Exposure to Formaldehyde Perturbs the Mouse Gut Microbiome
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Animals and Design
2.2. 16S Ribosomal RNA Gene Sequencing
2.3. High-Throughput 16S rRNA Gene Sequencing Data Analysis
2.4. Statistical and Bioinformatic Analysis
3. Results
3.1. Richness and Diversity of the Mouse Gut Microbiome
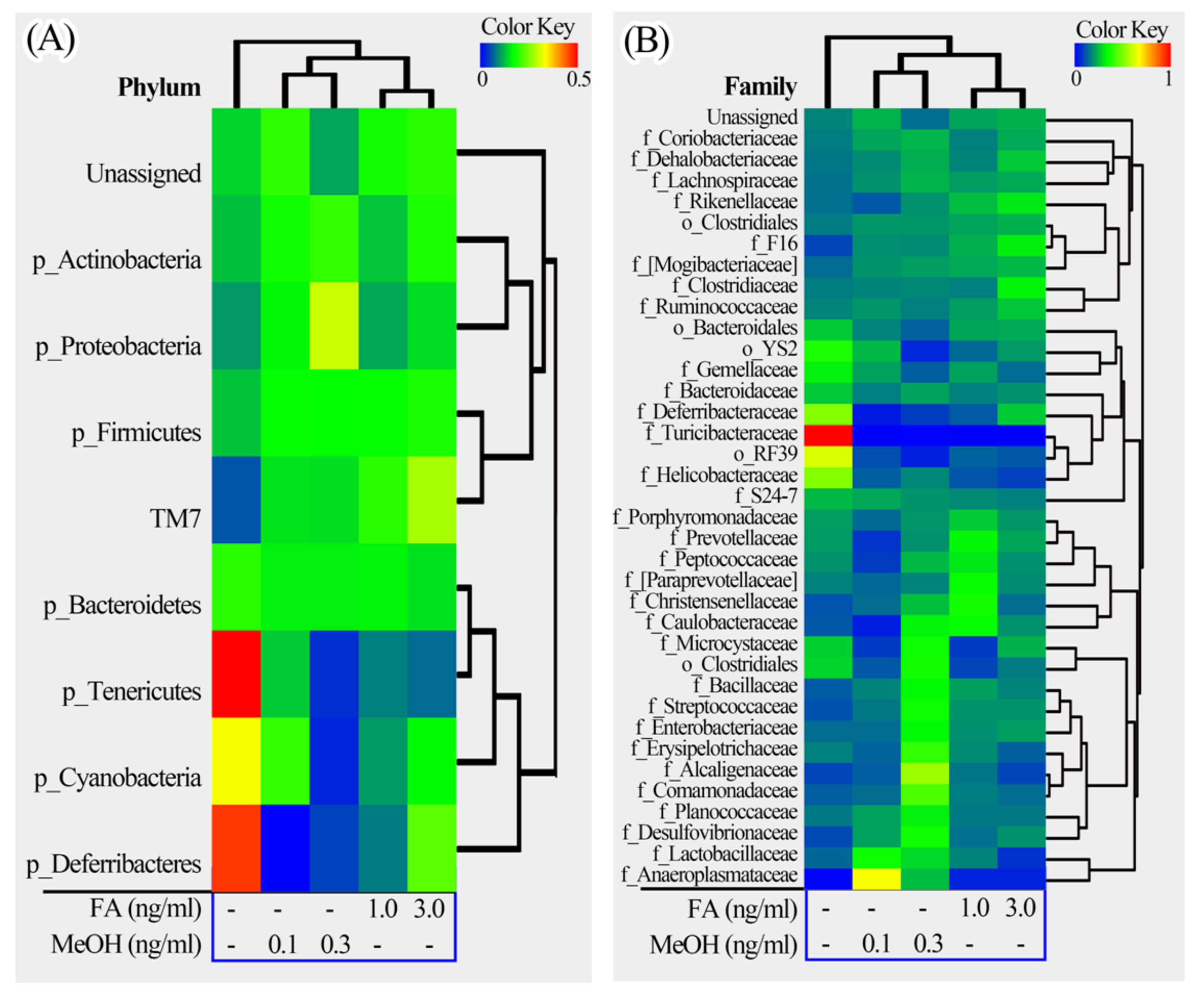
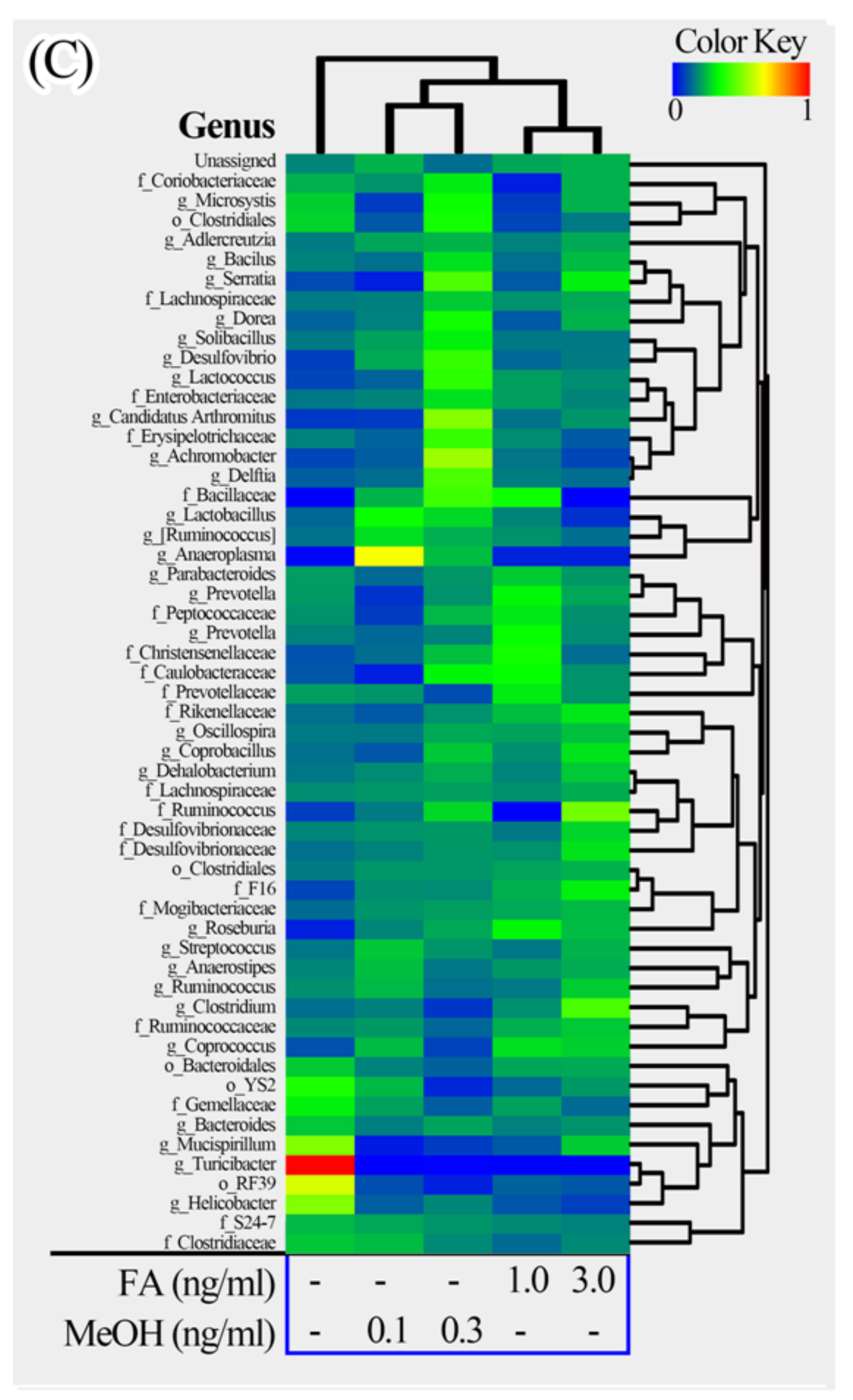
3.2. Mouse Gut Microbiota Composition Profiles
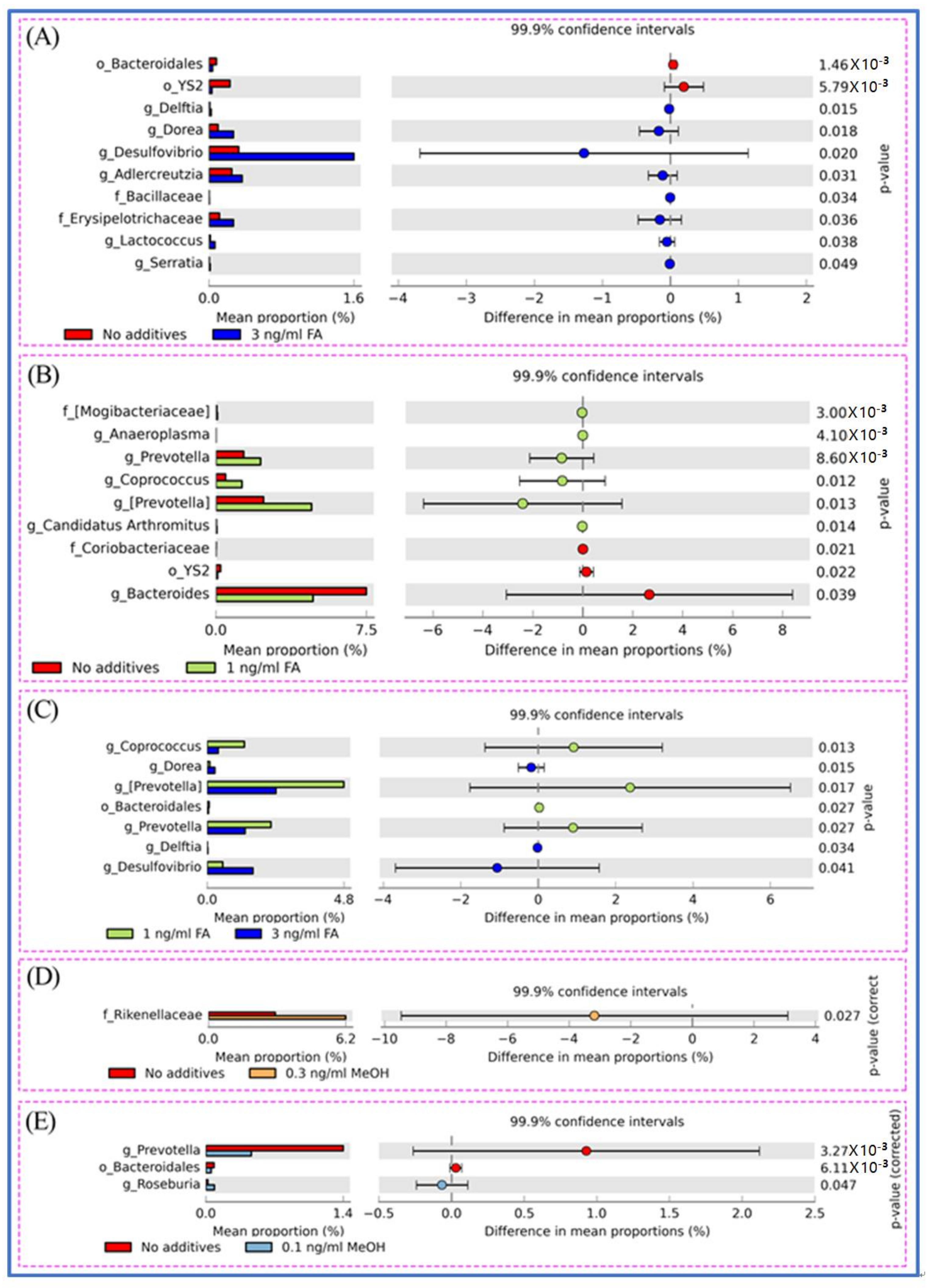
3.3. Microbial Taxa Changes Associated with FA Treatment
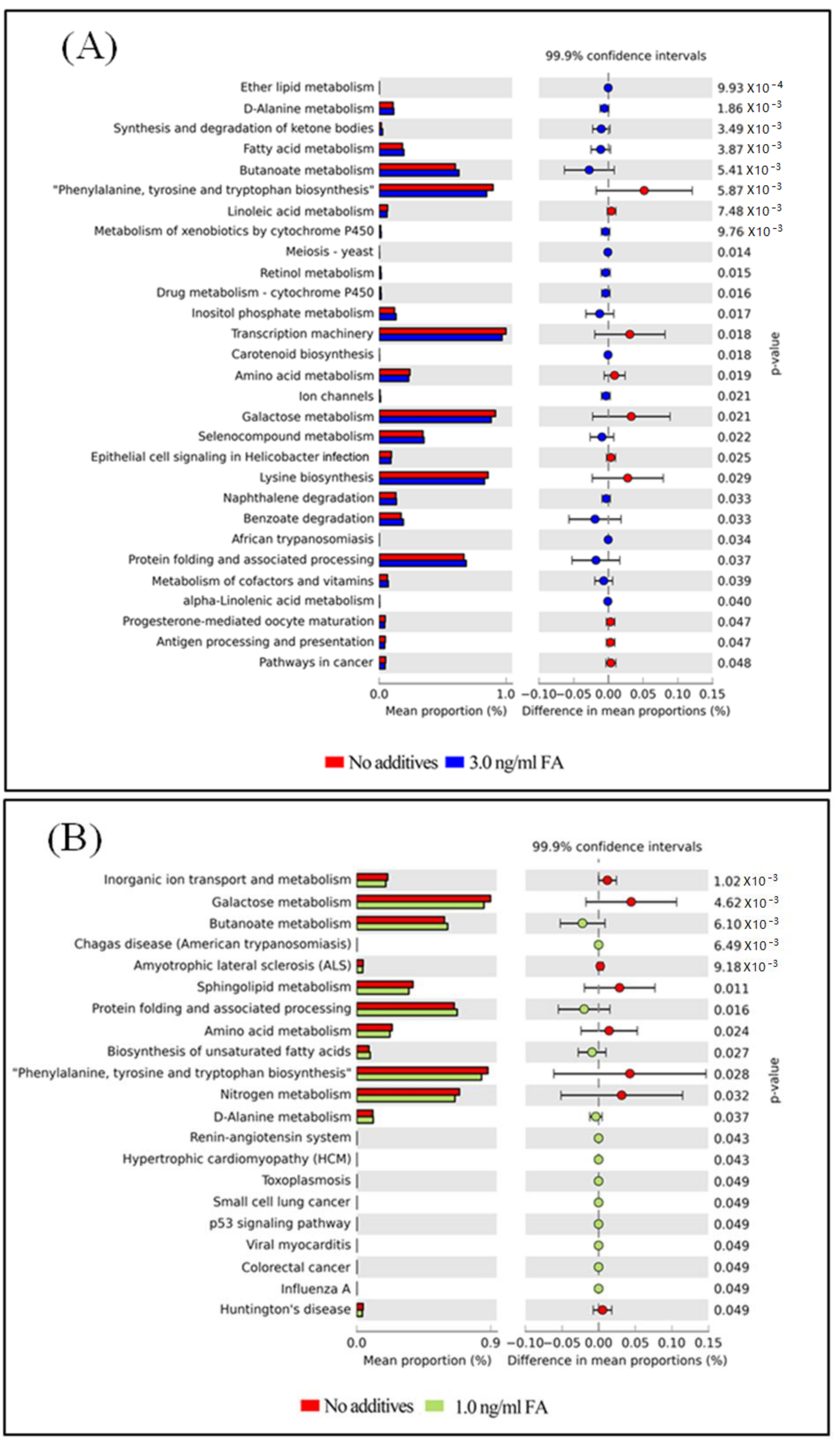
3.4. Functional Capacity Profiling Prediction of the Gut Microbiome
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ribière, C.; Peyret, P.; Parisot, N.; Darcha, C.; Déchelotte, P.J.; Barnich, N.; Peyretaillade, E.; Boucher, D. Oral exposure to environmental pollutant benzo[a]pyrene impacts the intestinal epithelium and induces gut microbial shifts in murine model. Sci. Rep. 2016, 6, 31027. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Delzenne, N.M. The gut microbiome as therapeutic target. Pharmacol. Ther. 2011, 130, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Peterson, D.A.; Gordon, J.I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 2006, 124, 837–848. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, A.M.; Shanahan, F. The gut flora as a forgotten organ. EMBO Rep. 2006, 7, 688–693. [Google Scholar] [CrossRef] [PubMed]
- Palmer, C.; Bik, E.M.; DiGiulio, D.B.; Relman, D.A.; Brown, P.O. Development of the human infant intestinal microbiota. PLoS ONE 2007, 5, e177. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, L.; Wang, J.; Wei, S.; Ni, Y. Host-microbial interactions and regulation of intestinal epithelial barrier function: From physiology to pathology. World J. Gastrointest. Pathophysiol. 2012, 3, 27–43. [Google Scholar] [CrossRef] [PubMed]
