Genome-Wide Analysis of the NAC Transcription Factor Gene Family Reveals Differential Expression Patterns and Cold-Stress Responses in the Woody Plant Prunus mume
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome-Wide Scanning and Classification
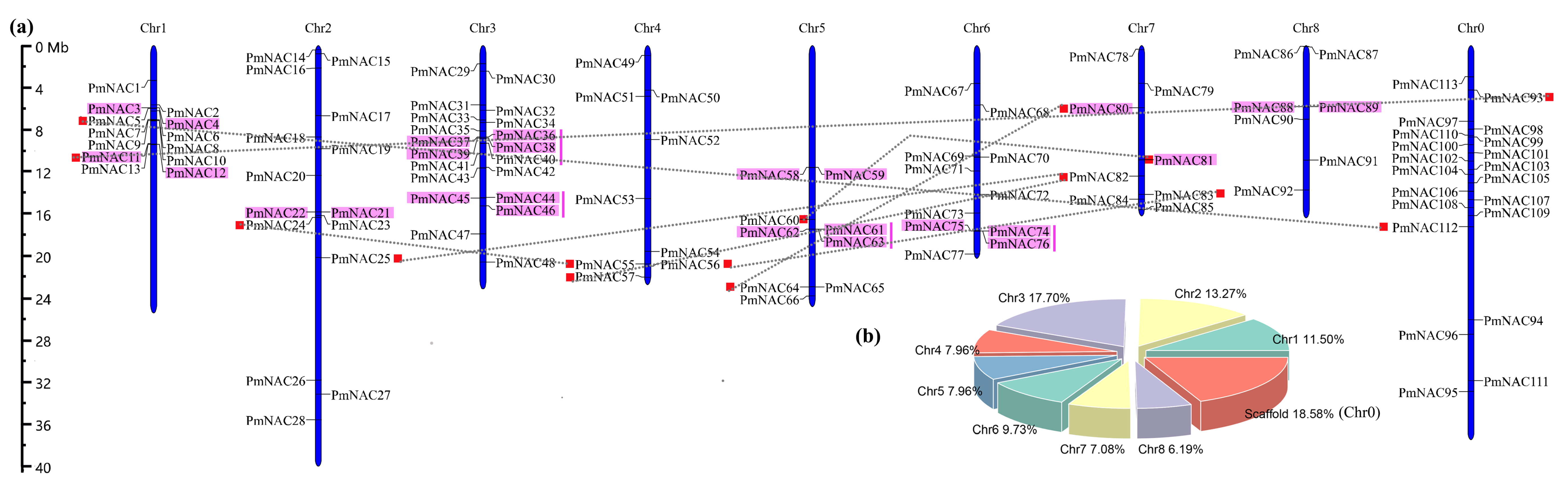
2.2. Chromosomal Location and Gene Structure Analyses
2.3. miR164 Target Site Prediction and Membrane-Bound PmNACs Analyses
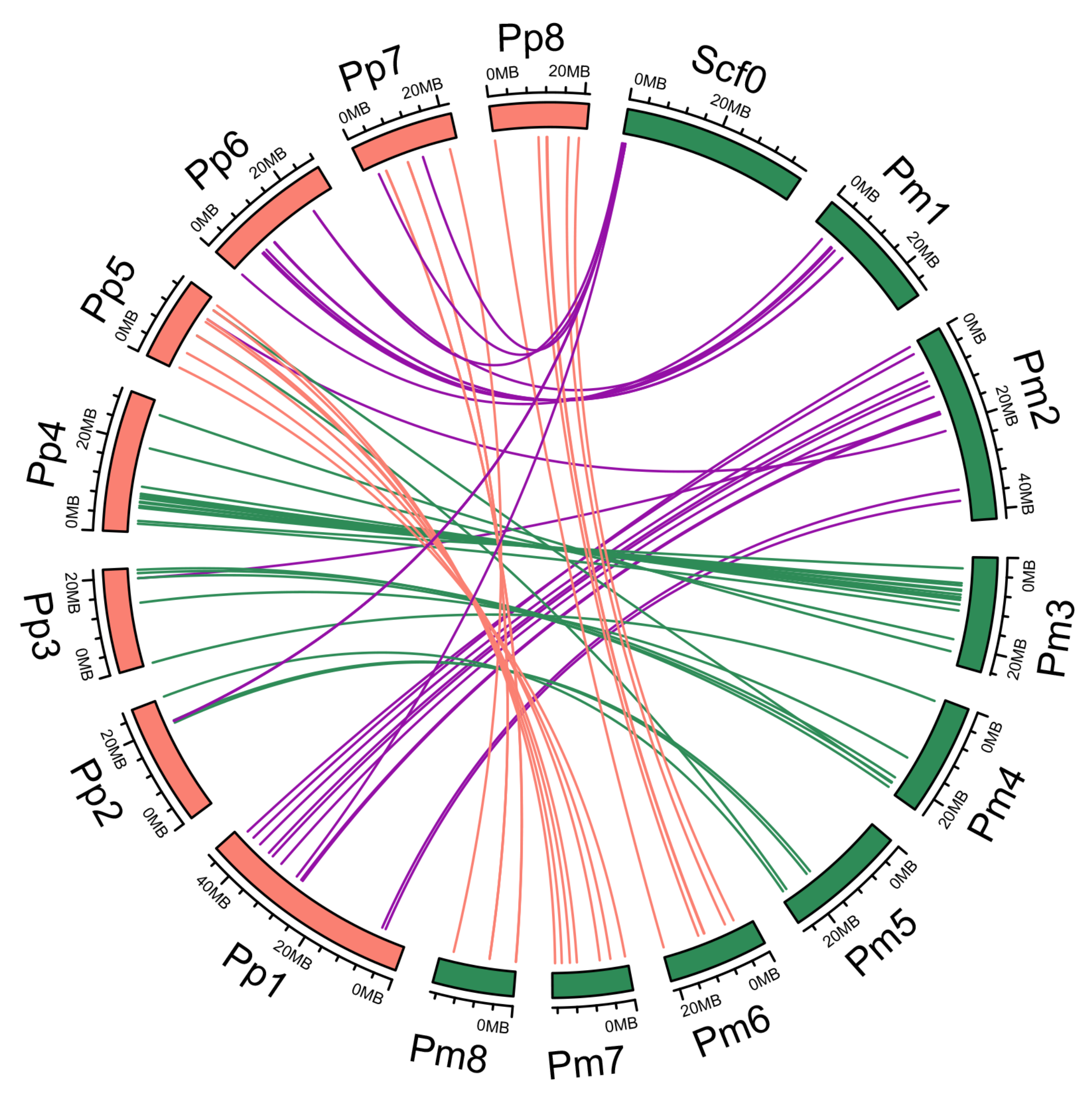
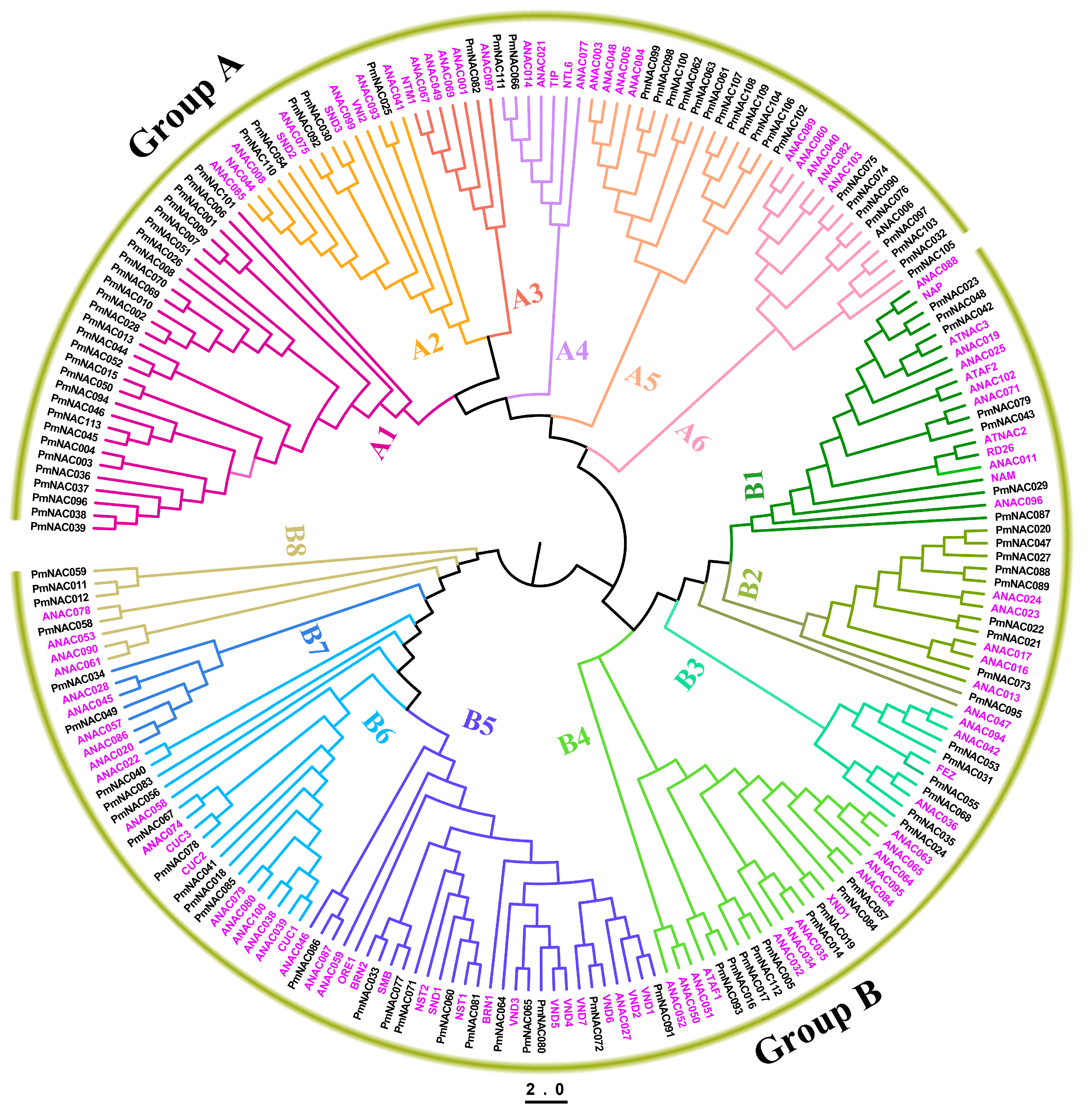
2.4. Phylogenetic and Synteny Analysis
2.5. Plant, Tissues and Stress Treatment
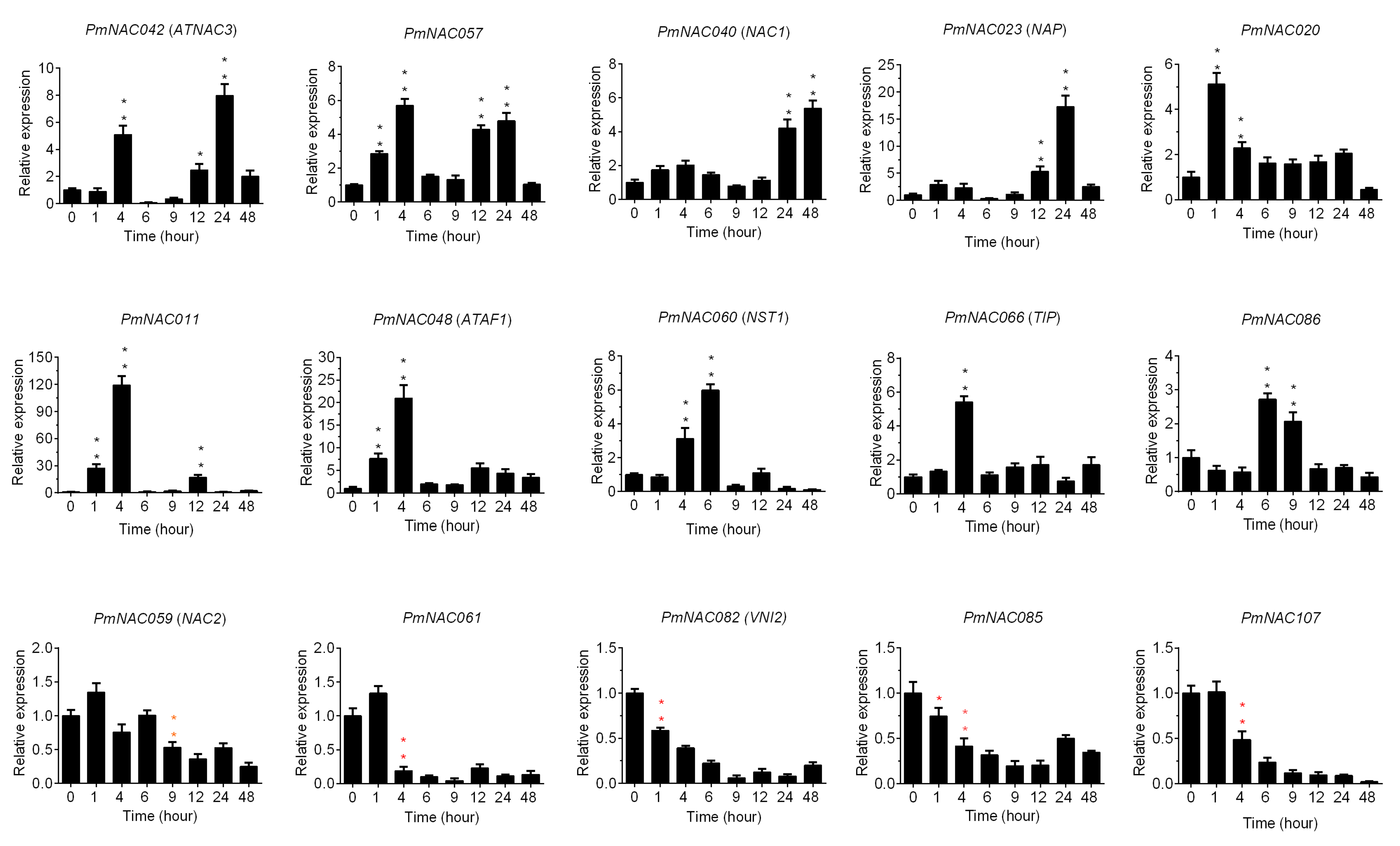
2.6. RNA Extraction and qRT-PCR
3. Results
3.1. Identification and Annotation of PmNAC Genes
3.2. Synteny Relationships and Duplication of PmNAC Genes
