Microarray Profile of Long Noncoding RNA and Messenger RNA Expression in a Model of Alzheimer’s Disease
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal and Tissue Preparation
2.2. RNA Extraction
2.3. RNA Labeling and Array Hybridization
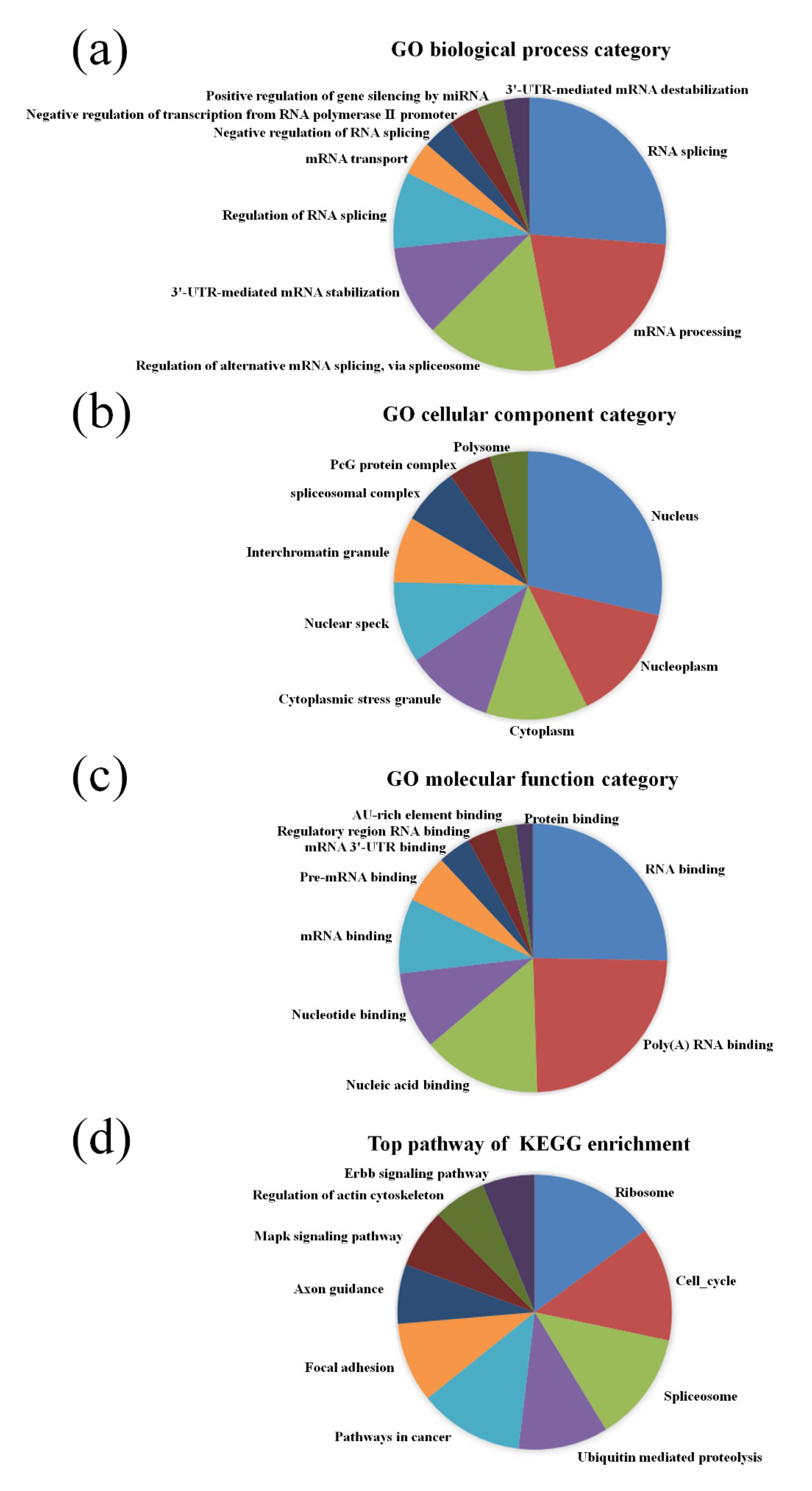
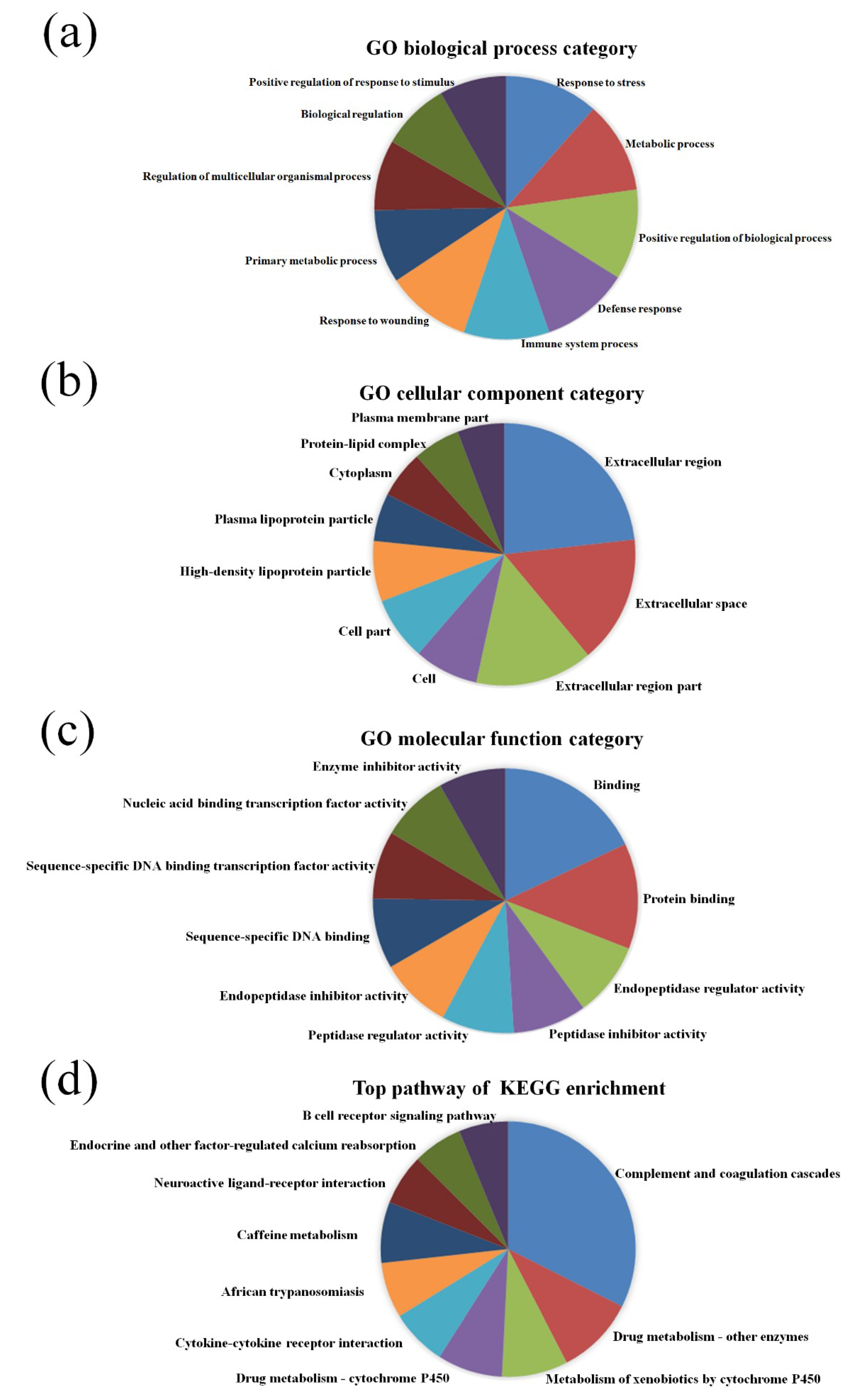
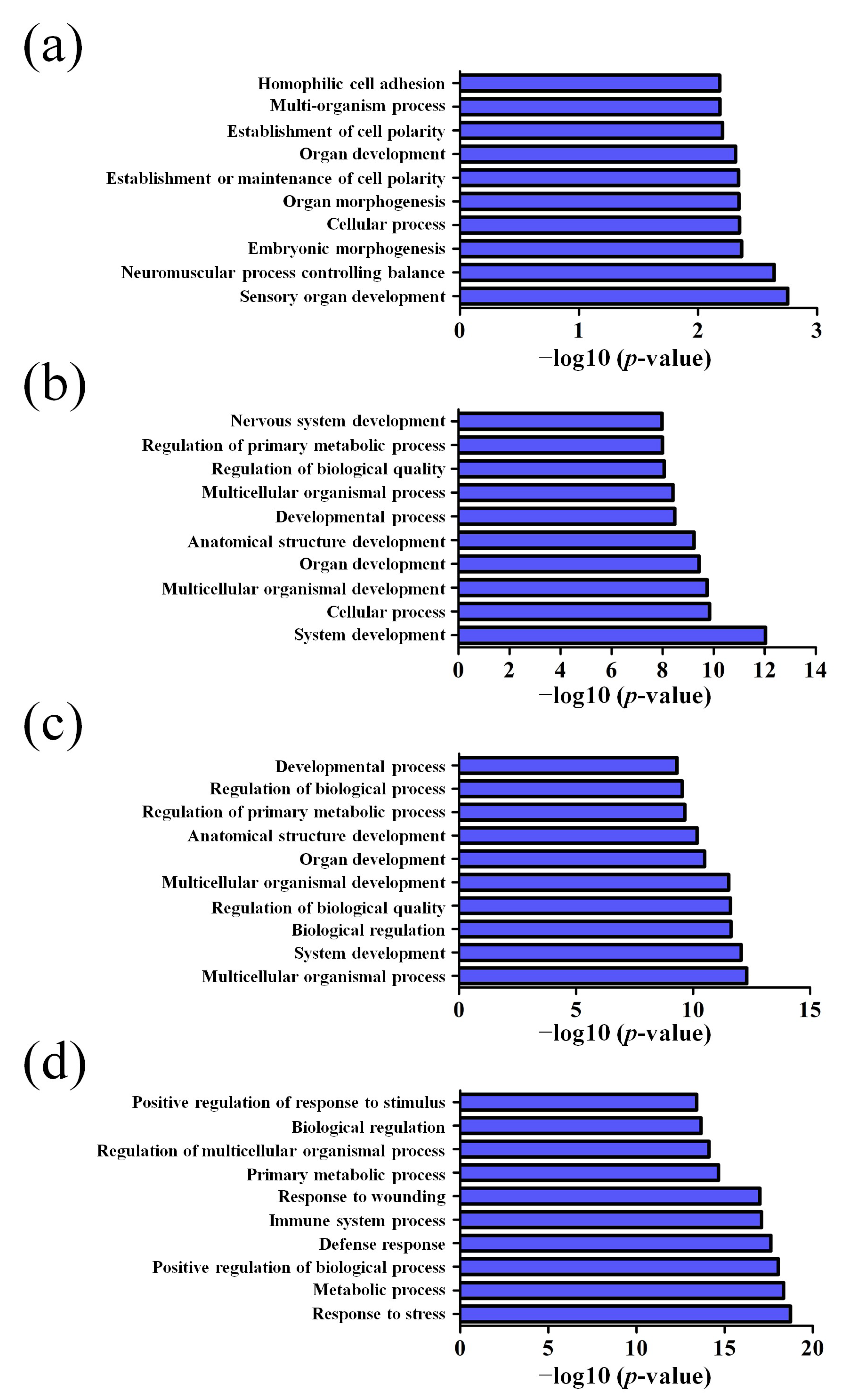
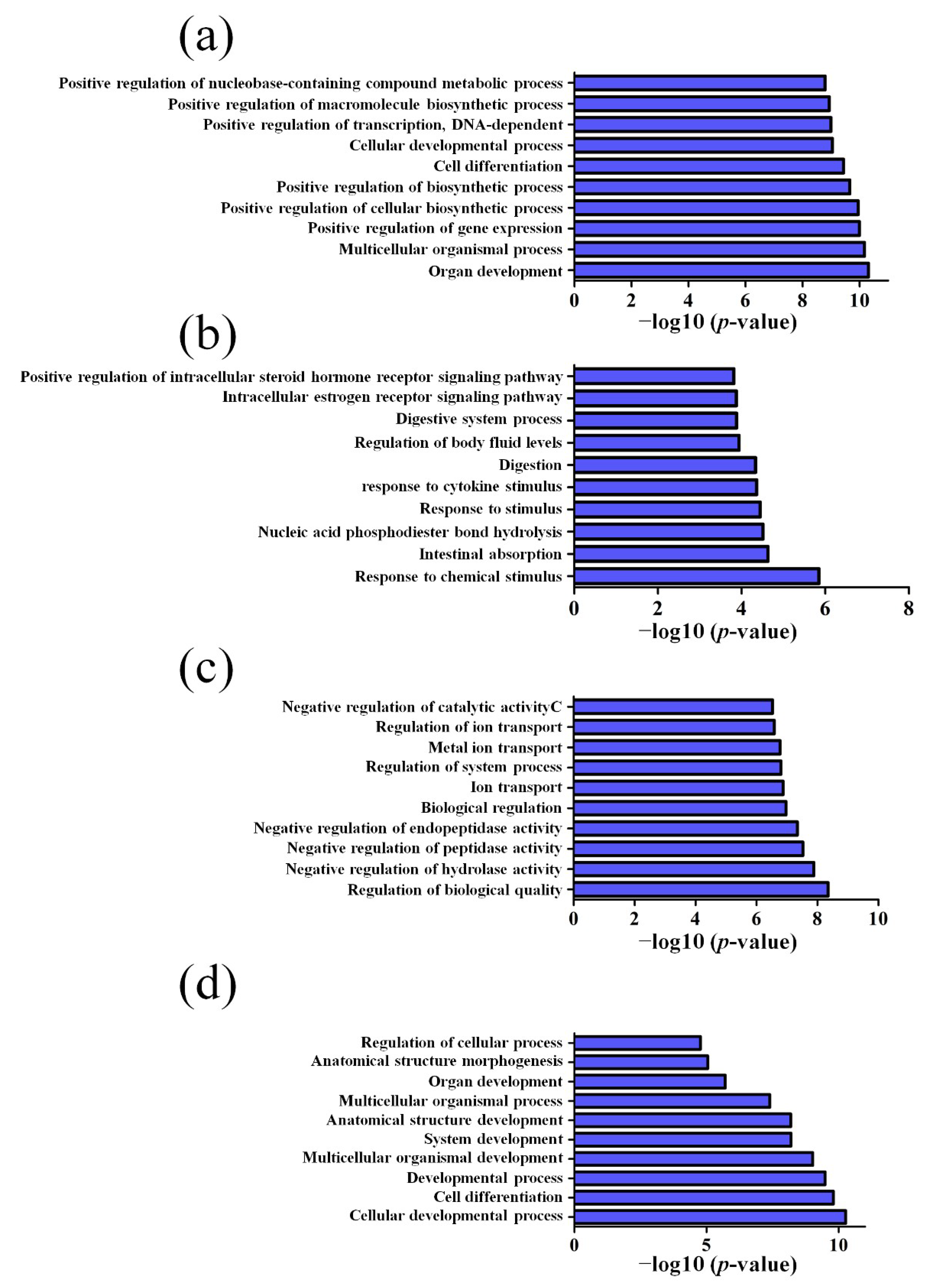
2.4. GO and Pathway Analysis
2.5. lncRNA–mRNA–miRNA Co-Expression Network Construction
2.6. Statistical Analysis
3. Results
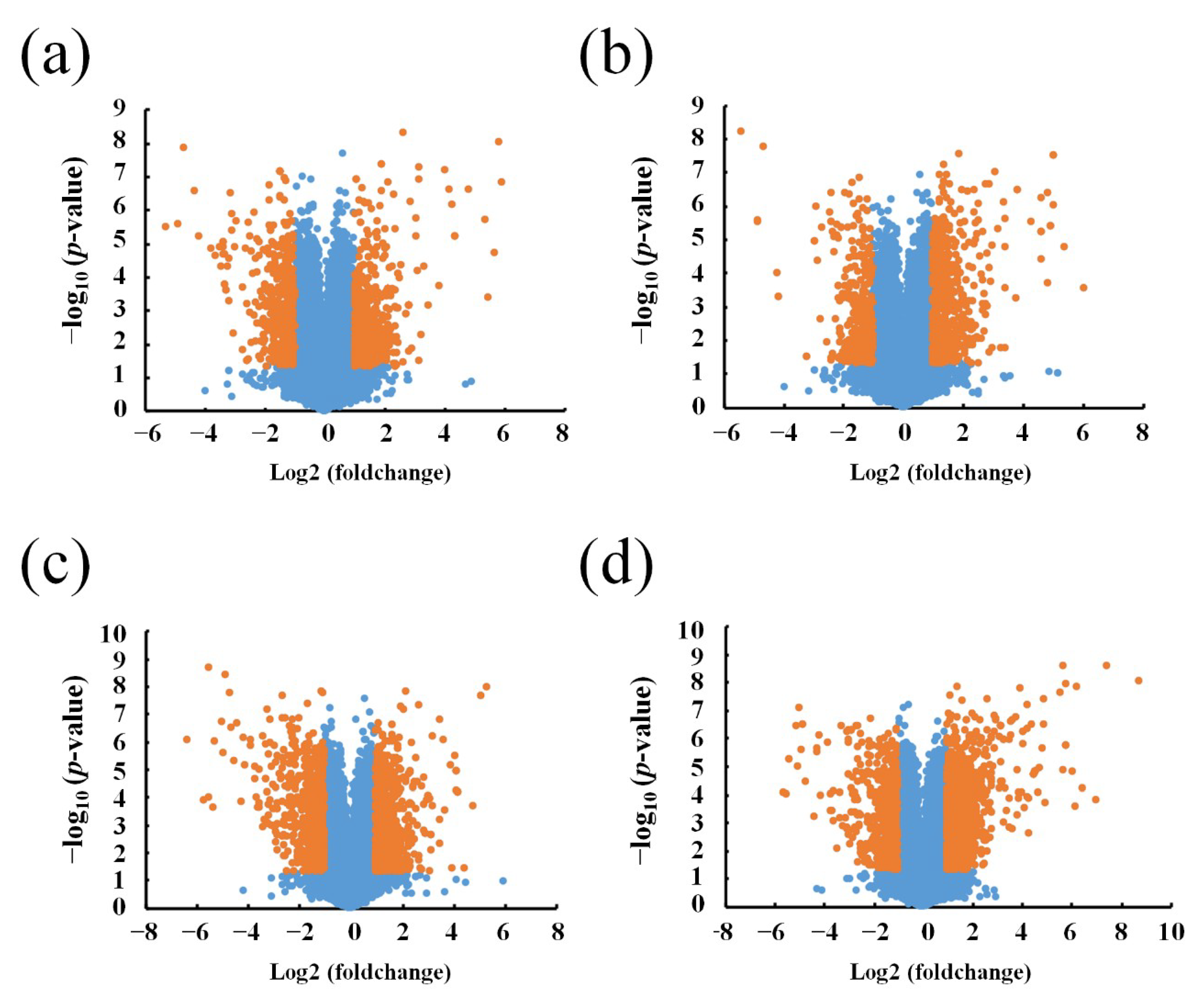
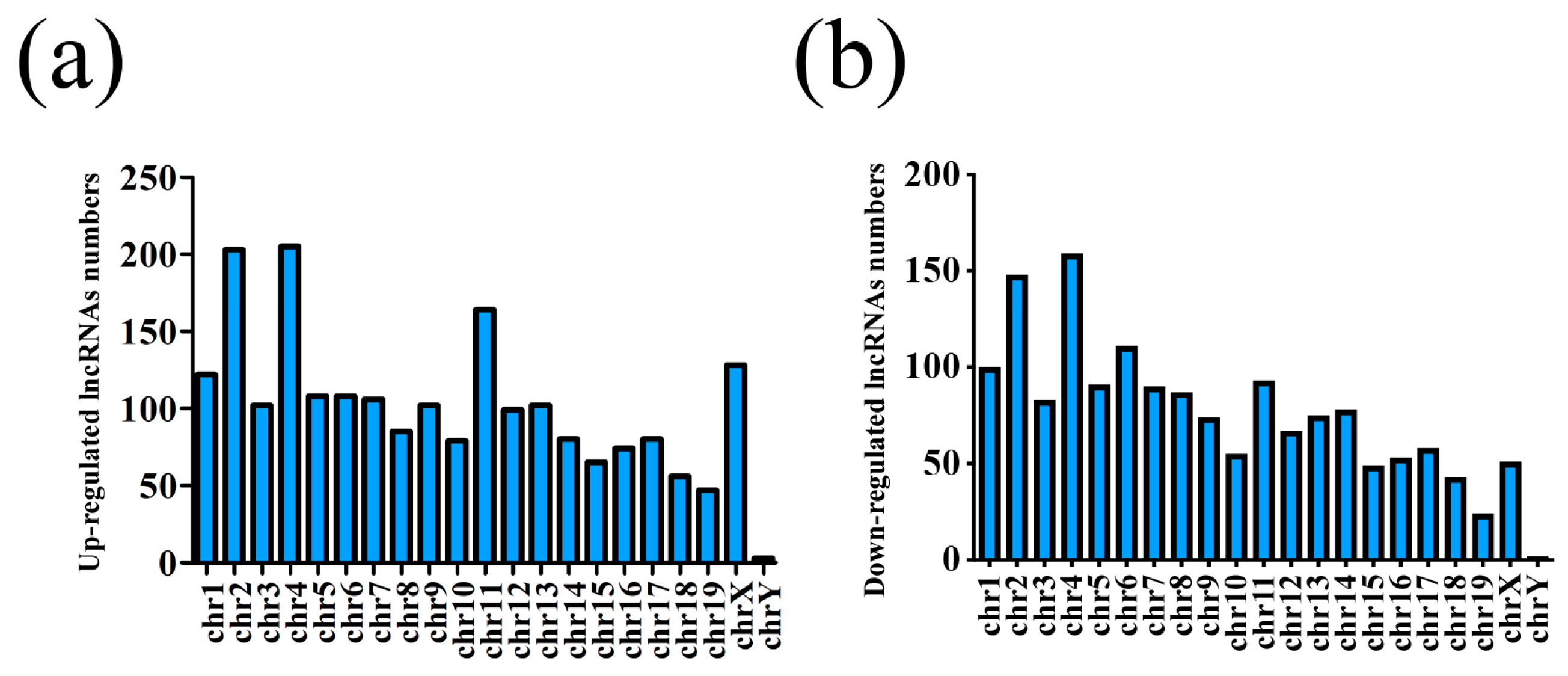
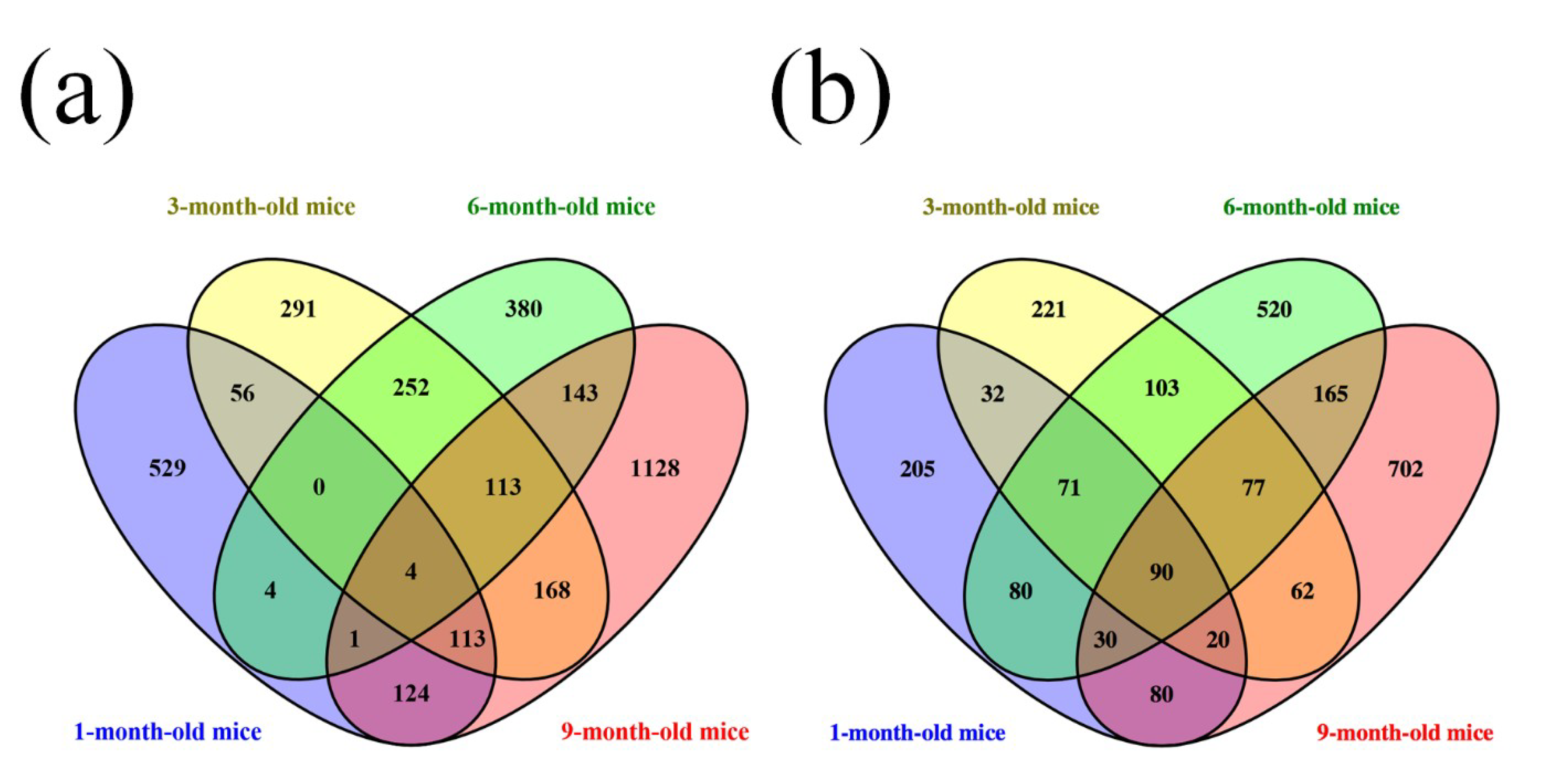
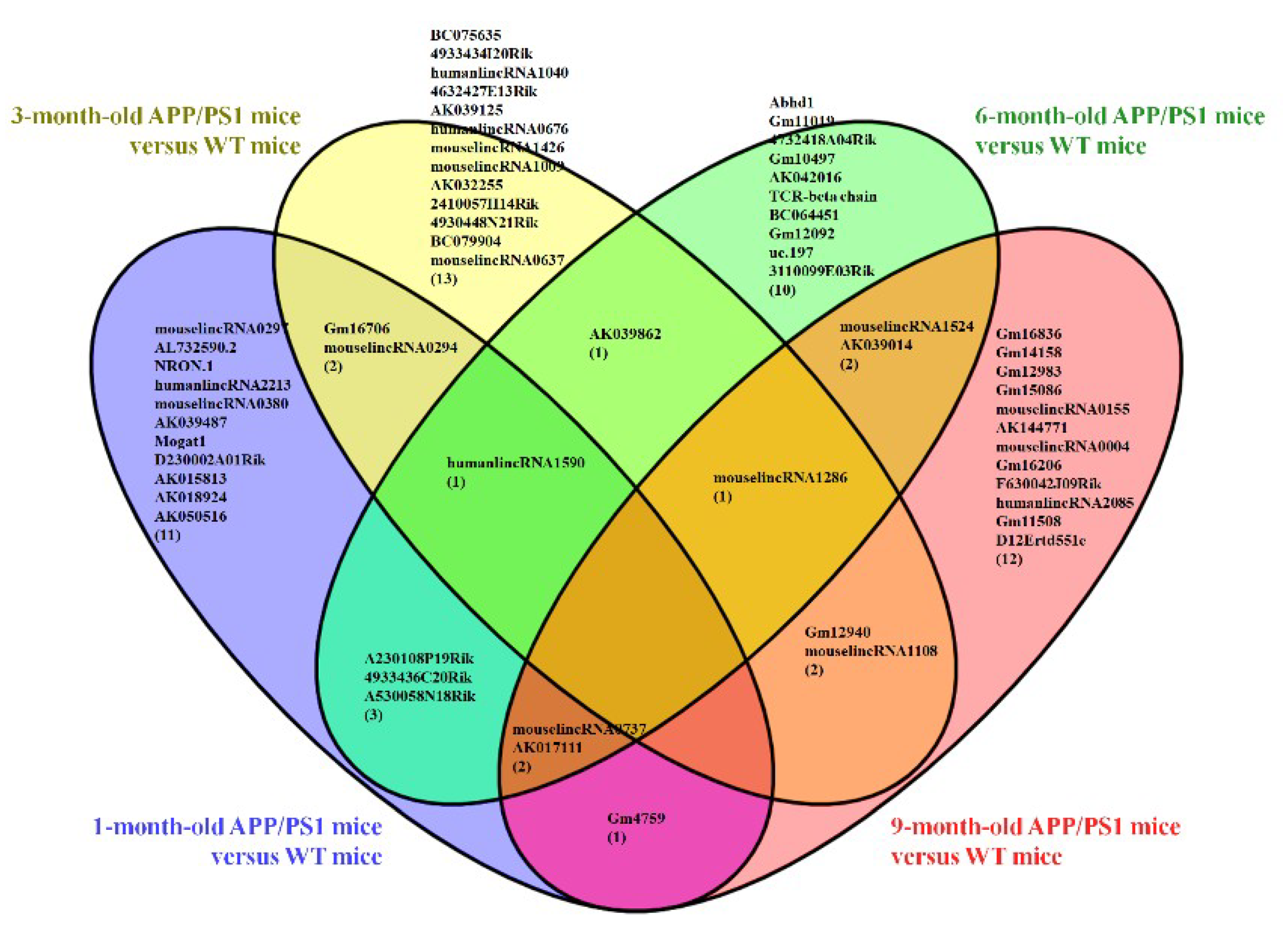
3.1. Different Expression Profile of lncRNA and mRNA
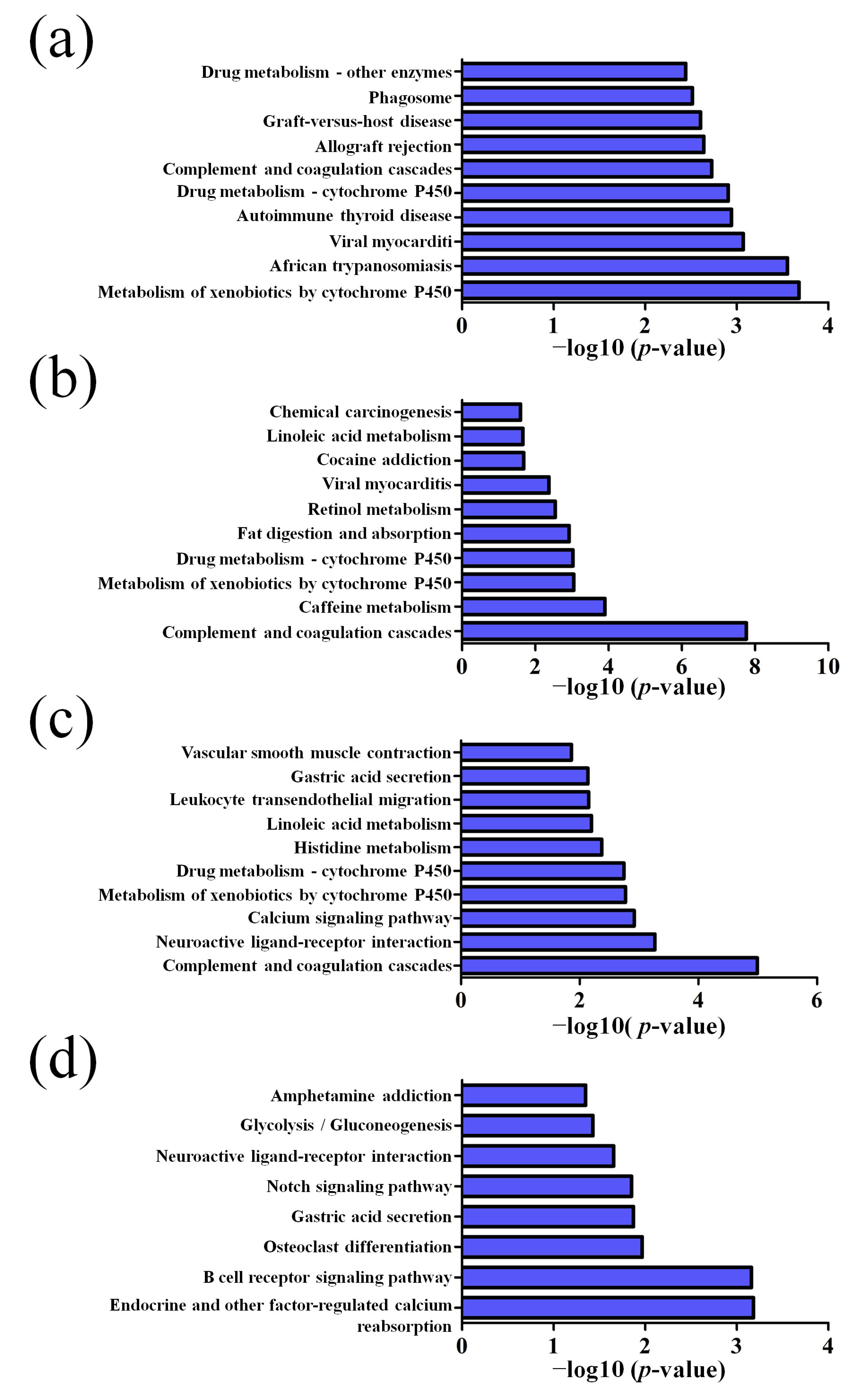