- Young, V.B.; Britton, R.A.; Schmidt, T.M. The human microbiome and infectious diseases: Beyond Koch. Interdiscip. Perspect. Infect. Dis. 2008, 2008, 296873. [Google Scholar] [CrossRef] [PubMed]
- Vos, W.M.; Vos, E.A. Role of the intestinal microbiome in health and disease: From correlation to causation. Nutr. Rev. 2012, 70, S45–S56. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Klipfell, E.; Bennett, B.J.; Koeth, R.; Levison, B.S.; DuGar, B.; Feldstein, A.E.; Britt, E.B.; Fu, X.; Chung, Y.M.; et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011, 472, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Bäckhed, F.; Turnbaugh, P.; Lozupone, C.A.; Knight, R.D.; Gordon, J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 2005, 102, 11070–11075. [Google Scholar] [CrossRef] [PubMed]
- Larsen, N.; Vogensen, F.K.; van den Berg, F.W.; Nielsen, D.S.; Andreasen, A.S.; Pedersen, B.K.; Al-Soud, W.A.; Sørensen, S.J.; Hansen, L.H.; Jakobsen, M. Gut microbiota in human adults with Type 2 diabetes differs from non-diabetic adults. PLoS ONE 2010, 5, e9085. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Abo, R.P.; Schlieper, K.A.; Graffam, M.E.; Levine, S.; Wishnok, J.S.; Swenberg, J.A.; Tannenbaum, S.R.; Fox, J.G. Arsenic exposure perturbs the gut microbiome and its metabolic profile in mice: An integrated metagenomics and metabolomics analysis. Environ. Health Perspect. 2014, 122, 284–291. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Huang, K.; Chen, S.; Qi, X.; He, X.; Cheng, W.H.; Luo, Y.; Xia, K.; Xu, W. Combination of metagenomics and culture-based methods to study the interaction between ochratoxin A and gut microbiota. Toxicol. Sci. 2014, 141, 314–326. [Google Scholar] [CrossRef] [PubMed]
- Chi, L.; Gao, B.; Bian, X.; Tu, P.; Ru, H.; Lu, K. Manganese-induced sex-specific gut microbiome perturbations in C57BL/6 mice. Toxicol. Appl. Pharmacol. 2017, 331, 142–153. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Nichols, R.G.; Correll, J.; Murray, I.A.; Tanaka, N.; Smith, P.B.; Hubbard, T.D.; Sebastian, A.; Albert, I.; Hatzakis, E.; et al. Persistent organic pollutants modify gut microbiota-host metabolic homeostasis in mice through aryl hydrocarbon receptor activation. Environ. Health Perspect. 2015, 123, 679–688. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Bian, X.; Mahbub, R.; Lu, K. Sex-specific effects of organophosphate diazinon on the gut microbiome and its metabolic functions. Environ. Health Perspect. 2017, 125, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Bai, Y.; Duong, A.; Martyn, T.S.; Li, L.; Zhang, L. Formaldehyde in China: Production, consumption, exposure levels, and health effects. Environ. Int. 2009, 35, 1210–1224. [Google Scholar] [CrossRef] [PubMed]
- Aydin, S.; Canpinar, H.; Ülkü, Ü.; Güç, D.; Çolakoǧlu, M.; Kars, A.; Başaran, N. Assessment of immunotoxicity and genotoxicity in workers exposed to low concentrations of formaldehyde. Arch. Toxicol. 2013, 87, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Tulpule, K.; Dringen, R. Formaldehyde in brain: An overlooked player in neurodegeneration? J. Neurochem. 2013, 127, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Gerin, F.; Erman, H.; Erboga, M.; Sener, U.; Yilmaz, A.; Seyhan, H.; Gurel, A. The effects of ferulic acid against oxidative stress and inflammation in formaldehyde-induced hepatotoxicity. Inflammation 2016, 39, 1377–1386. [Google Scholar] [CrossRef] [PubMed]
- Ramos, C.O.; Nardeli, C.R.; Campos, K.K.; Pena, K.B.; Machado, D.F.; Bandeira, A.C.; de Paula Costa, G.; Talvani, A.; Bezerra, F.S. The exposure to formaldehyde causes renal dysfunction, inflammation and redox imbalance in rats. Exp. Toxicol. Pathol. 2017, 69, 367–372. [Google Scholar] [CrossRef] [PubMed]
- IARC. Formaldehyde, 2-Butoxyethanol and 1-Tert-Butoxypropan-2-Ol. Monographs on the Evaluation of Carcinogenic Risks to Humans; World Health Organization: Geneva, Switzerland, 2006; Available online: https://www.ncbi.nlm.nih.gov/pubmed/17366697 (accessed on 3 April 2018).
