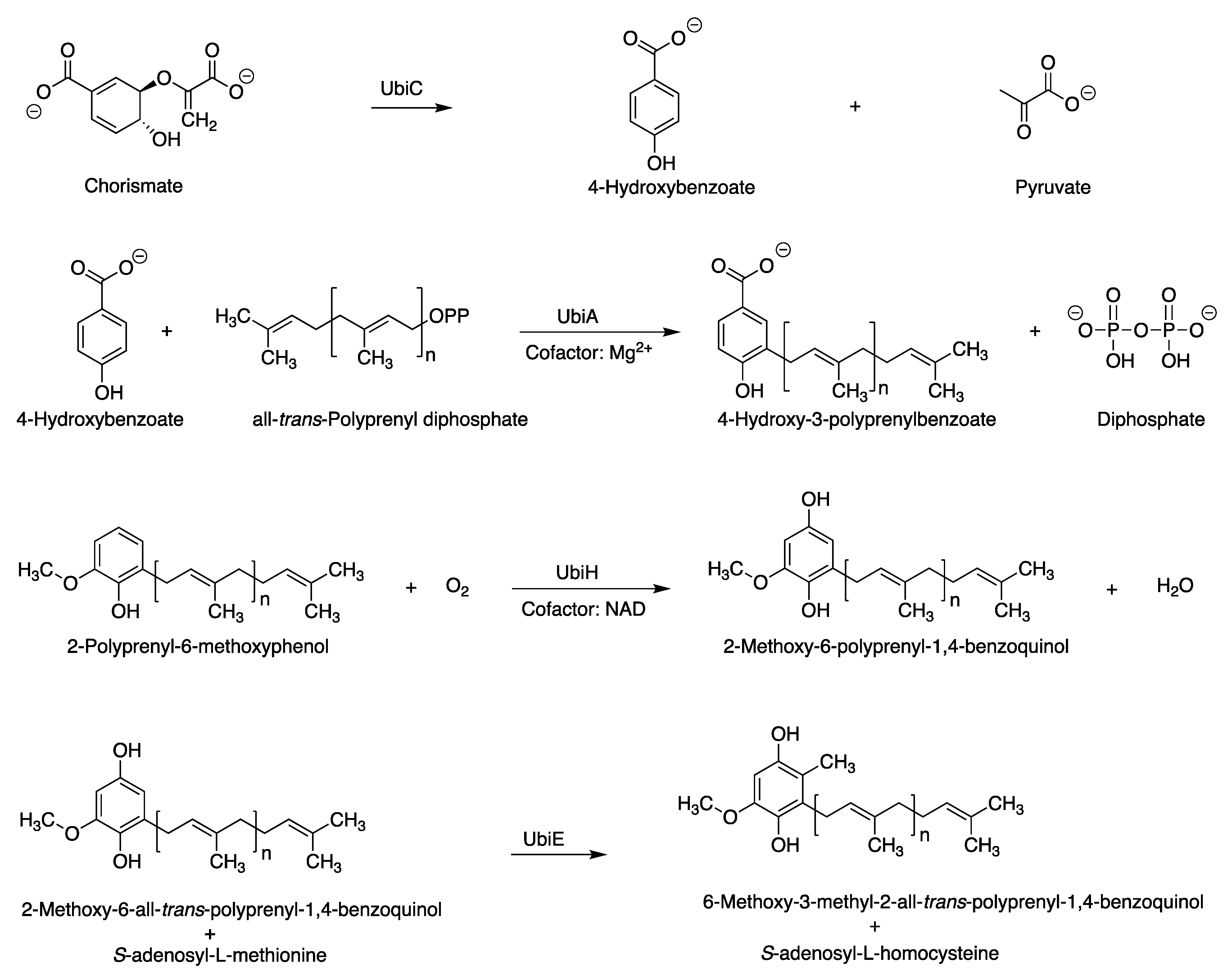
3.1. Putative Tolypodiols BGC
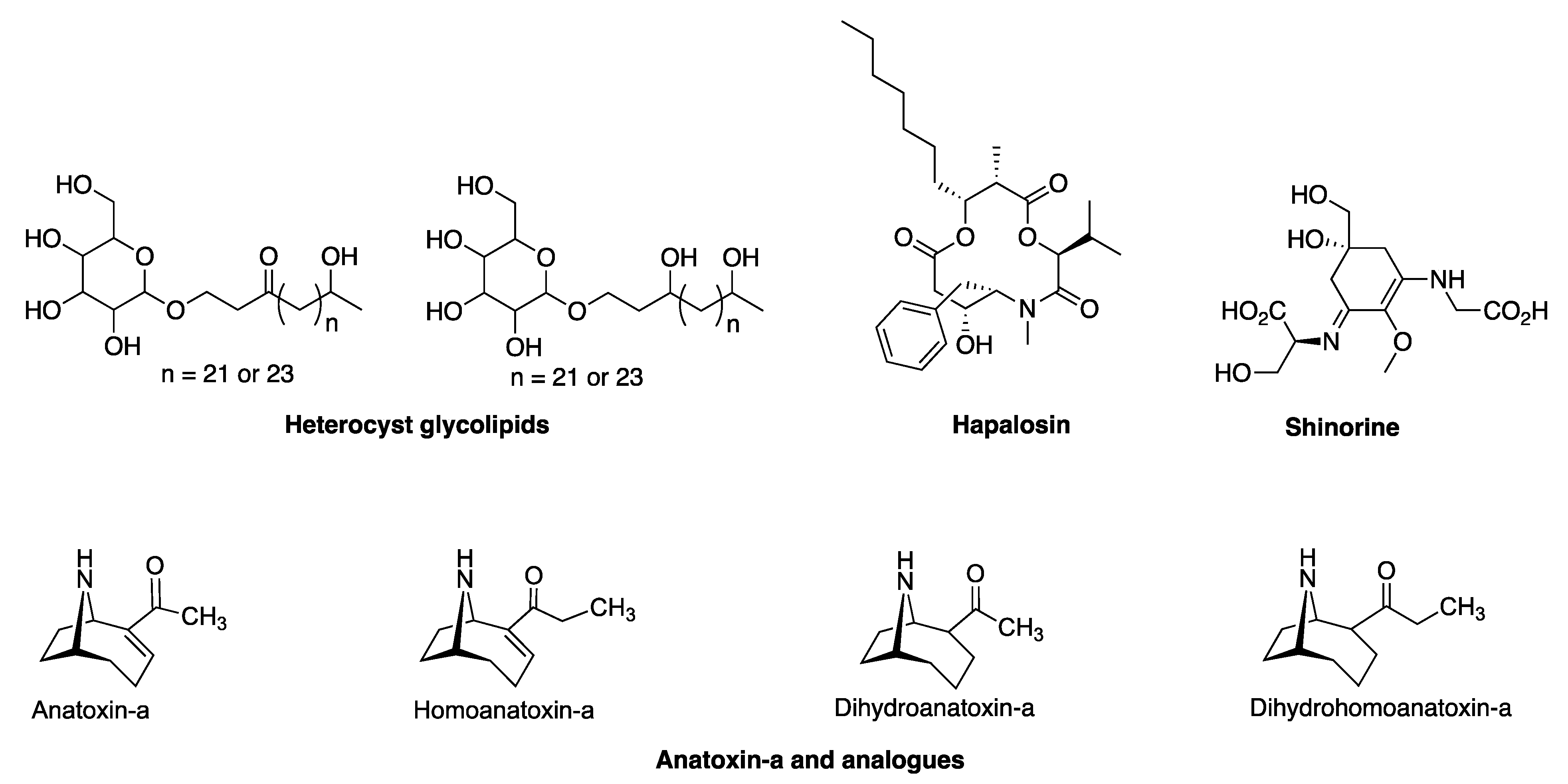
3.2. BGCs for Diverse Natural Products
3.2.1. Hapalosin BGC
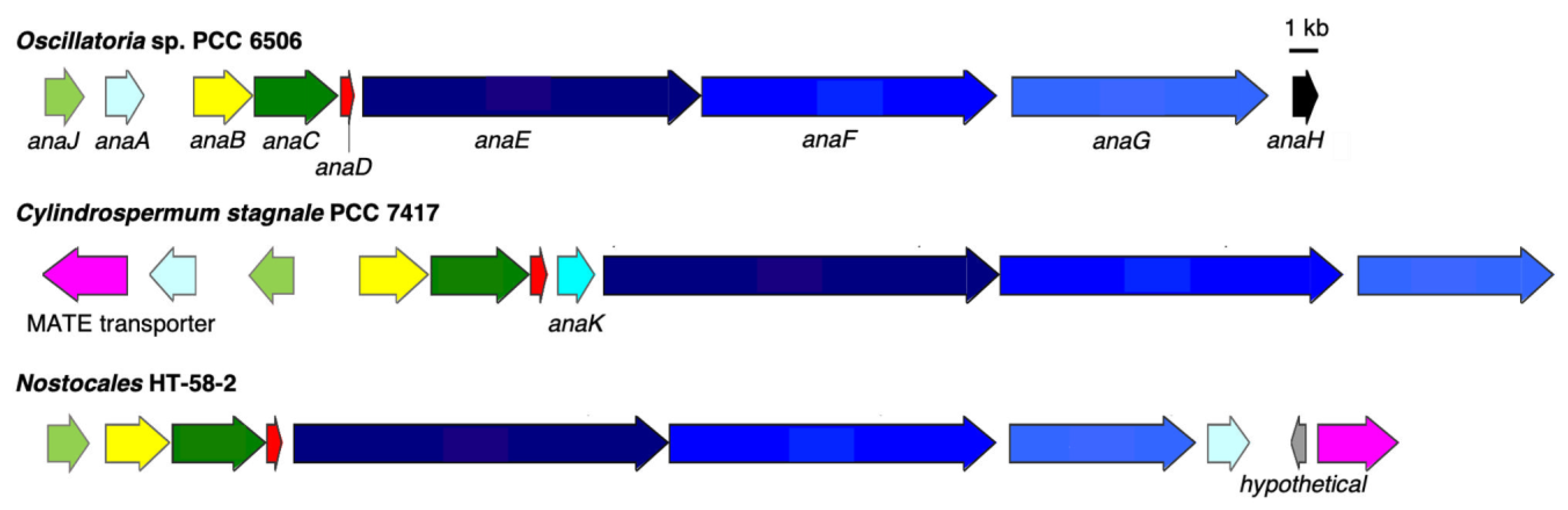
3.2.2. Anatoxin-a/Homoanatoxin-a BGC
3.2.3. Hexose-Shinorine BGC
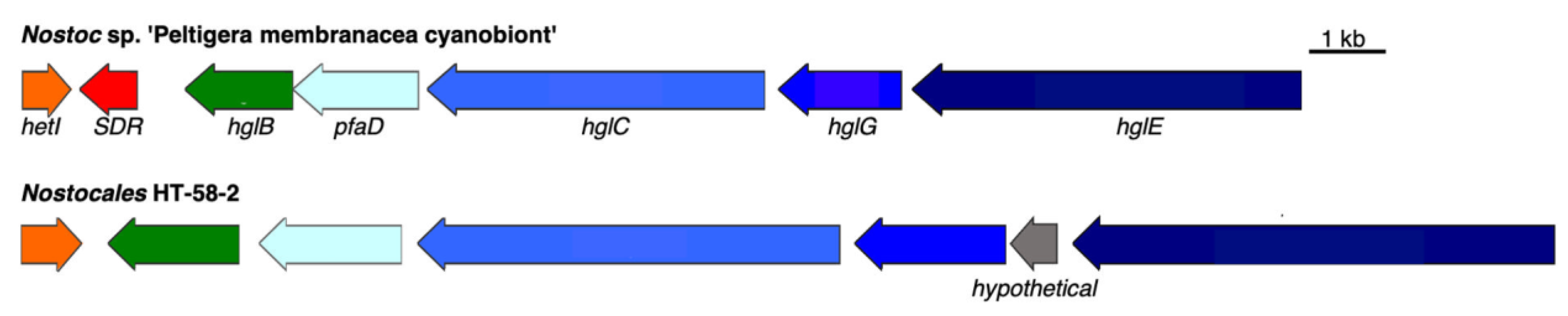
3.2.4. Heterocyst Glycolipid BGC
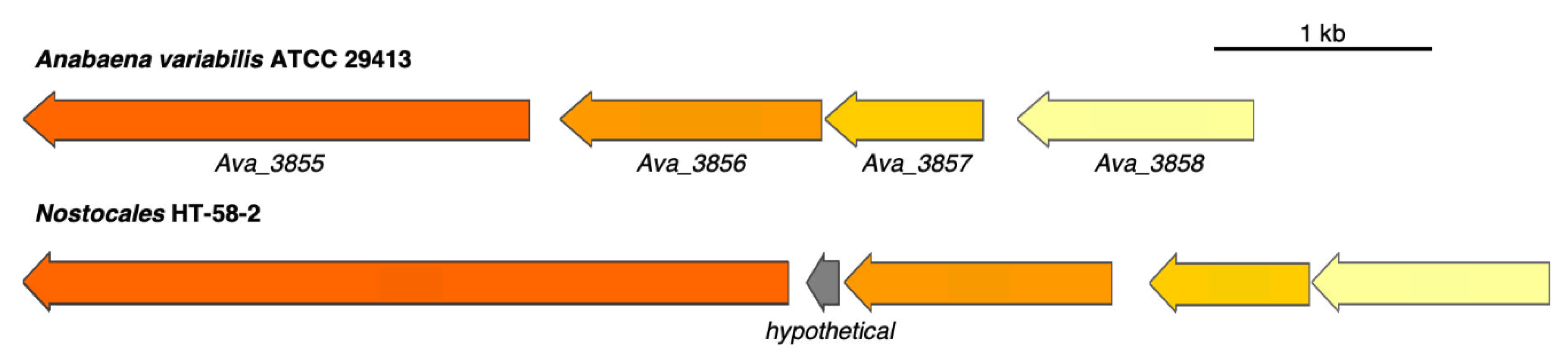
3.3. Putative BGC for Nitrogen Fixation
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Prinsep, M.R.; Caplan, F.R.; Moore, R.E.; Patterson, G.M.L.; Smith, C.D. Tolyporphin, a Novel Multidrug Resistance Reversing Agent from the Blue-green Alga Tolypothrix nodosa. J. Am. Chem. Soc. 1992, 114, 385–387. [Google Scholar] [CrossRef]
- Patterson, G.M.L.; Baldwin, C.L.; Bolis, C.M.; Caplan, F.R.; Karuso, H.; Larsen, L.K.; Levine, I.A.; Moore, R.E.; Nelson, C.S.; Tschappat, K.D.; et al. Antineoplastic Activity of Cultured Blue-Green Algae (Cyanophyta). J. Phycol. 1991, 27, 530–536. [Google Scholar] [CrossRef]
- Smith, C.D.; Prinsep, M.R.; Caplan, F.R.; Moore, R.E.; Patterson, G.M.L. Reversal of Multiple Drug Resistance by Tolyporphin, a Novel Cyanobacterial Natural Product. Oncol. Res. 1994, 6, 211–218. [Google Scholar]
- Morlière, P.; Mazière, J.-C.; Santus, R.; Smith, C.D.; Prinsep, M.R.; Stobbe, C.C.; Fenning, M.C.; Golberg, J.L.; Chapman, J.D. Tolyporphin: A Natural Product from Cyanobacteria with Potent Photosensitizing Activity against Tumor Cells in Vitro and in Vivo. Cancer Res. 1998, 58, 3571–3578. [Google Scholar] [PubMed]
- Prinsep, M.R.; Appleton, T.G.; Hanson, G.R.; Lane, I.; Smith, C.D.; Puddick, J.; Fairlie, D.P. Tolyporphin Macrocycles from the Cyanobacterium Tolypothrix nodosa Selectively Bind Copper and Silver and Reverse Multidrug Resistance. Inorg. Chem. 2017, 56, 5577–5585. [Google Scholar] [CrossRef] [PubMed]
- Prinsep, M.R.; Patterson, G.M.L.; Larsen, L.K.; Smith, C.D. Further Tolyporphins from the Blue-Green Alga Tolypothrix nodosa. Tetrahedron 1995, 51, 10523–10530. [Google Scholar] [CrossRef]
- Prinsep, M.R.; Patterson, G.M.L.; Larsen, L.K.; Smith, C.D. Tolyporphins J and K, Two Further Porphinoid Metabolites from the Cyanobacterium Tolypothrix nodosa. J. Nat. Prod. 1998, 61, 1133–1136. [Google Scholar] [CrossRef]
