Diatom-Derived Polyunsaturated Aldehydes Are Unlikely to Influence the Microbiota Composition of Laboratory-Cultured Diatoms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Treatments
2.2. Algal Culture
2.3. Nitrate Determination
2.4. DNA Extraction
2.5. Sequence Analysis
2.6. Alpha and Beta Diversity Analysis
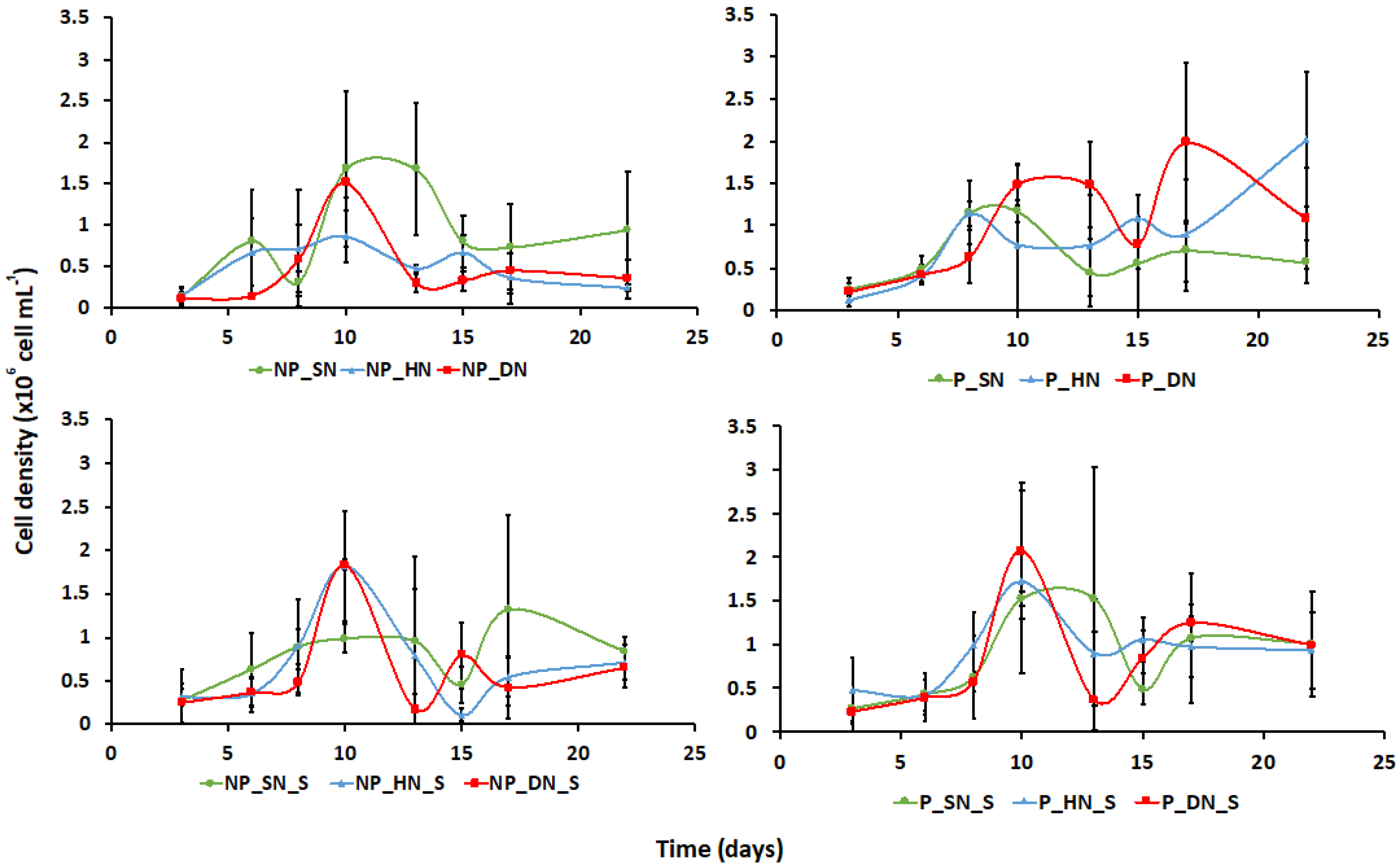
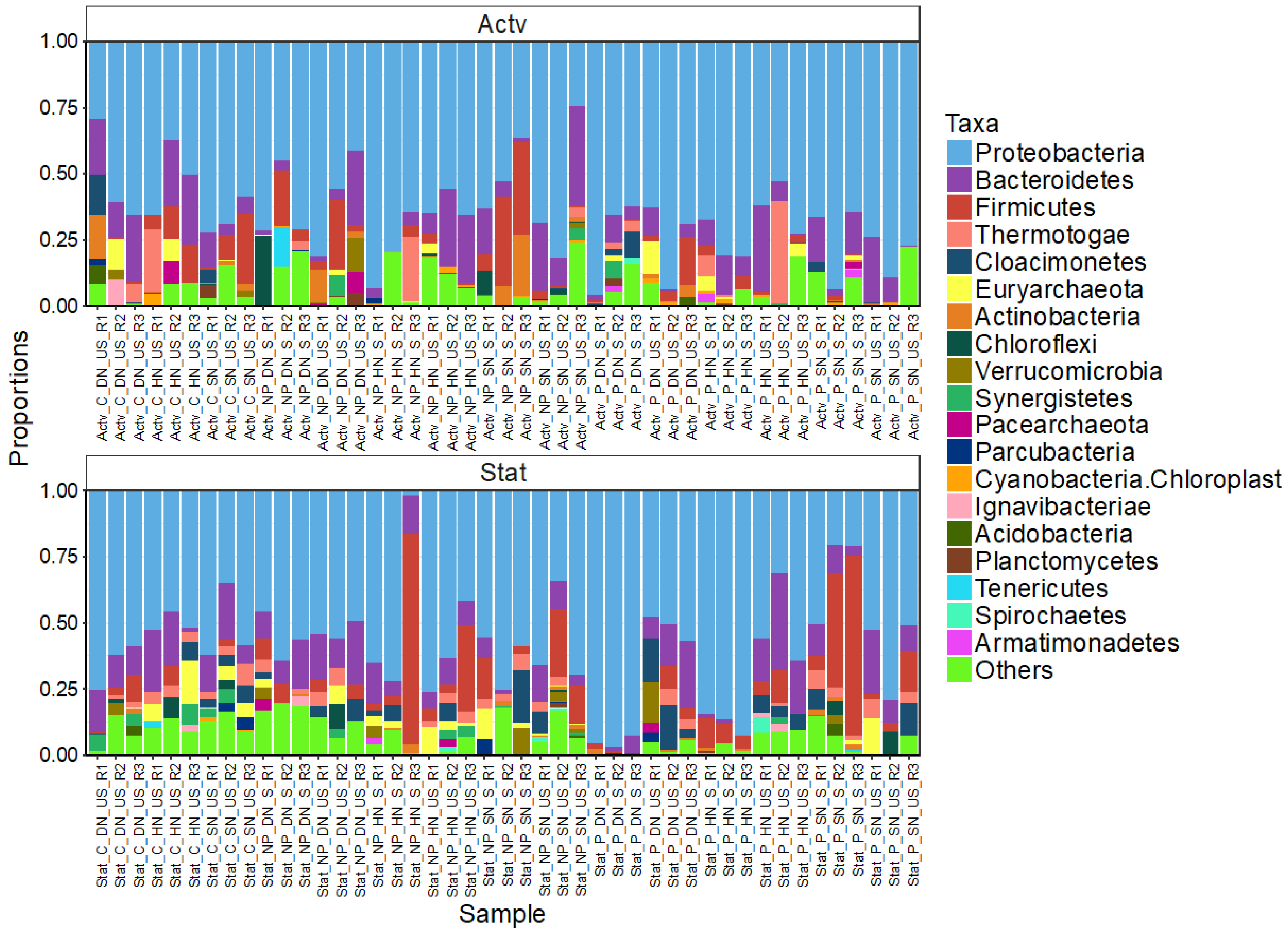
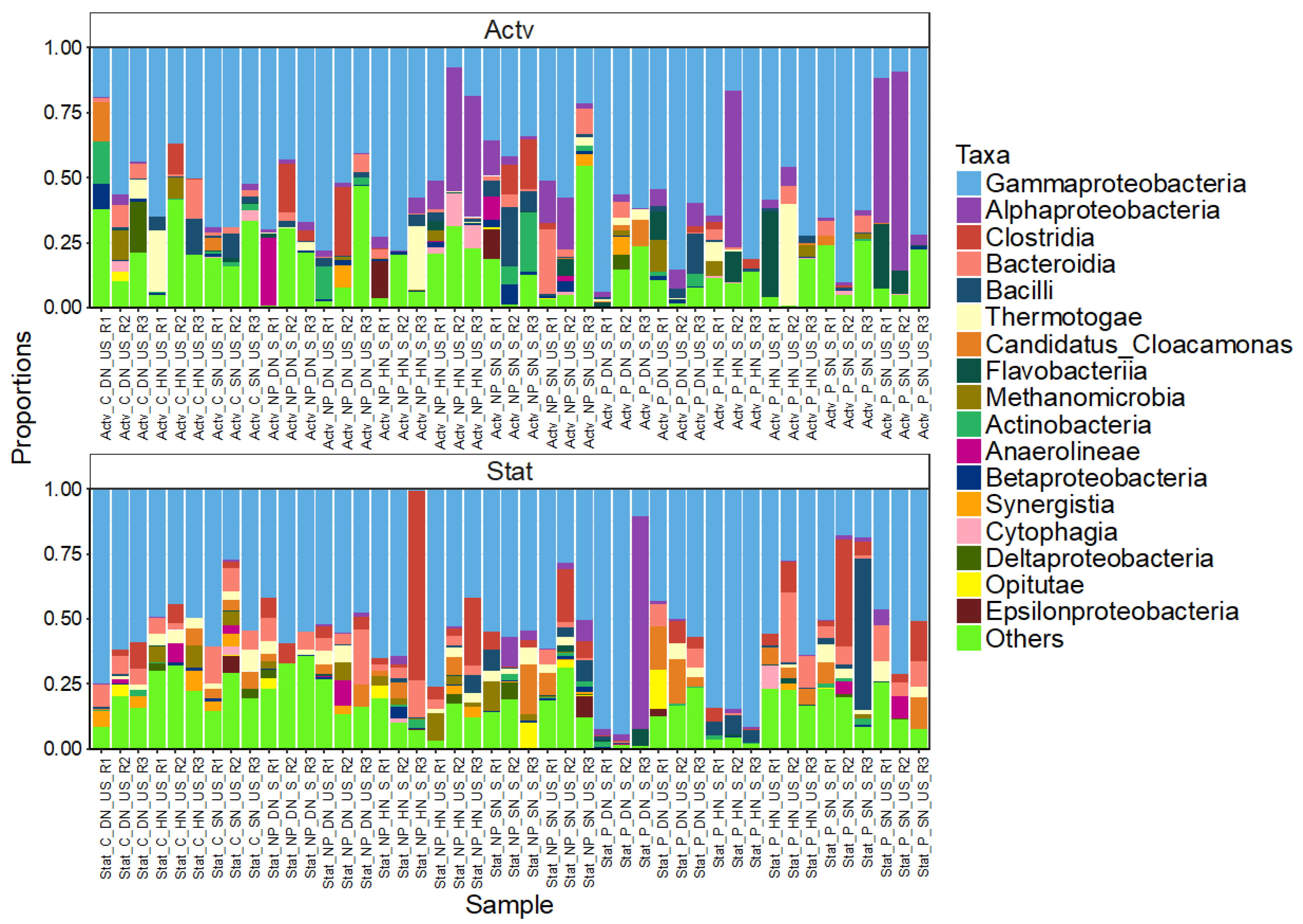
3. Results
3.1. Culture Growth
3.2. Sequencing Analysis
3.3. Community Richness and Diversity Indices
3.4. Bacterial Community Composition
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sarthou, G.; Timmermans, K.R.; Blain, S.; Tréguer, P. Growth physiology and fate of diatoms in the ocean: A review. J. Sea Res. 2005, 53, 25–42. [Google Scholar] [CrossRef]
- Azam, F.; Malfatti, F. Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 2007, 5, 782–791. [Google Scholar] [CrossRef]
- Ribalet, F.; Intertaglia, L.; Lebaron, P.; Casotti, R. Differential effect of three polyunsaturated aldehydes on marine bacterial isolates. Aquat. Toxicol. 2008, 86, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Sison-Mangus, M.P.; Jiang, S.; Kudela, R.M.; Mehic, S. Phytoplankton-associated bacterial community composition and succession during toxic diatom bloom and non-bloom events. Front. Microbiol. 2016, 7, 1433. [Google Scholar] [CrossRef] [PubMed]
- Amin, S.A.; Parker, M.S.; Armbrust, E.V. Interactions between diatoms and bacteria. Microbiol. Mol. Biol. Rev. 2012, 76, 667–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cirri, E.; Pohnert, G. Algae−bacteria interactions that balance the planktonic microbiome. New Phytol. 2019, 223, 100–106. [Google Scholar] [CrossRef] [Green Version]
- Worden, A.Z.; Follows, M.J.; Giovannoni, S.J.; Wilken, S.; Zimmerman, A.E.; Keeling, P.J. Rethinking the marine carbon cycle: Factoring in the multifarious lifestyles of microbes. Science 2015, 347, 1257594. [Google Scholar] [CrossRef] [Green Version]
- Meyer, N.; Rettner, J.; Werner, M.; Werz, O.; Pohnert, G. Algal oxylipins mediate the resistance of diatoms against algicidal bacteria. Mar. Drugs 2018, 16, 486. [Google Scholar] [CrossRef] [Green Version]
- Meyer, N.; Bigalke, A.; Kaulfuß, A.; Pohnert, G. Strategies and ecological roles of algicidal bacteria. Fems Microbiol. Rev. 2017, 41, 880–899. [Google Scholar] [CrossRef] [Green Version]