- IARC. Monographs on the Evaluation of Carcinogenic Risks to Humans-Formaldehyde; World Health Organization: Geneva, Switzerland, 2012; 100, Part F; pp. 401–436. Available online: http://www.docin.com/p-542499166.html (accessed on 30 March 2018).
- Tan, T.; Zhang, Y.; Luo, W.H.; Lv, J.H.; Han, C.H.; Hamlin, J.N.R.; Luo, H.J.; Li, H.; Wan, Y.; Yang, X.; Song, W.H.; Tong, Z.Q. Formaldehyde induces diabetes-associated cognitive impairments. FASEB J. 2018. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Steinmaus, C.; Eastmond, D.A.; Xin, X.K.; Smith, M.T. Formaldehyde exposure and leukemia: A new meta-analysis and potential mechanisms. Mutat. Res. Rev. 2009, 681, 150–168. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Wen, H.; Yuan, L.; McHale, C.M.; Li, R.; Wang, K.; Yuan, J.; Yang, X.; Zhang, L. Formaldehyde induces toxicity in mouse bone marrow and hematopoietic stem/progenitor cells and enhances benzene-induced adverse effects. Arch. Toxicol. 2017, 91, 921–933. [Google Scholar] [CrossRef] [PubMed]
- Wen, H.; Yuan, L.; Wei, C.; Zhao, Y.; Qian, Y.; Ma, P.; Ding, S.; Yang, X.; Wang, X. Effects of combined exposure to formaldehyde and benzene on immune cells in the blood and spleen in Balb/c mice. Environ. Toxicol. Pharmacol. 2016, 45, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 8, 1–10. Available online: http://picrust.github.io/picrust/ (accessed on 29 March 2018). [CrossRef] [PubMed] [Green Version]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Gonzalez Pena, A.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010. Available online: http://qiime.org (accessed on 29 March 2018). [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA cene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E.; et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, F.J.; Blanchet, G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. 2018. Available online: https://CRAN.R-project.org/package=vegan (accessed on 29 March 2018).