3.3. Phylogenetic Analysis of PmNACs
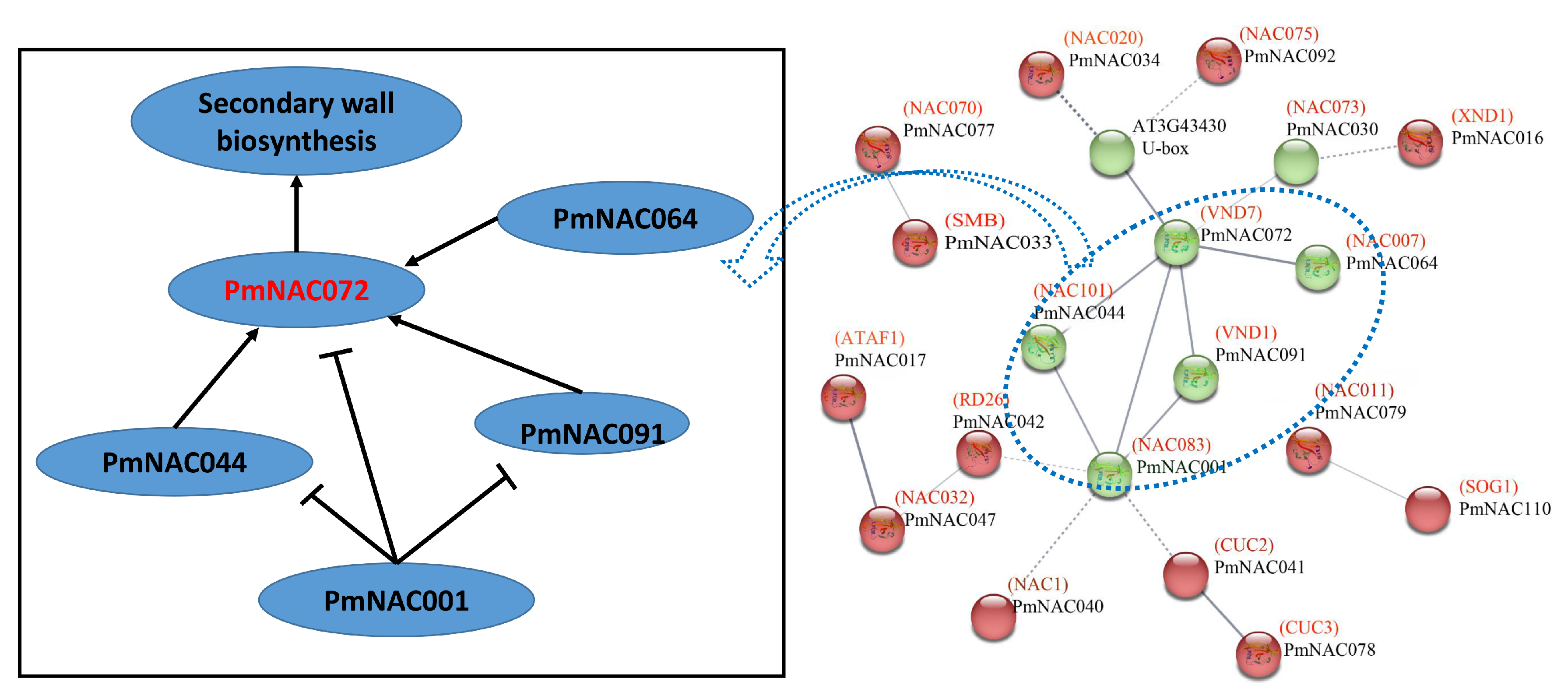
3.4. Conserved Motif, Gene Structure and Protein Interaction Analysis
3.5. Membrane-Bound Analysis and miR164 Target Site Prediction
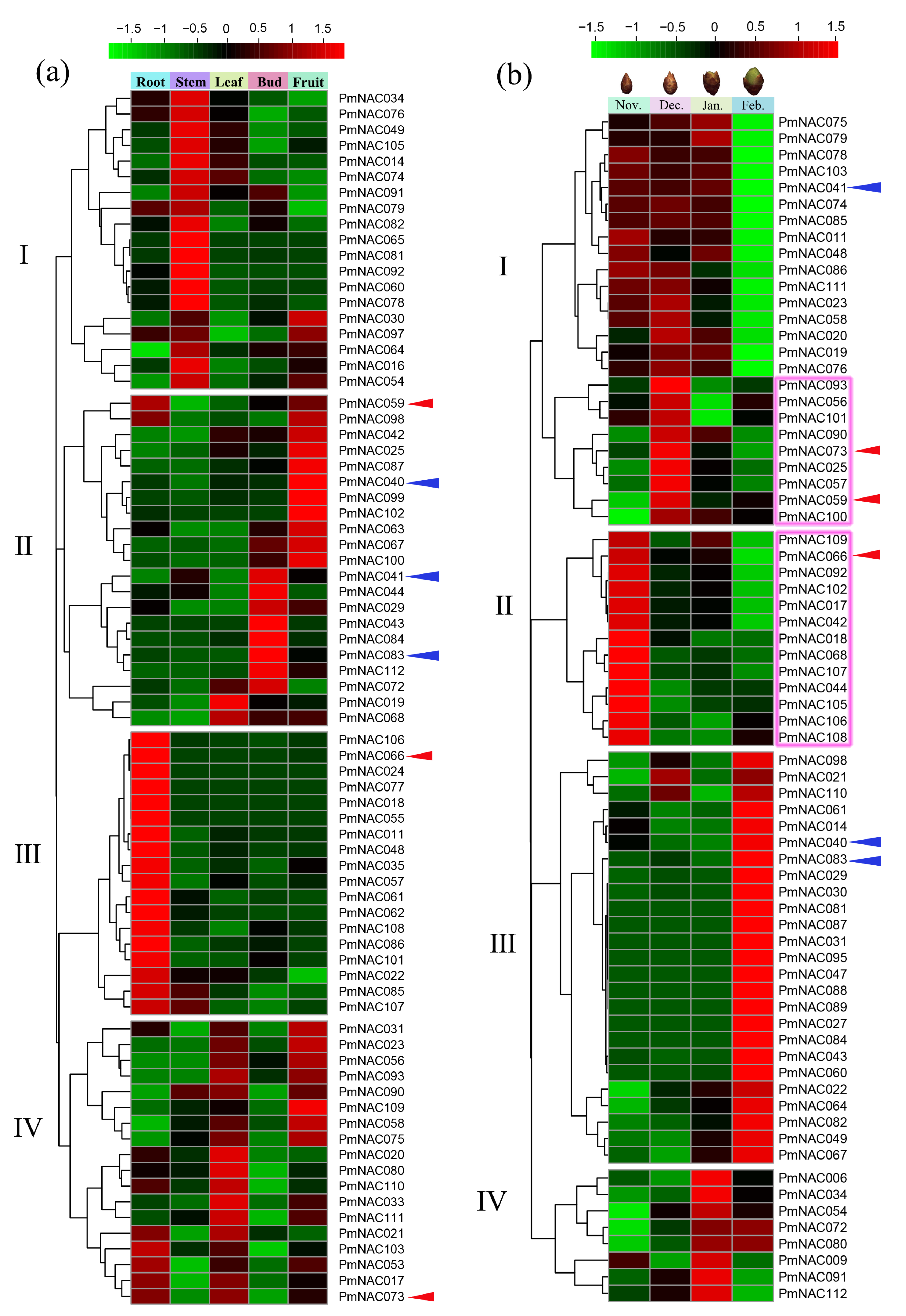
3.6. Expression Pattern Analysis by Transcriptome
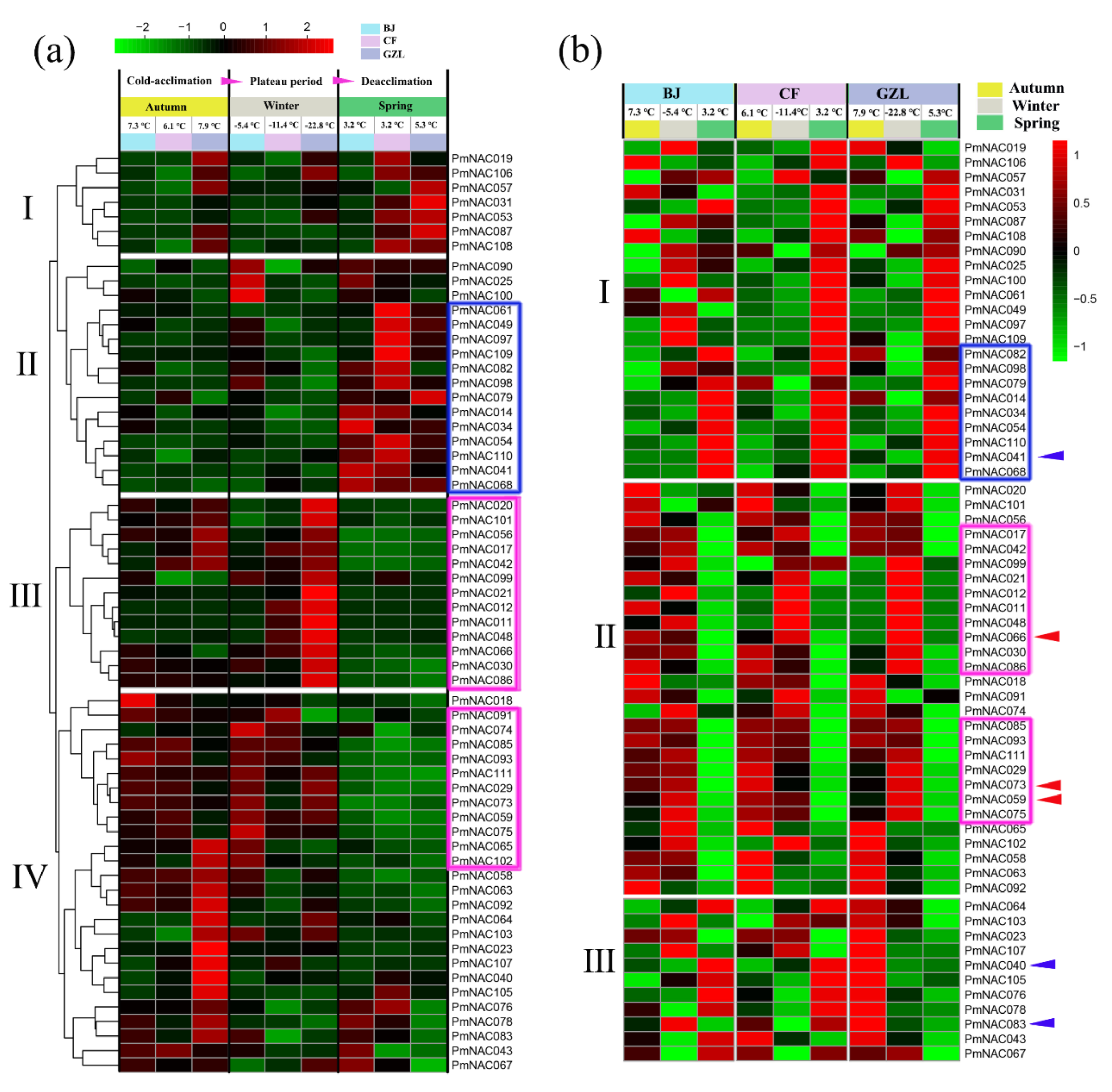
3.7. Cold Response Patterns of PmNACs during Cold Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Xu, B.; Ohtani, M.; Yamaguchi, M.; Toyooka, K.; Wakazaki, M.; Sato, M.; Kubo, M.; Nakano, Y.; Sano, R.; Hiwatashi, Y.; et al. Contribution of NAC transcription factors to plant adaptation to land. Science 2014, 343, 1505–1508. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Stewart, C.N. Plant synthetic promoters and transcription factors. Curr. Opin. Biotechnol. 2016, 37, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Kong, L.; Luo, J.; Gao, G. Planttfdb 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P.; et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003, 10, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Aida, M.; Ishida, T.; Fukaki, H.; Fujisawa, H.; Tasaka, M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 1997, 9, 841. [Google Scholar] [CrossRef] [PubMed]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Ernst, H.A.; Nina Olsen, A.; Skriver, K.; Larsen, S.; Lo Leggio, L. Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 2004, 5, 297. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Wang, Q.; Xiong, L.; Lou, Z. A structural view of the conserved domain of rice stress-responsive NAC1. Protein Cell 2011, 2, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Kim, S.-G.; Park, C.-M. Membrane-bound transcription factors in plants. Trends Plant Sci. 2008, 13, 550–556. [Google Scholar] [CrossRef] [PubMed]
- Xin-Jian, H.; Rui-Ling, M.; Wan-Hong, C.; Zhi-Gang, Z.; Jin-Song, Z.; Shou-Yi, C. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 2005, 44, 903–916. [Google Scholar] [CrossRef]
- Chen, X.; Cheng, J.; Chen, L.; Zhang, G.; Huang, H.; Zhang, Y.; Xu, L. Auxin-independent NAC pathway acts in response to explant-specific wounding and promotes root tip emergence during de novo root organogenesis in Arabidopsis. Plant Physiol. 2016, 170, 2136. [Google Scholar] [CrossRef] [PubMed]