- Thornton, D.C.O. Dissolved organic matter (DOM) release by phytoplankton in the contemporary and future ocean. Eur. J. Phycol. 2014, 49, 20–46. [Google Scholar] [CrossRef] [Green Version]
- d’Ippolito, G.; Nuzzo, G.; Sardo, A.; Manzo, E.; Gallo, C.; Fontana, A. Lipoxygenases and lipoxygenase products in marine diatoms. Methods Enzymol. 2018, 605, 69–100. [Google Scholar]
- Gallo, C.; D’Ippolito, G.; Nuzzo, G.; Sardo, A.; Fontana, A. Autoinhibitory sterol sulfates mediate programmed cell death in a bloom-forming marine diatom. Nat. Commun. 2017, 8, 1292. [Google Scholar] [CrossRef]
- Russo, E.; d’Ippolito, G.; Fontana, A.; Sarno, D.; D’Alelio, D.; Busseni, G.; Ianora, A.; von Elert, E.; Carotenuto, Y. Density-dependent oxylipin production in natural diatom communities: Possible implications for plankton dynamics. ISME J. 2020, 14, 164–177. [Google Scholar] [CrossRef]
- Stonik, V.; Stonik, I. Low-molecular-weight metabolites from diatoms: Structures, biological roles and biosynthesis. Mar. Drugs 2015, 13, 3672–3709. [Google Scholar] [CrossRef] [Green Version]
- Adolph, S.; Poulet, S.A.; Pohnert, G. Synthesis and biological activity of α,β,γ,δ-unsaturated aldehydes from diatoms. Tetrahedron 2003, 59, 3003–3008. [Google Scholar] [CrossRef]
- Ianora, A.; Bentley, M.G.; Caldwell, G.S.; Casotti, R.; Cembella, A.D.; Engström-Öst, J.; Halsband, C.; Sonnenschein, E.; Legrand, C.; Llewellyn, C.A.; et al. The relevance of marine chemical ecology to plankton and ecosystem function: An emerging field. Mar. Drugs 2011, 9, 1625–1648. [Google Scholar] [CrossRef] [Green Version]
- Miralto, A.; Barone, G.; Romano, G.; Poulet, S.A.; Ianora, A.; Russo, G.L.; Buttino, I.; Mazzarella, G.; Laablr, M.; Cabrini, M.; et al. The insidious effect of diatoms on copepod reproduction. Nature 1999, 402, 173–176. [Google Scholar] [CrossRef]
- Ianora, A.; Miralto, A.; Poulet, S.A.; Carotenuto, Y.; Buttino, I.; Romano, G.; Casotti, R.; Pohnert, G.; Wichard, T.; Colucci-D’Amato, L.; et al. Aldehyde suppression of copepod recruitment in blooms of a ubiquitous planktonic diatom. Nature 2004, 429, 403–407. [Google Scholar] [CrossRef]
- Caldwell, G.S. The influence of bioactive oxylipins from marine diatoms on invertebrate reproduction and development. Mar. Drugs 2009, 7, 367–400. [Google Scholar] [CrossRef] [Green Version]