- Jiang, X.P.; Weitz, J.S.; Dushoff, J. A non-negative matrix factorization framework for identifying modular patterns in metagenomic profile data. J. Math. Biol. 2012, 64, 697–711. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Wang, Y.; Liu, Z.; Cheng, H.; Xue, Y. HemI: A toolkit for illustrating heatmaps. PLoS ONE 2014, 9, e111988. [Google Scholar] [CrossRef] [PubMed]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef] [PubMed]
- Dimova, L.G.; Zlatkov, N.; Verkade, H.J.; Uhlin, B.E.; Tietge, U.J.F. High-cholesterol diet does not alter gut microbiota composition in mice. Nutr. Metab. 2017, 14, 15. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.J.; Eum, S.Y.; Rampersaud, E.; Daunert, S.; Abreu, M.T.; Toborek, M. Exercise attenuates PCB-induced changes in the mouse gut microbiome. Environ. Health Perspect. 2013, 121, 725–730. [Google Scholar] [CrossRef] [PubMed]
- Breton, J.; Massart, S.; Vandamme, P.; Brandt, E.D.; Pot, B.; Foligné, B. Ecotoxicology inside the gut: Impact of heavy metals on the mouse microbiome. BMC Pharmacol. Toxicol. 2013, 14, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, T.L.A.; Vieira-Silva, S.; Liston, A.; Raes, J. How informative is the mouse for human gut microbiota research? Dis. Models Mech. 2015, 8, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Everard, A.; Lazarevic, V.; Gaïa, N.; Johansson, M.; Ståhlman, M.; Backhed, F.; Delzenne, N.M.; Schrenzel, J.; Francois, P.; Cani, P.D. Microbiome of prebiotic-treated mice reveals novel targets involved in host response during obesity. ISME J. 2014, 8, 2116–2130. [Google Scholar] [CrossRef] [PubMed]
- Schwab, C.; Berry, D.; Rauch, I.; Rennisch, I.; Ramesmayer, J.; Hainzl, E.; Heider, S.; Decker, T.; Kenner, L.; Müller, M.; et al. Longitudinal study of murine microbiota activity and interactions with the host during acute inflammation and recovery. ISME J. 2014, 8, 1101–1114. [Google Scholar] [CrossRef] [PubMed]
- Pedrós-Alió, C. The rare bacterial biosphere. Ann. Rev. Mar. Sci. 2012, 4, 449–466. [Google Scholar] [CrossRef] [PubMed]
- Hawley, C.K.; Harsch, H.H. Gastric outlet obstruction as a late complication of formaldehyde ingestion: A case report. Am. J. Gastroenterol. 1999, 94, 2289–2291. [Google Scholar] [CrossRef] [PubMed]