- Gurr, J.R.; Dai, J.; Philbin, C.S.; Sartain, H.T.; O’Donnell, T.J.; Yoshida, W.Y.; Rheingold, A.L.; Williams, P.G. Tolyporphins L–R: Unusual Tetrapyrroles from a Brasilonema sp. of Cyanobacterium. J. Org. Chem. 2020, 85, 318–326. [Google Scholar] [CrossRef]
- Hughes, R.-A.; Zhang, Y.; Zhang, R.; Williams, P.G.; Lindsey, J.S.; Miller, E.S. Genome Sequence and Composition of a Tolyporphin-Producing Cyanobacterium-Microbial Community. Appl. Environ. Microbiol. 2017, 83, e01068-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hughes, R.-A.; Jin, X.; Zhang, Y.; Zhang, R.; Tran, S.; Williams, P.G.; Lindsey, J.S.; Miller, E.S. Genome Sequence, Metabolic Properties and Cyanobacterial Attachment of Porphyrobacter sp. HT-58-2 Isolated from a Filamentous Cyanobacterium-Microbial Consortium. Microbiology 2018, 164, 1229–1239. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, R.; Hughes, R.-A.; Dai, J.; Gurr, J.R.; Williams, P.G.; Miller, E.S.; Lindsey, J.S. Quantitation of Tolyporphins, Diverse Tetrapyrrole Secondary Metabolites with Chlorophyll-Like Absorption, from a Filamentous Cyanobacterium–Microbial Community. Phytochem. Anal. 2018, 29, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Barnhart-Dailey, M.; Zhang, Y.; Zhang, R.; Anthony, S.M.; Aaron, J.S.; Miller, E.S.; Lindsey, J.S.; Timlin, J.A. Cellular Localization of Tolyporphins, Unusual Tetrapyrroles, in a Microbial Photosynthetic Community Determined using Hyperspectral Confocal Fluorescence Microscopy. Photosynth. Res. 2019, 141, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Prinsep, M.R.; Thomson, R.A. Tolypodiol, an Antiinflammatory Diterpenoid from the Cyanobacterium Tolypothrix nodosa. J. Nat. Prod. 1996, 59, 786–788. [Google Scholar] [CrossRef] [PubMed]
- Gurr, J.R.; O’Donnell, T.J.; Luo, Y.; Yoshida, W.Y.; Hall, M.L.; Mayer, A.M.S.; Sun, R.; Williams, P.G. 6-Deoxy- and 11-Hydroxytolypodiols: Meroterpenoids from the Cyanobacterium HT-58-2. J. Nat. Prod. 2020, 83, 1691–1695. [Google Scholar] [CrossRef] [PubMed]
- Tidgewell, K.; Clark, B.R.; Gerwick, W.H. The Natural Products Chemistry of Cyanobacteria. In Comprehensive Natural Products II Chemistry and Biology; Mander, L., Lui, H.-W., Eds.; Elsevier: Oxford, UK, 2010; Volume 2, pp. 141–188. ISBN 978-008-045-382-8. [Google Scholar]
- Leão, P.N.; Engene, N.; Antunes, A.; Gerwick, W.H.; Vasconcelos, V. The Chemical Ecology of Cyanobacteria. Nat. Prod. Rep. 2012, 29, 372–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Müller, R.; Wohlleben, W.; et al. AntiSMASH 3.0–A Comprehensive Resource for the Genome Mining of Biosynthetic Gene Clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef] [Green Version]