- Russo, E.; Ianora, A.; Carotenuto, Y. Re-shaping marine plankton communities: Effects of diatom oxylipins on copepods and beyond. Mar. Biol. 2019, 166, 9. [Google Scholar] [CrossRef]
- Vardi, A.; Formiggini, F.; Casotti, R.; De Martino, A.; Ribalet, F.; Miralto, A.; Bowler, C. A stress surveillance system based on calcium and nitric oxide in marine diatoms. PLoS Biol. 2006, 4, 0411–0419. [Google Scholar] [CrossRef] [Green Version]
- Leflaive, J.; Ten-Hage, L. Chemical interactions in diatoms: Role of polyunsaturated aldehydes and precursors. New Phytol. 2009, 184, 794–805. [Google Scholar] [CrossRef]
- Ribalet, F.; Vidoudez, C.; Cassin, D.; Pohnert, G.; Ianora, A.; Miralto, A.; Casotti, R. High plasticity in the production of diatom-derived polyunsaturated aldehydes under nutrient limitation: Physiological and ecological implications. Protist 2009, 160, 444–451. [Google Scholar] [CrossRef]
- Pepi, M.; Heipieper, H.J.; Balestra, C.; Borra, M.; Biffali, E.; Casotti, R. Toxicity of diatom polyunsaturated aldehydes to marine bacterial isolates reveals their mode of action. Chemosphere 2017, 177, 258–265. [Google Scholar] [CrossRef]
- Scholz, B.; Küpper, F.C.; Vyverman, W.; Ólafsson, H.G.; Karsten, U. Chytridiomycosis of marine diatoms-the role of stress physiology and resistance in parasite-host recognition and accumulation of defense molecules. Mar. Drugs 2017, 15, 26. [Google Scholar] [CrossRef] [Green Version]
- Smith, V.J.; Desbois, A.P.; Dyrynda, E.A. Conventional and unconventional antimicrobials from fish, marine invertebrates and micro-algae. Mar. Drugs 2010, 8, 1213–1262. [Google Scholar] [CrossRef] [Green Version]
- Bartual, A.; Vicente-Cera, I.; Flecha, S.; Prieto, L. Effect of dissolved polyunsaturated aldehydes on the size distribution of transparent exopolymeric particles in an experimental diatom bloom. Mar. Biol. 2017, 164, 1131. [Google Scholar] [CrossRef]
- Edwards, B.R.; Bidle, K.D.; Van Mooy, B.A.S. Dose-dependent regulation of microbial activity on sinking particles by polyunsaturated aldehydes: Implications for the carbon cycle. Proc. Natl. Acad. Sci. USA 2015, 112, 5909–5914. [Google Scholar] [CrossRef] [Green Version]