3.2. GO and KEGG Pathway Analysis
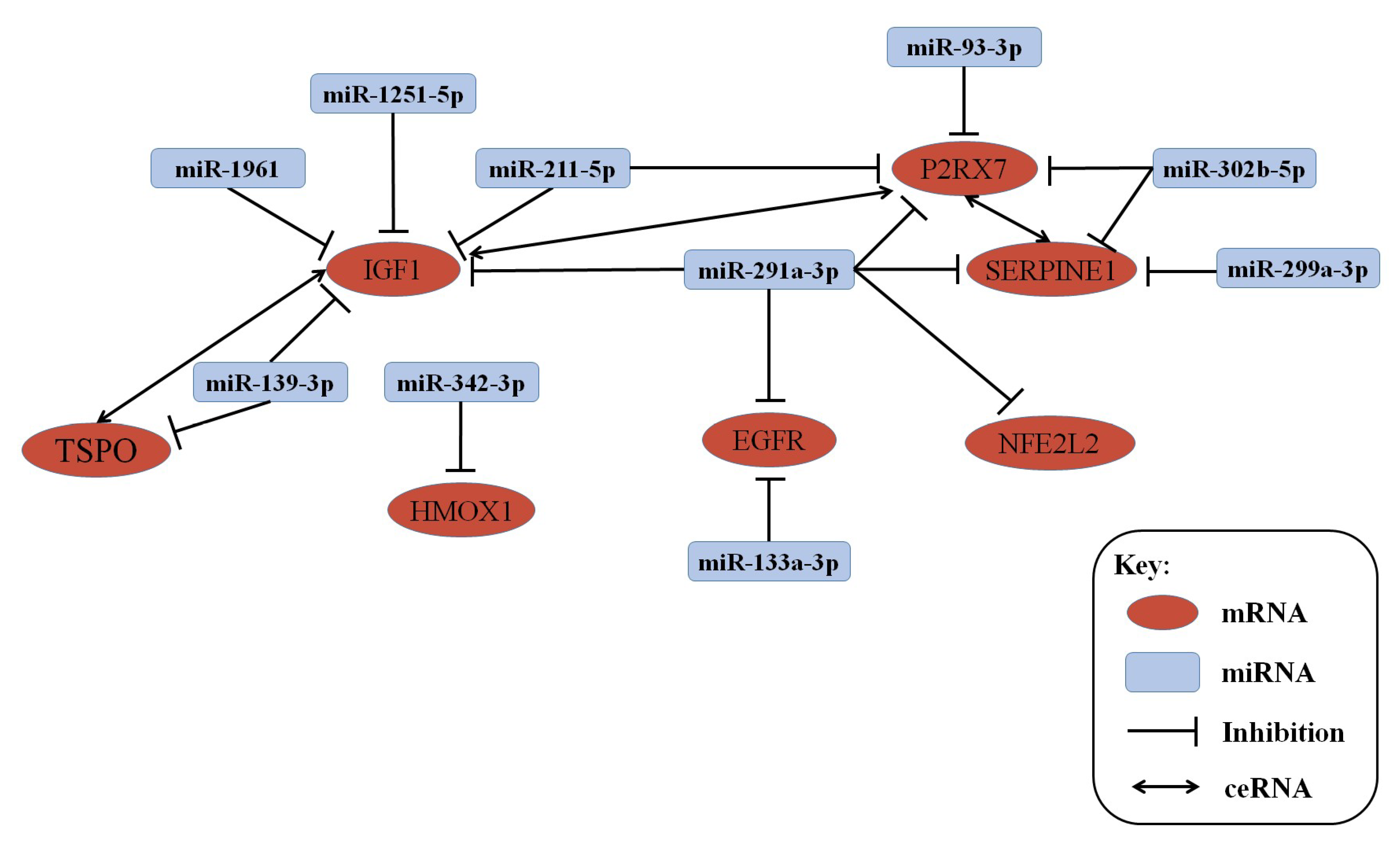
3.3. lncRNA–mRNA–miRNA Co-Expression Network
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ansari, S.A.; Satar, R.; Perveen, A.; Ashraf, G.M. Current opinion in Alzheimer’s disease therapy by nanotechnology-based approaches. Curr. Opin. Psychiatr. 2017, 30, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Alexiou, A.; Kamal, M.A.; Ashraf, G.M. The Alzheimer’s disease challenge. Front. Neurosci. 2019, 13, 768. [Google Scholar] [CrossRef] [PubMed]
- Mamun, A.A.; Uddin, M.S.; Mathew, B.; Ashraf, G.M. Toxic tau: Structural origins of tau aggregation in Alzheimer’s disease. Neural Regen. Res. 2020, 15, 1417–1420. [Google Scholar] [PubMed]