- Skinnider, M.A.; Merwin, N.J.; Johnston, C.W.; Magarvey, N.A. PRISM 3: Expanded Prediction of Natural Product Chemical Structures from Microbial Genomes. Nucleic Acids Res. 2017, 45, W49–W54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mungan, M.D.; Alanjary, M.; Blin, K.; Weber, T.; Medema, M.H.; Ziemert, N. ARTS 2.0: Feature Updates and Expansion of the Antibiotic Resistant Target Seeker for Comparative Genome Mining. Nucleic Acids Res. 2020, 48, W546–W552. [Google Scholar] [CrossRef]
- States, D.J.; Gish, W. Combined Use of Sequence Similarity and Codon Bias for Coding Region Identification. J. Comput. Biol. 1994, 1, 39–50. [Google Scholar] [CrossRef] [Green Version]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI Search and Sequence Analysis Tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [Green Version]
- Robert, X.; Gouet, P. Deciphering Key Features in Protein Structures with the New ENDscript Server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [Green Version]
- Rohmer, M. The Discovery of a Mevalonate-Independent Pathway for Isoprenoid Biosynthesis in Bacteria, Algae and Higher Plants. Nat. Prod. Rep. 1999, 16, 565–574. [Google Scholar] [CrossRef]
- Rohmer, M. Mevalonate-Independent Methylerythritol Phosphate Pathway for Isoprenoid Biosynthesis. Elucidation and Distribution. Pure Appl. Chem. 2003, 75, 375–387. [Google Scholar] [CrossRef] [Green Version]
- Young, I.G.; Stroobant, P.; MacDonald, C.G.; Gibson, F. Pathway for Ubiquinone Biosynthesis in Escherichia coli K-12: Gene-Enzyme Relationships and Intermediates. J. Bacteriol. 1973, 114, 42–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siebert, M.; Severin, K.; Heide, L. Formation of 4-Hydroxybenzoate in Escherichia coli: Characterization of the ubiC Gene and its Encoded Enzyme Chorismate Pyruvate-lyase. Microbiology 1994, 140, 897–904. [Google Scholar] [CrossRef] [Green Version]
- Melzer, M.; Heide, L. Characterization of Polyprenyldiphosphate: 4-Hydroxybenzoate Polyprenyltransferase from Escherichia coli. Biochim. Biophys. Acta 1994, 1212, 93–102. [Google Scholar] [CrossRef]