- Sidhu, K.S.; Sidhu, J.S. An alleged poisoning with methanol and formaldehyde. Vet. Hum. Toxicol. 1999, 41, 237–242. [Google Scholar] [PubMed]
- Kamata, E.; Nakadate, M.; Uchida, O.; Ogawa, Y.; Kaneko, T.; Kurokawa, Y. Effects of formaldehyde vapor on the nasal cavity and lungs of F-344 rats. J. Environ. Pathol. Toxicol. Oncol. 1996, 15, 1–8. [Google Scholar] [PubMed]
- Casset, A.; Purohit, A.; Marchand, C.L.; Le, S.C.; Donnay, C.; Uring-Lambert, B.; Bahram, S.; Pauli, G. The bronchial response to inhaled formaldehyde. Revue des Maladies Respiratoires 2006, 23, 3S25–3S34. [Google Scholar] [PubMed]
- Tang, X.Q.; Zhuang, Y.Y.; Zhang, P.; Fang, H.R.; Zhou, C.F.; Gu, H.F.; Zhang, H.; Wang, C.Y. Formaldehyde impairs learning and memory involving the disturbance of hydrogen sulfide generation in the hippocampus of rats. J. Mol. Neurosci. 2013, 49, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Strubelt, O.; Younes, M.; Pentz, R.; Kuhnel, W. Mechanistic study on formaldehyde-induced hepatotoxicity. J. Toxicol. Environ. Health 1989, 27, 351–366. [Google Scholar] [CrossRef] [PubMed]
- Lima, L.F.; Murta, G.L.; Bandeira, A.C.; Nardeli, C.R.; Lima, W.G.; Bezerra, F.S. Short-term exposure to formaldehyde promotes oxidative damage and inflammation in the trachea and diaphragm muscle of adult rats. Ann. Anat. 2015, 202, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Bakar, E.; Ulucam, E.; Cerkezkayabekir, A. Investigation of the protective effects of proanthocyanidin and vitamin E against the toxic effect caused by formaldehyde on the liver tissue. Environ. Toxicol. 2015, 30, 1406–1415. [Google Scholar] [CrossRef] [PubMed]
- Lukens, J.R.; Gurung, P.; Vogel, P.; Johnson, G.R.; Carter, R.A.; McGoldrick, D.J.; Bandi, S.R.; Calabrese, C.R.; Walle, L.V.; Lamkanfi, M.; et al. Dietary modulation of the microbiome affects autoinflammatory disease. Nature 2014, 516, 246–249. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Ling, Z.; Zhang, Y.; Mao, H.; Ma, Z.; Yin, Y.; Wang, W.; Tang, W.; Tan, Z.; Shi, J.; et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav. Immun. 