- Adolph, S.; Bach, S.; Blondel, M.; Cueff, A.; Moreau, M.; Pohnert, G.; Poulet, S.A.; Wichard, T.; Zuccaro, A. Cytotoxicity of diatom-derived oxylipins in organisms belonging to different phyla. J. Exp. Biol. 2004, 207, 2935–2946. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Ho, A.Y.T.; Yin, K.; Yuan, X.; Anderson, D.M.; Lee, J.H.W.; Harrison, P.J. Temporal and spatial variations in nutrient stoichiometry and regulation of phytoplankton biomass in Hong Kong waters: Influence of the Pearl River outflow and sewage inputs. Mar. Pollut. Bull. 2008, 57, 335–348. [Google Scholar] [CrossRef] [Green Version]
- Yin, K.; Harrison, P.J. Nitrogen over enrichment in subtropical Pearl River estuarine coastal waters: Possible causes and consequences. Cont. Shelf Res. 2008, 28, 1435–1442. [Google Scholar] [CrossRef]
- Ribalet, F.; Wichard, T.; Pohnert, G.; Ianora, A.; Miralto, A.; Casotti, R. Age and nutrient limitation enhance polyunsaturated aldehyde production in marine diatoms. Phytochemistry 2007, 68, 2059–2067. [Google Scholar] [CrossRef] [PubMed]
- Lauritano, C.; Andersen, J.H.; Hansen, E.; Albrigtsen, M.; Escalera, L.; Esposito, F.; Helland, K.; Hanssen, K.Ø.; Romano, G.; Ianora, A. Bioactivity screening of microalgae for antioxidant, anti-inflammatory, anticancer, anti-diabetes, and antibacterial activities. Front. Mar. Sci. 2016, 3, 1405. [Google Scholar] [CrossRef] [Green Version]
- Vidoudez, C.; Pohnert, G. Growth phase-specific release of polyunsaturated aldehydes by the diatom Skeletonema marinoi. J. Plankton Res. 2008, 30, 1305–1313. [Google Scholar] [CrossRef] [Green Version]
- Bartual, A.; Ortega, M.J. Temperature differentially affects the persistence of polyunsaturated aldehydes in seawater. Environ. Chem. 2013, 10, 403–408. [Google Scholar] [CrossRef]
- Morillo-García, S.; Valcaŕcel-Pérez, N.; Cózar, A.; Ortega, M.J.; Maciás, D.; Ramiŕez-Romero, E.; García, C.M.; Echevarría, F.; Bartual, A. Potential polyunsaturated aldehydes in the Strait of Gibraltar under two tidal regimes. Mar. Drugs 2014, 12, 1438–1459. [Google Scholar] [CrossRef] [Green Version]
- Bartual, A.; Arandia-Gorostidi, N.; Cózar, A.; Morillo-Garciá, S.; Ortega, M.J.; Vidal, M.; Cabello, A.M.; Gonzaĺez-Gordillo, J.I.; Echevarría, F. Polyunsaturated aldehydes from large phytoplankton of the Atlantic Ocean surface (42° N to 33° S). Mar. Drugs 2014, 12, 682–699. [Google Scholar] [CrossRef] [Green Version]
- Cózar, A.; Morillo-García, S.; Ortega, M.J.; Li, Q.P.; Bartual, A. Macroecological patterns of the phytoplankton production of polyunsaturated aldehydes. Sci. Rep. 2018, 8, 12282. [Google Scholar] [CrossRef]