- Lee, P.T.; Hsu, A.Y.; Ha, H.T.; Clarke, C.F. A C-Methyltransferase Involved in Both Ubiquinone and Menaquinone Biosynthesis: Isolation and Identification of the Escherichia coli ubiE Gene. J. Bacteriol. 1997, 179, 1748–1754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stratmann, K.; Burgoyne, D.L.; Moore, R.E.; Patterson, G.M.L.; Smith, C.D. Hapalosin, a Cyanobacterial Cyclic Depsipeptide with Multidrug-Resistance Reversing Activity. J. Org. Chem. 1994, 59, 7219–7226. [Google Scholar] [CrossRef]
- D’Agostino, P.M.; Gulder, T.A.M. Direct Pathway Cloning Combined with Sequence- and Ligation-Independent Cloning for Fast Biosynthetic Gene Cluster Refactoring and Heterologous Expression. ACS Synth. Biol. 2018, 7, 1702–1708. [Google Scholar] [CrossRef]
- Micallef, M.L.; D’Agostino, P.M.; Sharma, D.; Viswanathan, R.; Moffitt, M.C. Genome Mining for Natural Product Biosynthetic Gene Clusters in the Subsection V Cyanobacteria. BMC Genom. 2015, 16, 669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Codd, G.; Bell, S.; Kaya, K.; Ward, C.; Beattie, K.; Metcalf, J. Cyanobacterial Toxins, Exposure Routes and Human Health. Eur. J. Phycol. 1999, 34, 405–415. [Google Scholar] [CrossRef]
- Rastogi, R.P.; Madamwar, D.; Incharoensakdi, A. Bloom Dynamics of Cyanobacteria and Their Toxins: Environmental Health Impacts and Mitigation Strategies. Front Microbiol. 2015, 6, 1254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devlin, J.P.; Edwards, O.E.; Gorham, P.R.; Hunter, N.R.; Pike, R.K.; Stavric, B. Anatoxin-a, A Toxic Alkaloid from Anabaena flos-aquae NRC-44h. Can. J. Chem. 1977, 55, 1367–1371. [Google Scholar] [CrossRef]
- Hemscheidt, T.; Rapala, J.; Sivonen, K.; Skulberg, O.M. Biosynthesis of Anatoxin-a in Anabaena flos-aquae and Homoanatoxin-a in Oscillatoria formosa. J. Chem. Soc. Chem. Commun. 1995, 13, 1361–1362. [Google Scholar] [CrossRef]
- Carmichael, W.W.; Gorham, P.R.; Biggs, D.F. Two Laboratory Case Studies on the Oral Toxicity to Calves of the Freshwater Cyanophyte (Blue-green Alga) Anabaena flos-aquae NRC-44-1. Can. Vet. J. 1977, 18, 71–75. [Google Scholar] [PubMed]
- Rogers, E.H.; Hunter, E.S., III; Moser, V.C.; Phillips, P.M.; Herkovits, J.; Muñoz, L.; Hall, L.L.; Chernoff, N. Potential Developmental Toxicity of Anatoxin-a, A Cyanobacterial Toxin. J. Appl. Toxicol. 2005, 25, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Cadel-Six, S.; Iteman, I.; Peyraud-Thomas, C.; Mann, S.; Ploux, O.; Méjean, A. Identification of a Polyketide Synthase Coding Sequence Specific for Anatoxin-a-Producing Oscillatoria Cyanobacteria. Appl. Environ. Microbiol. 2009, 75, 4909–4912. [Google Scholar] [CrossRef] [Green Version]