2015, 48, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Figliuolo, V.R.; dos Santos, L.M.; Abalo, A.; Nanini, H.; Santos, A.; Brittes, N.M.; Bernardazzi, C.; de Souza, H.S.; Vieira, L.Q.; Coutinho-Silva, R.; et al. Sulfate-reducing bacteria stimulate gut immune responses and contribute to inflammation in experimental colitis. Life Sci. 2017, 189, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Frankel, A.E.; Coughlin, L.A.; Kim, J.; Froehlich, T.W.; Xie, Y.; Frenkel, E.P.; Koh, A.Y. Metagenomic shotgun sequencing and unbiased metabolomic profiling identify specific human gut microbiota and metabolites associated with immune checkpoint therapy efficacy in melanoma patients. Neoplasia 2017, 19, 848–855. [Google Scholar] [CrossRef] [PubMed]
- Maruo, T.; Sakamoto, M.; Ito, C.; Toda, T.; Benno, Y. Adlercreutzia equolifaciens gen. nov., sp. nov., an equol-producing bacterium isolated from human faeces, and emended description of the genus Eggerthella. Int. J. Syst. Evol. Microbiol. 2008, 58, 1221–1227. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Pan, M.; Li, D.; Yin, Y.; Jiang, T.; Fang, S.; Wang, G.; Zhao, J.; Zhang, H.; Chen, W. Metagenomic insights into the effects of oligosaccharides on the microbial composition of fecal contents in constipated mice. J. Funct. Foods 2017, 38, 486–496. [Google Scholar] [CrossRef]
- Thompson, C.L.; Vier, R.; Mikaelyan, A.; Wienemann, T.; Brune, A. ‘Candidatus Arthromitus’ revised: Segmented filamentous bacteria in arthropod guts are members of Lachnospiraceae. Environ. Microbiol. 2012, 14, 1454–1465. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Sistla, S.; Dhodapkar, R.; Parija, S.C. Fatal Delftia acidovorans infection in an immunocompetent patient with empyema. Asian Pac. J. Trop. Biomed. 2012, 2, 923–924. [Google Scholar] [CrossRef]
- Marin, L.; Rowan, R.; Mantilla, A.; Olupona, B.; MacIntyre, A. Lower-extremity infections caused by Serratia marcescens: A report of three cases and a literature review. J. Am. Podiat. Med. Assoc. 2017, 107, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Schwalm, N.D.; Groisman, E.A. Navigating the gut buffet: Control of polysaccharide utilization in Bacteroides spp. Trends Microbiol. 