- Bartual, A.; Morillo-García, S.; Ortega, M.J.; Cózar, A. First report on vertical distribution of dissolved polyunsaturated aldehydes in marine coastal waters. Mar. Chem. 2018, 204, 1–10. [Google Scholar] [CrossRef]
- Guillard, R.R.; Ryther, J.H. Studies of marine planktonic diatoms: I. Cyclotella nana Hustedt, and Detonula confervacea (Cleve) Gran. Can. J. Microbiol. 1962, 8, 229–240. [Google Scholar] [CrossRef]
- Caldwell, G.S. Diatom Mediated Disruption of Invertebrate Reproduction and Development; Newcastle University: Newcastle, UK, 2004. [Google Scholar]
- Collos, Y.; Mornet, F.; Sciandra, A.; Waser, N.; Larson, A.; Harrison, P.J. An optical method for the rapid measurement of micromolar concentrations of nitrate in marine phytoplankton cultures. J. Appl. Phycol. 1999, 11, 179–184. [Google Scholar] [CrossRef]
- Kozich, J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torondel, B.; Ensink, J.H.J.; Gundogdu, O.; Ijaz, U.Z.; Parkhill, J.; Abdelahi, F.; Nguyen, V.A.; Sudgen, S.; Gibson, W.; Walker, A.W.; et al. Assessment of the influence of intrinsic environmental and geographical factors on the bacterial ecology of pit latrines. Microb. Biotechnol. 2016, 9, 209–223. [Google Scholar] [CrossRef] [Green Version]
- Rooney-Varga, J.N.; Giewat, M.W.; Savin, M.C.; Sood, S.; LeGresley, M.; Martin, J.L. Links between phytoplankton and bacterial community dynamics in a coastal marine environment. Microb. Ecol. 2005, 49, 163–175. [Google Scholar] [CrossRef]
- d’Ippolito, G.; Romano, G.; Iadicicco, O.; Miralto, A.; Ianora, A.; Cimino, G.; Fontana, A. New birth-control aldehydes from the marine diatom Skeletonema costatum: Characterization and biogenesis. Tetrahedron Lett. 2002, 43, 6133–6136. [Google Scholar] [CrossRef]
- Paul, C.; Reunamo, A.; Lindehoff, E.; Bergkvist, J.; Mausz, M.A.; Larsson, H.; Richter, H.; Wängberg, S.-Å.; Leskinen, P.; Båmstedt, U.; et al. Diatom derived polyunsaturated aldehydes do not structure the planktonic microbial community in a mesocosm study. Mar. Drugs 2012, 10, 775–792. [Google Scholar] [CrossRef]
- Riemann, L.; Steward, G.F.; Azam, F. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl. Environ. Microbiol. 2000, 66, 578–587. [Google Scholar] [CrossRef] [Green Version]