- Méjean, A.; Mann, S.; Maldiney, T.; Vassiliadis, G.; Lequin, O.; Ploux, O. Evidence that Biosynthesis of the Neurotoxic Alkaloids Anatoxin-a and Homoanatoxin-a in the Cyanobacterium Oscillatoria PCC 6506 Occurs on a Modular Polyketide Synthase Initiated by L-Proline. J. Am. Chem. Soc. 2009, 131, 7512–7513. [Google Scholar] [CrossRef]
- Méjean, A.; Dalle, K.; Paci, G.; Bouchonnet, S.; Mann, S.; Pichon, V.; Ploux, O. Dihydroanatoxin-a Is Biosynthesized from Proline in Cylindrospermum stagnale PCC 7417: Isotopic Incorporation Experiments and Mass Spectrometry Analysis. J. Nat. Prod. 2016, 79, 1775–1782. [Google Scholar] [CrossRef]
- Kellmann, R.; Mihali, T.K.; Jeon, Y.J.; Pickford, R.; Pomati, F.; Neilan, B.A. Biosynthetic Intermediate Analysis and Functional Homology Reveal a Saxitoxin Gene Cluster in Cyanobacteria. Appl. Environ. Microbiol. 2008, 74, 4044–4053. [Google Scholar] [CrossRef] [Green Version]
- Rastogi, R.P.; Incharoensakdi, A. Characterization of UV-screening Compounds, Mycosporine-like Amino Acids, and Scytonemin in the Cyanobacterium Lyngbya sp. CU2555. FEMS Microbiol. Ecol. 2014, 87, 244–256. [Google Scholar] [CrossRef] [Green Version]
- Sinha, R.P.; Singh, S.P.; Häder, D.-P. Database on Mycosporines and Mycosporine-like Amino Acids (MAAs) in Fungi, Cyanobacteria, Macroalgae, Phytoplankton and Animals. J. Photochem. Photobiol. B Biol. 2007, 89, 29–35. [Google Scholar] [CrossRef]
- Hartmann, A.; Murauer, A.; Ganzera, M. Quantitative Analysis of Mycosporine-like Amino Acids in Marine Algae by Capillary Electrophoresis with Diode-array Detection. J. Pharm. Biomed. Anal. 2017, 138, 153–157. [Google Scholar] [CrossRef]
- Singh, S.P.; Sinha, R.P.; Klisch, M.; Häder, D.-P. Mycosporine-like Amino Acids (MAAs) Profile of a Rice-Field Cyanobacterium Anabaena doliolum as Influenced by PAR and UVR. Planta 2008, 229, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.J.; Ferran, G.-P. An ATP-Grasp Ligase Involved in the Last Biosynthetic Step of the Iminomycosporine Shinorine in Nostoc punctiforme ATCC 29133. J. Bacteriol. 2011, 193, 5923–5928. [Google Scholar] [CrossRef] [Green Version]