- Uddin, M.S.; Mamun, A.A.; Labu, Z.K.; Hidalgo-Lanussa, O.; Barreto, G.E.; Ashraf, G.M. Autophagic dysfunction in Alzheimer’s disease: Cellular and molecular mechanistic approaches to halt Alzheimer’s pathogenesis. J. Cell Physiol. 2019, 234, 8094–8112. [Google Scholar] [CrossRef] [PubMed]
- 2018 Alzheimer’s disease facts and figures. Alzheimer Dement. 2018, 14, 367–429. [CrossRef]
- Gasiorowski, K.; Brokos, B.; Leszek, J.; Tarasov, V.V.; Ashraf, G.M.; Aliev, G. Insulin resistance in alzheimer disease: p53 and micrornas as important players. Curr. Top Med. Chem. 2017, 17, 1429–1437. [Google Scholar] [CrossRef]
- Ashraf, G.M.; Tabrez, S.; Jabir, N.R.; Firoz, C.K.; Ahmad, S.; Hassan, I.; Alexiou, A.; Kamal, M.A. An overview on global trends in nanotechnological approaches for alzheimer therapy. Curr. Drug Metab. 2015, 16, 719–727. [Google Scholar] [CrossRef]
- Shi, C.; Zhang, L.; Qin, C. Long non-coding RNAs in brain development, synaptic biology, and Alzheimer’s disease. Brain Res. Bull. 2017, 132, 160–169. [Google Scholar] [CrossRef]
- Wang, K.L.; Chen, F.X.; He, D.D.; Li, Y.; Fu, J. Dissection of functional lncRNAs in Alzheimer’s disease by construction and analysis of lncRNA-mRNA networks based on competitive endogenous RNAs. Biochem. Biophys. Res. Commun. 2017, 485, 569–576. [Google Scholar] [CrossRef]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Huang, Y.; Chen, J.; Chi, H.; Yu, Z.; Wang, J.; Chen, C. Attenuated ability of BACE1 to cleave the amyloid precursor protein via silencing long noncoding RNA BACE1AS expression. Mol. Med. Rep. 2014, 10, 1275–1281. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zheng, L.; Jiang, A.; Mo, Y.; Gong, Q. Identification of the biological affection of long noncoding RNA BC200 in Alzheimer’s disease. Neuroreport 2018, 29, 1061–1067. [Google Scholar] [CrossRef] [PubMed]
- Ciarlo, E.; Massone, S.; Penna, I.; Nizzari, M.; Gigoni, A.; Dieci, G.; Russo, C.; Florio, T.; Cancedda, R.; Pagano, A. An intronic ncRNA-dependent regulation of SORL1 expression affecting Aβ formation is upregulated in post-mortem Alzheimer’s disease brain samples. Dis. Model. Mech. 2013, 6, 424–433. [Google Scholar] [CrossRef] [PubMed]
- Massone, S.; Vassallo, I.; Fiorino, G.; Castelnuovo, M.; Barbieri, F.; Borghi, R.; Tabaton, M.; Robello, M.; Gatta, E.; Russo, C.; et al. 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol. Dis. 2011, 41, 308–317. [Google Scholar] [CrossRef]
- Massone, S.; Ciarlo, E.; Vella, S.; Nizzari, M.; Florio, T.; Russo, C.; Cancedda, R.; Pagano, A. NDM29, a RNA polymerase III-dependent non coding RNA, promotes amyloidogenic processing of APP and amyloid b secretion. Biochim. Biophys. Acta 2012, 1823, 1170–1177. [Google Scholar] [CrossRef]
- Parenti, R.; Paratore, S.; Torrisi, A.; Cavallaro, S. A natural antisense transcript against Rad18, specifically expressed in neurons and upregulated during b-amyloid-induced apoptosis. Eur. J. Neurosci. 2007, 26, 2444–2457. [Google Scholar] [CrossRef]
- Sosinska, P.; Mikula-Pietrasik, J.; Ksiazek, K. The double-edged sword of long non-coding RNA: The role of human brain-specific BC200 RNA in translational control, neurodegenerative diseases, and cancer. Mutation research. Rev. Mutat. Res. 2015, 766, 58–67. [Google Scholar] [CrossRef]
- Shen, Z.; Lei, J.; Li, X.; Wang, Z.; Bao, X.; Wang, R. Multifaceted assessment of the APP/PS1 mouse model for Alzheimer’s disease: Applying MRS, DTI, and ASL. Brain Res. 2018, 1698, 114–120. [Google Scholar] [CrossRef]
- Gene Ontology. Available online: http://www.geneontology.org (accessed on 6 January 2017).
- Kyoto Encyclopedia of Genes and Genomes. Available online: http://www.genome.jp/kegg/ (accessed on 15 January 2017).
- miRWalk. Available online: http://www.ma.uni-heidelberg.de/ (accessed on 25 January 2017).
- TargetScan. Available online: http://www.targetscan.org/ (accessed on 25 January 2017).
- miRBase. Available online: http://www.mirbase.org/ (accessed on 25 January 2017).
- miRanda. Available online: http://www.microrna.org/ (accessed on 25 January 2017).