- Grossart, H.-P.; Levold, F.; Allgaier, M.; Simon, M.; Brinkhoff, T. Marine diatom species harbour distinct bacterial communities. Environ. Microbiol. 2005, 7, 860–873. [Google Scholar] [CrossRef]
- Simek, K.; Hornak, K.; Jezbera, J.; Nedoma, J.; Znachor, P.; Hejzlar, J.; Sed’a, J. Spatio-temporal patterns of bacterioplankton production and community composition related to phytoplankton composition and protistan bacterivory in a dam reservoir. Aquat. Microb. Ecol. 2008, 51, 249–262. [Google Scholar] [CrossRef] [Green Version]
- Lauritano, C.; Romano, G.; Roncalli, V.; Amoresano, A.; Fontanarosa, C.; Bastianini, M.; Braga, F.; Carotenuto, Y.; Ianora, A. New oxylipins produced at the end of a diatom bloom and their effects on copepod reproductive success and gene expression levels. Harmful Algae 2016, 55, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Ribalet, F.; Bastianini, M.; Vidoudez, C.; Acri, F.; Berges, J.; Ianora, A.; Miralto, A.; Pohnert, G.; Romano, G.; Wichard, T.; et al. Phytoplankton cell lysis associated with polyunsaturated aldehyde release in the northern Adriatic Sea. PLoS ONE 2014, 9, e85947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prieto, A.; Barber-Lluch, E.; Hernández-Ruiz, M.; MartÍnez-GarcÍa, S.; Fernández, E.; Teira, E. Assessing the role of phytoplankton–bacterioplankton coupling in the response of microbial plankton to nutrient additions. J. Plankton Res. 2016, 38, 55–63. [Google Scholar] [CrossRef]
- Logares, R.; Audic, S.; Bass, D.; Bittner, L.; Boutte, C.; Christen, R.; Claverie, J.-M.; Decelle, J.; Dolan, J.R.; Dunthorn, M.; et al. Patterns of rare and abundant marine microbial eukaryotes. Curr. Biol. 2014, 24, 813–821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wear, E.K.; Carlson, C.A.; James, A.K.; Brzezinski, M.A.; Windecker, L.A.; Nelson, C.E. Synchronous shifts in dissolved organic carbon bioavailability and bacterial community responses over the course of an upwelling-driven phytoplankton bloom. Limnol. Oceanogr. 2015, 60, 657–677. [Google Scholar] [CrossRef]
- Zheng, X.; Xiao, L.; Ren, J.; Yang, L. The effect of a Microcystis aeruginosa bloom on the bacterioplankton community composition of Lake Xuanwa. J. Freshw. Ecol. 2008, 23, 297–304. [Google Scholar] [CrossRef] [Green Version]