- Balskus, E.P.; Walsh, C.T. The Genetic and Molecular Basis for Sunscreen Biosynthesis in Cyanobacteria. Science 2010, 329, 1653–1656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Llewellyn, C.A.; Greig, C.; Silkina, A.; Kultschar, B.; Hitchings, M.D.; Farnham, G. Mycosporine-like Amino Acid and Aromatic Amino Acid Transcriptome Response to UV and Far-Red Light in the Cyanobacterium Chlorogloeopsis fritschii PCC 6912. Sci. Rep. 2020, 10, 20638. [Google Scholar] [CrossRef]
- Ho, M.-Y.; Bryant, D.A. Global Transcriptional Profiling of the Cyanobacterium Chlorogloeopsis fritschii PCC 9212 in Far-Red Light: Insights into the Regulation of Chlorophyll d Synthesis. Front. Microbiol. 2019, 10, 465. [Google Scholar] [CrossRef] [PubMed]
- Wörmer, L.; Cirés, S.; Velázquez, D.; Quesada, A.; Hinrichs, K.-U. Cyanobacterial Heterocyst Glycolipids in Cultures and Environmental Samples: Diversity and Biomarker Potential. Limnol. Oceanogr. 2012, 57, 1775–1788. [Google Scholar] [CrossRef]
- Kampa, A.; Gagunashvili, A.N.; Gulder, T.A.M.; Morinaka, B.I.; Daolio, C.; Godejohann, M.; Miao, V.P.W.; Piel, J.; Andrésson, Ó. Metagenomic Natural Product Discovery in Lichen Provides Evidence for a Family of Biosynthetic Pathways in Diverse Symbioses. Proc. Natl. Acad. Sci. USA 2013, 110, 3129–3137. [Google Scholar] [CrossRef] [Green Version]
- Dos Santos, P.C.; Fang, Z.; Mason, S.W.; Setubal, J.C.; Dixon, R. Distribution of Nitrogen Fixation and Nitrogenase-like Sequences amongst Microbial Genomes. BMC Genom. 2012, 13, 162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thiel, T.; Pratte, B.S. Regulation of Three Nitrogenase Gene Clusters in the Cyanobacterium Anabaena variabilis ATCC 29413. Life 2014, 4, 944–967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aráoz, R.; Molgó, J.; Tandeau de Marsac, N. Neurotoxic Cyanobacterial Toxins. Toxicon 2010, 56, 813–828. [Google Scholar] [CrossRef] [PubMed]
- Gaysina, L.A.; Saraf, A.; Singh, P. Cyanobacteria in Diverse Habitats. In Cyanobacteria—From Basic Science to Applications; Mishra, A.K., Tiwari, D.N., Rai, A.N., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 1–28. ISBN 978-012-814-667-5. [Google Scholar]
- Rastogi, R.P.; Sinha, R.P. Biotechnological and Industrial Significance of Cyanobacterial Secondary Metabolites. Biotechnol. Adv. 2009, 27, 521–539. [Google Scholar] [CrossRef]