- Wang, L.L.; Min, L.; Guo, Q.D.; Zhang, J.X.; Jiang, H.L.; Shao, S.; Xing, J.G.; Yin, L.L.; Liu, J.H.; Liu, R. Profiling microRNA from Brain by Microarray in a Transgenic Mouse Model of Alzheimer’s Disease. BioMed Res. Int. 2017, 8030369. [Google Scholar] [CrossRef]
- Uddin, M.S.; Kabir, M.T.; Al Mamun, A.; Abdel-Daim, M.M.; Barreto, G.E.; Ashraf, G.M. APOE and Alzheimer’s Disease: Evidence Mounts that Targeting APOE4 may Combat Alzheimer’s Pathogenesis. Mol. Neurobiol. 2019, 56, 2450–2465. [Google Scholar] [CrossRef] [PubMed]
- Al Mamun, A.; Uddin, M.S.; Kabir, M.T.; Khanum, S.; Sarwar, M.S.; Mathew, B.; Rauf, A.; Ahmed, M.; Ashraf, G.M. Exploring the promise of targeting ubiquitin-proteasome system to combat alzheimer’s disease. Neurotox. Res. 2020. [Google Scholar] [CrossRef]
- Zhou, D.; Liu, H.; Li, C.; Wang, F.; Shi, Y.; Liu, L.; Zhao, X.; Liu, A.; Zhang, J.; Wang, C.; et al. Atorvastatin ameliorates cognitive impairment, Aβ1-42 production and Tau hyperphosphorylation in APP/PS1 transgenic mice. Metab. Brain Dis. 2016, 31, 693–703. [Google Scholar] [CrossRef]
- Delatour, B.; Guegan, M.; Volk, A.; Dhenain, M. In vivo MRI and histological evaluation of brain atrophy in APP/PS1 transgenic mice. Neurobiol. Aging 2006, 27, 835–847. [Google Scholar] [CrossRef] [PubMed]
- Poisnel, G.; Dhilly, M.; Moustie, O.; Delamare, J.; Abbas, A.; Guilloteau, D.; Louisa, B. PET imaging with [18F] AV-45 in an APP/PS1-21 murine model of amyloid plaque deposition. Neurobiol. Aging 2012, 33, 2561–2571. [Google Scholar] [CrossRef] [PubMed]
- Lagadec, S.; Rotureau, L.; Hemar, A.; Macrez, N.; Delcasso, S.; Jeantet, Y.; Yoon, H.C. Early temporal short-term memory deficits in double transgenic APP/PS1 mice. Neurobiol. Aging 2012, 33, e201–e211. [Google Scholar] [CrossRef]
- Ferguson, A.S.; Sarkar, S.; Schmued, C.L. Longitudinal behavioral changes in the APP/PS1 transgenic Alzheimer’s disease model. Behav. Brain Res. 2013, 242C, 125–134. [Google Scholar] [CrossRef]
- Bao, J.; Zhang, H.; Hu, K.; Liu, H.; Wang, H.; Li, A.; Lin, F.; Zhang, L.; Sun, X.; Du, Z.; et al. Silencing DNA methyltransferase 1 (DNMT1) inhibits proliferation metastasis and invasion in ESCC by suppressing methylation of RASSF1A and DAPK. Oncotarget 2016, 7, 44129–44141. [Google Scholar] [CrossRef]
- West, R.L.; Lee, J.M.; Maroun, L.E. Hypomethylation of the Amyloid Precursor Protein Gene in the Brain of an Alzheimer’s Disease Patient. J. Mol. Neurosci. Lett. 1995, 6, 141–146. [Google Scholar] [CrossRef]
- Fuso, A.; Seminara, L.; Cavallaro, R.A.; Anselmi, F.; Scarpa, S. S-adenosylmethionine/homocysteine cycle alterations modify DNA methylation status with consequent deregulation of PS1 and BACE and beta-amyloid production. Mol. Cell. Neurosci. 2005, 28, 195–204. [Google Scholar] [CrossRef]
- Yang, B.; Xia, Z.A.; Zhong, B.; Xiong, X.; Sheng, C.; Wang, Y.; Gong, W.; Cao, Y.; Wang, Z.; Peng, W. Distinct hippocampal expression profiles of long non-coding RNAs in an Alzheimer’s disease model. Mol. Neurobiol. 2017, 54, 4833–4846. [Google Scholar] [CrossRef] [PubMed]
- Frater, J.; Lie, D.; Bartlett, P.; McGrath, J.J. Insulin-like Growth Factor 1 (IGF-1) as a marker of cognitive decline in normal ageing: A review. Ageing Res. Rev. 2018, 42, 14–27. [Google Scholar] [CrossRef] [PubMed]
- Sperlagh, B.; Illes, P. P2X7 receptor: An emerging target in central nervous system diseases. Trends Pharmacol. Sci. 2014, 35, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.E. Numb and Numblike control cell number during vertebrate neurogenesis. Trends Neurosci. 2003, 26, 395–396. [Google Scholar] [CrossRef]
- Diaz-Hernandez, J.; Gomez-Villafuertes, R. In vivo P2X7 inhibition reduces amyloid plaques in Alzheimer’sdisease through GSK3 and secretases. Neurobiol. Aging 2012, 33, 1816–1828. [Google Scholar] [CrossRef]
- Cabezas, R.; Baez-Jurado, E.; Hidalgo-Lanussa, O.; Echeverria, V.; Ashraf, G.M.; Sahebkar, A.; Barreto, G.E. Growth Factors and Neuroglobin in Astrocyte Protection Against Neurodegeneration and Oxidative Stress. Mol. Neurobiol. 2019, 56, 2352. [Google Scholar] [CrossRef]
- Repalli, J. Translocator Protein (TSPO) Role in Aging and Alzheimer’s Disease. Curr. Aging Sci. 2012, 7, 168–175. [Google Scholar] [CrossRef]
- Barker, R.; Kehoe, P.G.; Love, S. Activators and inhibitors of the plasminogen system in Alzheimer’s disease. J. Cell. Mol. Med. 2012, 16, 865–876. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Zeng, L.; Jiang, H.; Li, Z.; Liu, R. Microarray Profile of Long Noncoding RNA and Messenger RNA Expression in a Model of Alzheimer’s Disease. Life 2020, 10, 64. https://doi.org/10.3390/life10050064
Wang L, Zeng L, Jiang H, Li Z, Liu R. Microarray Profile of Long Noncoding RNA and Messenger RNA Expression in a Model of Alzheimer’s Disease. Life. 2020; 10(5):64. https://doi.org/10.3390/life10050064
Chicago/Turabian StyleWang, Linlin, Li Zeng, Hailun Jiang, Zhuorong Li, and Rui Liu. 2020. "Microarray Profile of Long Noncoding RNA and Messenger RNA Expression in a Model of Alzheimer’s Disease" Life 10, no. 5: 64. https://doi.org/10.3390/life10050064
APA StyleWang, L., Zeng, L., Jiang, H., Li, Z., & Liu, R. (2020). Microarray Profile of Long Noncoding RNA and Messenger RNA Expression in a Model of Alzheimer’s Disease. Life, 10(5), 64. https://doi.org/10.3390/life10050064