- Wear, E.K.; Carlson, C.A.; Windecker, L.A.; Brzezinski, M.A. Roles of diatom nutrient stress and species identity in determining the short- and long-term bioavailability of diatom exudates to bacterioplankton. Mar. Chem. 2015, 177, 335–348. [Google Scholar] [CrossRef]
- Logue, J.B.; Langenheder, S.; Andersson, A.F.; Bertilsson, S.; Drakare, S.; Lanzen, A.; Lindstrom, E.S. Freshwater bacterioplankton richness in oligotrophic lakes depends on nutrient availability rather than on species-area relationships. ISME J. 2012, 6, 1127–1136. [Google Scholar] [CrossRef] [Green Version]
- Zakharova, Y.R.; Galachyants, Y.P.; Kurilkina, M.I.; Likhoshvay, A.V.; Petrova, D.P.; Shishlyannikov, S.M.; Ravin, N.V.; Mardanov, A.V.; Beletsky, A.V.; Likhoshway, Y.V. The structure of microbial community and degradation of diatoms in the deep near-bottom layer of Lake Baikal. PLoS ONE 2013, 8, e59977. [Google Scholar] [CrossRef] [Green Version]
- Riemann, L.; Winding, A. Community dynamics of free-living and particle-associated bacterial assemblages during a freshwater phytoplankton bloom. Microb. Ecol. 2001, 42, 274–285. [Google Scholar] [CrossRef] [PubMed]
- Naviner, M.; Bergé, J.P.; Durand, P.; Le Bris, H. Antibacterial activity of the marine diatom Skeletonema costatum against aquacultural pathogens. Aquaculture 1999, 174, 15–24. [Google Scholar] [CrossRef]
- González-Davis, O.; Ponce-Rivas, E.; Sánchez-Saavedra, M.D.P.; Muñoz-Márquez, M.E.; Gerwick, W.H. Bioprospection of microalgae and cyanobacteria as biocontrol agents against Vibrio campbellii and their use in white shrimp Litopenaeus vannamei culture. J. World Aquac. Soc. 2012, 43, 387–399. [Google Scholar] [CrossRef]
- Lauritano, C.; Martín, J.; De La Cruz, M.; Reyes, F.; Romano, G.; Ianora, A. First identification of marine diatoms with anti-tuberculosis activity. Sci. Rep. 2018, 8, 2284. [Google Scholar] [CrossRef] [Green Version]
- Balestra, C.; Alonso-Sáez, L.; Gasol, J.M.; Casotti, R. Group-specific effects on coastal bacterioplankton of polyunsaturated aldehydes produced by diatoms. Aquat. Microb. Ecol. 2011, 63, 123–131. [Google Scholar] [CrossRef] [Green Version]
- Shao, K.; Zhang, L.; Wang, Y.; Yao, X.; Tang, X.; Qin, B.; Gao, G. The responses of the taxa composition of particle-attached bacterial community to the decomposition of Microcystis blooms. Sci. Total Environ. 2014, 488–489, 236–242. [Google Scholar] [CrossRef]
- Kirchman, D.L. The ecology of Cytophaga–Flavobacteria in aquatic environments. Fems Microb. Ecol. 2002, 39, 91–100. [Google Scholar] [CrossRef]