- Gibson, F. The Elusive Branch-Point Compound of Aromatic Amino Acid Biosynthesis. Trends Biochem. Sci. 1999, 24, 36–38. [Google Scholar] [CrossRef]
- Medema, M.H.; Fischbach, M.A. Computational Approaches to Natural Product Discovery. Nat. Chem. Biol. 2015, 11, 639–648. [Google Scholar] [CrossRef]







| HT-58-2 Accession | Aligned Protein (Gene) |
|---|---|
| WP_015211021 | 2-polyprenyl-6-methoxyphenol hydroxylase (ubiH) b |
| ARV62772 | 4-hydroxybenzoate polyprenyltransferase (ubiA) |
| ARV57253 | chorismate lyase (ubiC) |
| ARV57254 | 1-deoxy-D-xylulose-5-phosphate synthase (dxs) |
| ARV57255 | 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate synthase (ispG) |
| ARV62773 | geranylgeranyl pyrophosphate synthase (crtE) |
| ARV57256 | hypothetical protein |
| ARV57257 | phenylpropionate dioxygenase or ring-hydroxylating dioxygenase (hcaE) |
| ARV57258 | ubiquinone/menaquinone biosynthesis C-methylase (ubiE) |
| ARV57259 | 4-Hydroxy-3-methylbut-2-enyl diphosphate reductase (ispH) |
| ARV57260 | cytochrome P450 |
| ARV62774 | 4-diphosphocytidyl-2C-methyl-D-erythritol kinase (ispE) |
| ARV62775 | dephospho-CoA kinase (coaE) |
| ARV57261 | 1-deoxy-D-xylulose 5-phosphate reductoisomerase (dxr) |
| ARV57262 | Dienelactone hydrolase |
| No. | Region (bp) | Length (nt) | Type | Similar Known Cluster |
|---|---|---|---|---|
| 1 | 308,169–329,626 | 21,458 | lassopeptide | |
| 2 | 344,287–393,016 | 49,759 | NRPS a | vioprolides, xenoamicins, cyanopeptin |
| 3 | 855,604–866,222 | 10,619 | RiPP-like b | |
| 4 * | 958,778–1,011,938 | 53,161 | hglE-KS c, T1PKS d | heterocyst glycolipids |
| 5 | 1,080,296–1,132,473 | 52,178 | T1PKS | carbamidocyclophanes |
| 6 | 1,225,583–1,267,343 | 41,761 | phosphonate, terpene | |
| 7 | 1,620,824–1,643,303 | 22,480 | lassopeptide | |
| 8 | 1,748,080–1,798,143 | 50,064 | NRPS | hapalosin |
| 9 * | 2,268,098–2,291,911 | 240,666 | NRPS, T1PKS | hapalosin |
| 10 | 2,303,699–2,508,763 | 20,507 | NRPS, T1PKS | nostopeptolide A2 |
| 11 * | 2,683,411–2,768,967 | 85,557 | T1PKS | anatoxin-a |
| 12 | 3,624,228–3,644,311 | 20,084 | terpene | |
| 13 | 3,794,851–3,850,264 | 55,414 | T1PKS | chondrochloren A |
| 14 | 4,005,987–4,016,220 | 10,234 | bacteriocin | |
| 15 * | 4,105,987–4,016220 | 42,348 | NRPS | hexose-palythine-serine |
| 16 | 4,214,166–4,256,226 | 41,891 | bacteriocin | |
| 17 | 4,262,518–4,353,871 | 91,354 | NRPS, lanthipeptide | nostopeptolide A2 |
| 18 | 5,824,405–5,845,337 | 20,933 | terpene |