2017, 25, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Saunders, E.; Pukall, R.; Abt, B.; Lapidus, A.; Del-Rio, T.G.; Copeland, A.; Tice, H.; Cheng, J.F.; Lucas, S.; Chen, F.; et al. Complete genome sequence of Eggerthella lenta type strain (IPP VPI 0255T). Stand. Genom. Sci. 2009, 1, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Copeland, A.; Sikorski, J.; Lapidus, A.; Nolan, M.; Del-Rio, T.G.; Lucas, S.; Chen, F.; Tice, H.; Pitluck, S.; Cheng, J.F.; et al. Complete genome sequence of Atopobium parvulum type strain (IPP 1246T). Stand. Genom. Sci. 2009, 1, 1660–1673. [Google Scholar] [CrossRef] [PubMed]
- Göker, M.; Held, B.; Lucas, S.; Nolan, M.; Yasawong, M.; Glavina Del Rio, T.; Tice, H.; Cheng, J.F.; Bruce, D.; Detter, J.C.; et al. Complete genome sequence of Olsenella uli type strain (VPI D76D-27CT). Stand. Genom. Sci. 2010, 3, 76–84. [Google Scholar] [CrossRef] [PubMed]
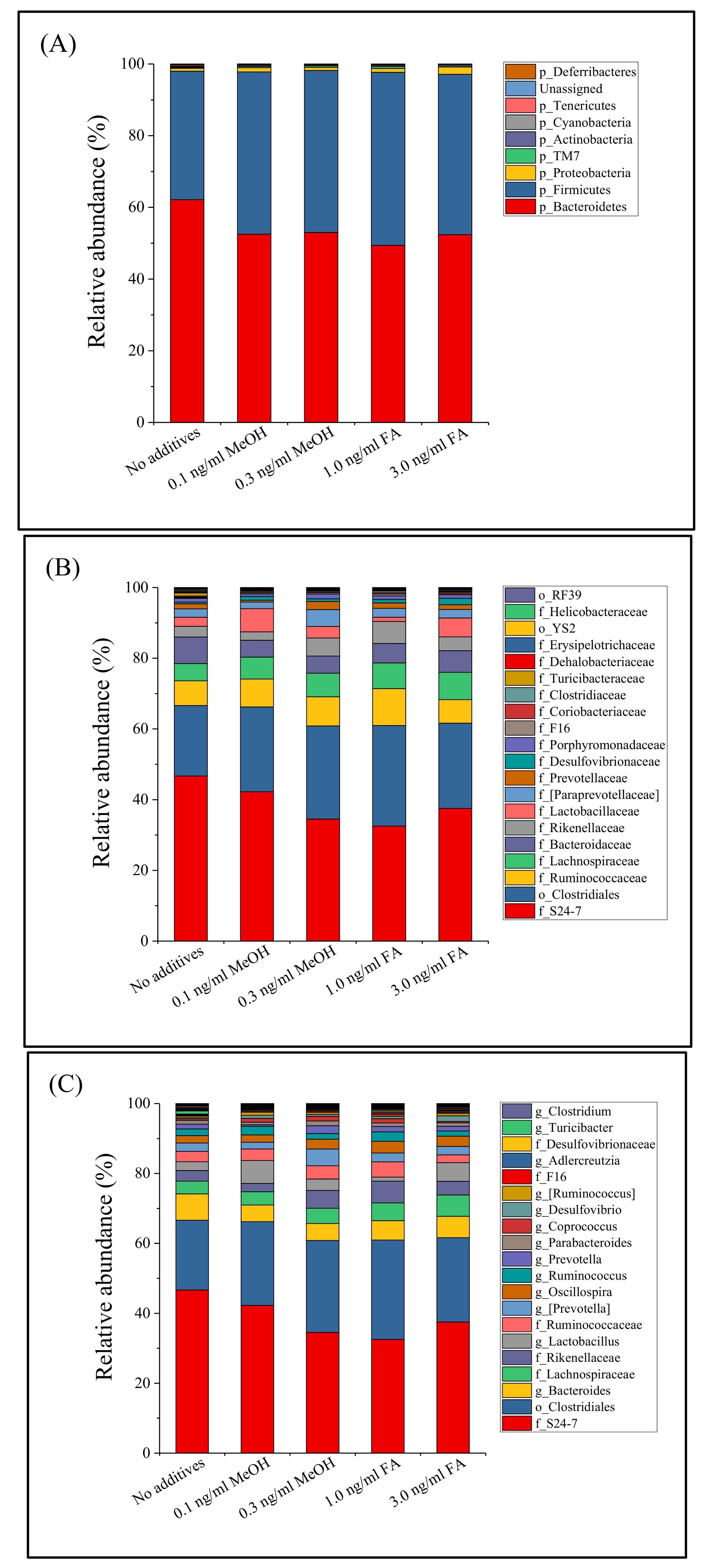
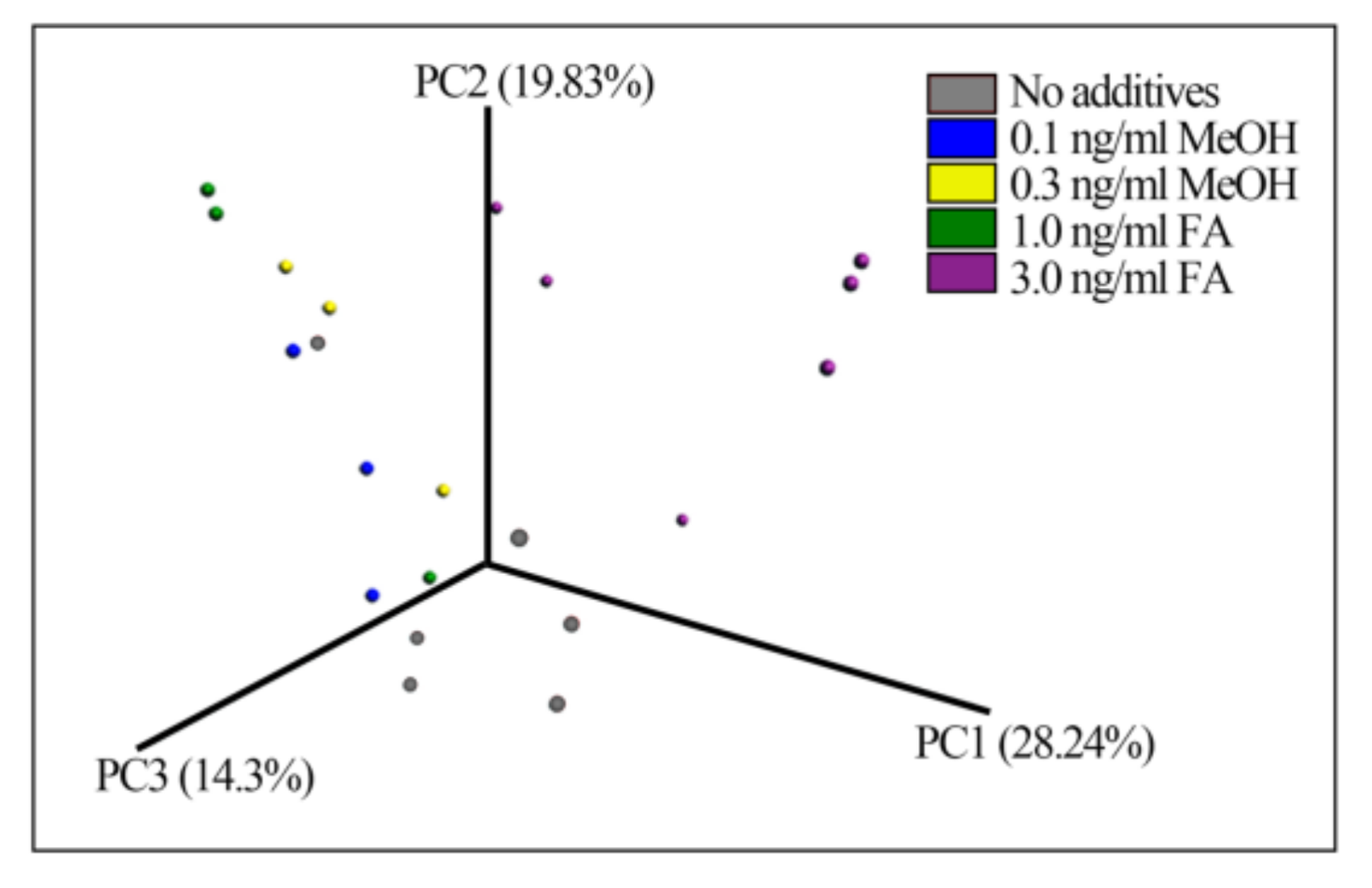
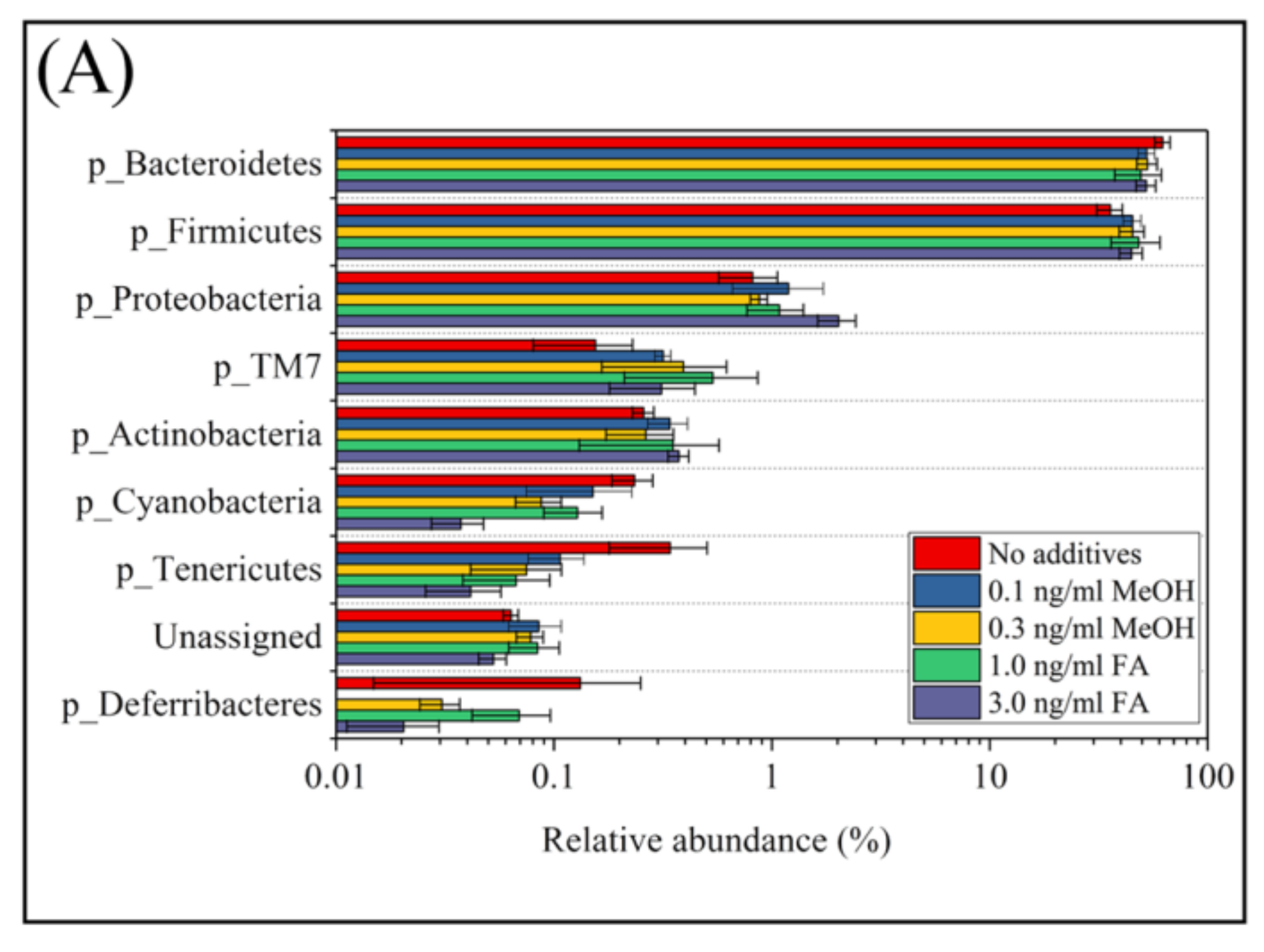
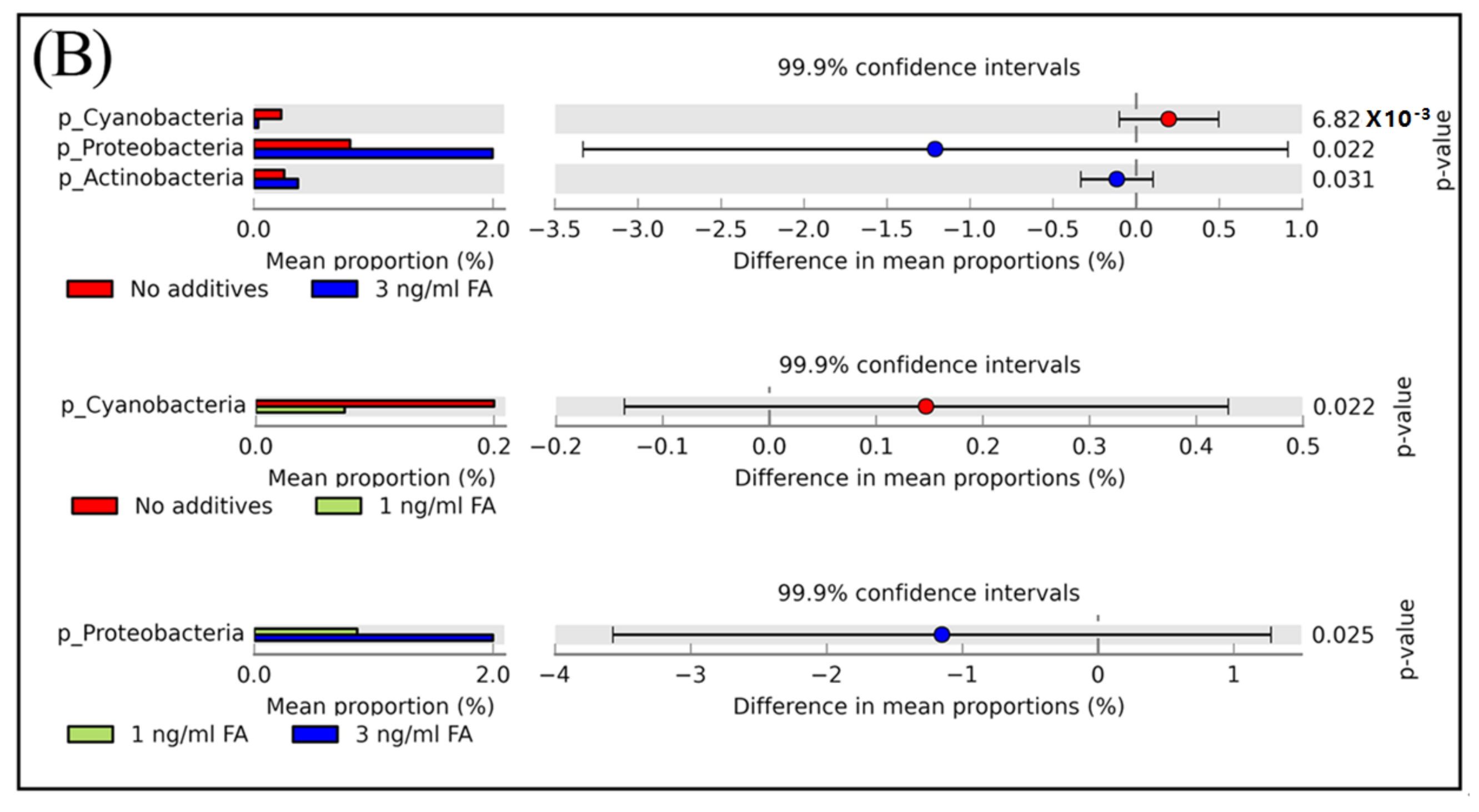

| Saline Control | 0.1 ng/mL MeOH | 0.3 ng/mL MeOH | 1.0 ng/mL FA | 3.0 ng/mL FA | p-Value * | |
|---|---|---|---|---|---|---|
| OTUs | 833.8 ± 33.7 | 892.7 ± 15.7 | 887.3 ± 26.6 | 881.6 ± 27.9 | 795.5 ± 69.5 | 0.046 |
| Chao1 | 929.2 ± 54.0 | 1011.8 ± 44.6 | 993.8 ± 35.6 | 1018.7 ± 21.0 | 915.3 ± 51.7 | 0.023 |
| PD | 35.8 ± 1.4 | 38.0 ± 0.5 | 37.9 ± 0.7 | 37.8 ± 1.0 | 35.4 ± 1.5 | 0.029 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, J.; Zhao, Y.; Jiang, X.; Li, R.; Xie, H.; Ge, L.; Xie, B.; Yang, X.; Zhang, L. Exposure to Formaldehyde Perturbs the Mouse Gut Microbiome. Genes 2018, 9, 192. https://doi.org/10.3390/genes9040192
Guo J, Zhao Y, Jiang X, Li R, Xie H, Ge L, Xie B, Yang X, Zhang L. Exposure to Formaldehyde Perturbs the Mouse Gut Microbiome. Genes. 2018; 9(4):192. https://doi.org/10.3390/genes9040192
Chicago/Turabian StyleGuo, Junhui, Yun Zhao, Xingpeng Jiang, Rui Li, Hao Xie, Leixin Ge, Bo Xie, Xu Yang, and Luoping Zhang. 2018. "Exposure to Formaldehyde Perturbs the Mouse Gut Microbiome" Genes 9, no. 4: 192. https://doi.org/10.3390/genes9040192