- Alonso-Saez, L.; Aristegui, J.; Pinhassi, J.; Gomez-Consarnau, L.; Gonzalez, J.M.; Vaque, D.; Agusti, S.; Gasol, J.M. Bacterial assemblage structure and carbon metabolism along a productivity gradient in the NE Atlantic Ocean. Aquat. Microb. Ecol. 2007, 46, 43–53. [Google Scholar] [CrossRef] [Green Version]
- Boone, D.R.; Castenholz, R.W.; Garrity, G.M. Bergey’s Manual of Systematic Bacteriology; The Archaea and the Deeply Branching and Phototrophic Bacteria; Springer: New York, NY, USA, 2001; Volume 1, p. 722. [Google Scholar]
- Bowman, J.S.; Vick-Majors, T.J.; Morgan-Kiss, R.; Takacs-Vesbach, C.; Ducklow, H.W.; Priscu, J.C. Microbial community dynamics in two polar extremes: The lakes of the McMurdo Dry Valleys and the West Antarctic Peninsula marine ecosystem. BioScience 2016, 66, 829–847. [Google Scholar] [CrossRef] [Green Version]
- Kamp, A.; Stief, P.; Bristow, L.A.; Thamdrup, B.; Glud, R.N. Intracellular nitrate of marine diatoms as a driver of anaerobic nitrogen cycling in sinking aggregates. Front. Microbiol. 2016, 7, 1669. [Google Scholar] [CrossRef]
- Lewis, J.; Harris, A.S.D.; Jones, K.J.; Edmonds, R.L. Long-term survival of marine planktonic diatoms and dinofiagellates in stored sediment samples. J. Plankton Res. 1999, 21, 343–354. [Google Scholar] [CrossRef] [Green Version]
- McQuoid, M.R.; Godhe, A.; Nordberg, K. Viability of phytoplankton resting stages in the sediments of a coastal Swedish fjord. Eur. J. Phycol. 2002, 37, 191–201. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eastabrook, C.L.; Whitworth, P.; Robinson, G.; Caldwell, G.S. Diatom-Derived Polyunsaturated Aldehydes Are Unlikely to Influence the Microbiota Composition of Laboratory-Cultured Diatoms. Life 2020, 10, 29. https://doi.org/10.3390/life10030029
Eastabrook CL, Whitworth P, Robinson G, Caldwell GS. Diatom-Derived Polyunsaturated Aldehydes Are Unlikely to Influence the Microbiota Composition of Laboratory-Cultured Diatoms. Life. 2020; 10(3):29. https://doi.org/10.3390/life10030029
Chicago/Turabian StyleEastabrook, Chloe L., Paul Whitworth, Georgina Robinson, and Gary S. Caldwell. 2020. "Diatom-Derived Polyunsaturated Aldehydes Are Unlikely to Influence the Microbiota Composition of Laboratory-Cultured Diatoms" Life 10, no. 3: 29. https://doi.org/10.3390/life10030029