| HT-58-2 Accession | Aligned Protein | E-Value | % Identity * |
|---|---|---|---|
| ARV58870 | HapA, AMP-dependent synthetase | 0.0 | 60.2% |
| ARV58846 | HapB, polyketide synthetase | 0.0 | 51.6% |
| ARV58869 | HapC, non-ribosomal peptide synthetase | 0.0 | 58.8% |
| ARV58868 | HapD, non-ribosomal peptide synthetase | 0.0 | 63.6% |
| ARV62947 | HapE, polyketide synthetase | 0.0 | 65.3% |
| HT-58-2 Accession | Aligned Protein | E-Value | % Identity * |
|---|---|---|---|
| ARV59080 | AnaJ, cyclase | 0.0 | 69.0% |
| ARV59087 | AnaA, thioesterase | 3e-157 | 82.8% |
| ARV59081 | AnaB, acyl-CoA dehydrogenase | 0.0 | 87.9% |
| ARV59082 | AnaC, proline adenylation | 0.0 | 87.7% |
| ARV59083 | AnaD, acyl carrier protein | 2e-51 | 87.4% |
| ARV59084 | AnaE, polyketide synthase | 0.0 | 86.7% |
| ARV59085 | AnaF, polyketide synthase | 0.0 | 82.2% |
| ARV59086 | AnaG, polyketide synthase | 0.0 | 83.9% |
| HT-58-2 Accession | Aligned Protein | E-Value | % Identity * |
|---|---|---|---|
| ARV60049 | 3-dehydroquinate synthase | 3e-149 | 78.7% |
| ARV60048 | SAM-dependent methyltransferase | 1e-109 | 74.2% |
| ARV60047 | ATP-grasp enzyme | 1e-215 | 79.0% |
| ARV60046 | NRPS | 0.0 | 72.9% |
| HT-58-2 Accession | Aligned Protein | E-Value | %Identity * |
|---|---|---|---|
| ARV57925 | HetI, 4′-phosphopantetheinyl transferase | 1e-125 | 71.2% |
| ARV57926 | HglB, thioester reductase | 0.0 | 73.0% |
| ARV62843 | PfaD, polyunsaturated fatty acid/polyketide biosynthesis protein | 0.0 | 79.5% |
| ARV57927 | HglC, beta-ketoacyl synthase | 0.0 | 63.6% |
| ARV57928 | HglG, polyketide synthase | 0.0 | 65.3% |
| ARV57929 | carboxypeptidase regulatory protein | N/A | N/A |
| ARV57930 | HglE, polyketide synthase | 0.0 | 66.8% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, X.; Miller, E.S.; Lindsey, J.S. Natural Product Gene Clusters in the Filamentous Nostocales Cyanobacterium HT-58-2. Life 2021, 11, 356. https://doi.org/10.3390/life11040356
Jin X, Miller ES, Lindsey JS. Natural Product Gene Clusters in the Filamentous Nostocales Cyanobacterium HT-58-2. Life. 2021; 11(4):356. https://doi.org/10.3390/life11040356
Chicago/Turabian StyleJin, Xiaohe, Eric S. Miller, and Jonathan S. Lindsey. 2021. "Natural Product Gene Clusters in the Filamentous Nostocales Cyanobacterium HT-58-2" Life 11, no. 4: 356. https://doi.org/10.3390/life11040356