- Rauf, M.; Arif, M.; Fisahn, J.; Xue, G.-P.; Balazadeh, S.; Mueller-Roeber, B. NAC transcription factor speedy hyponastic growth regulates flooding-induced leaf movement in Arabidopsis. Plant Cell 2013, 25, 4941–4955. [Google Scholar] [CrossRef] [PubMed]
- Souer, E.; van Houwelingen, A.; Kloos, D.; Mol, J.; Koes, R. The no apical meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 1996, 85, 159–170. [Google Scholar] [CrossRef]
- Christiansen, M.W.; Matthewman, C.; Podzimska-Sroka, D.; O’Shea, C.; Lindemose, S.; Møllegaard, N.E.; Holme, I.B.; Hebelstrup, K.; Skriver, K.; Gregersen, P.L. Barley plants over-expressing the NAC transcription factor gene HvNAC005 show stunting and delay in development combined with early senescence. J. Exp. Bot. 2016, 67, 5259–5273. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Hong, S.H.; Kim, Y.W.; Lee, I.H.; Jun, J.H.; Phee, B.-K.; Rupak, T.; Jeong, H.; Lee, Y.; Hong, B.S.; et al. Gene regulatory cascade of senescence-associated NAC transcription factors activated by ethylene-insensitive2-mediated leaf senescence signalling in Arabidopsis. J. Exp. Bot. 2014, 65, 4023–4036. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Lv, B.; Luo, L.; He, J.; Mao, C.; Xi, D.; Ming, F. The NAC-type transcription factor OsNAC2 regulates ABA-dependent genes and abiotic stress tolerance in rice. Sci. Rep. 2017, 7, 40641. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.P.; Kuo, C.H.; Lu, H.H.; Lo, H.S.; Yeh, K.W. The sweet potato NAC-domain transcription factor IbNAC1 is dynamically coordinated by the activator IbbHLH3 and the repressor IbbHLH4 to reprogram the defense mechanism against wounding. PLoS Genet. 2016, 12, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Delessert, C.; Kazan, K.; Wilson, I.W.; Van Der Straeten, D.; Manners, J.; Dennis, E.S.; Dolferus, R. The transcription factor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis. Plant J. 2005, 43, 745–757. [Google Scholar] [CrossRef] [PubMed]
- Toth, Z.; Winterhagen, P.; Kalapos, B.; Su, Y.; Kovacs, L.; Kiss, E. Expression of a grapevine NAC transcription factor gene is induced in response to powdery mildew colonization in salicylic acid-independent manner. Sci. Rep. 2016, 6, 30825. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Zhai, Q.; Deng, L.; Li, S.; Li, H.; Yan, L.; Huang, Z.; Wang, B.; Jiang, H.; Huang, T.; et al. Closely related NAC transcription factors of tomato differentially regulate stomatal closure and reopening during pathogen attack. Plant Cell 2014, 26, 3167–3184. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Wang, H.; Liu, S.; Li, Z.; Yang, X.; Yan, J.; Li, J.; Tran, L.-S.P.; Qin, F. A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat. Commun. 2015, 6, 8326. [Google Scholar] [CrossRef] [PubMed]
- Thirumalaikumar, V.P.; Vikas, D.; Nikolay, M.; Shawkat, A.; Rengin, O.; Ismail, T.; Bernd, M.-R.; Salma, B. NAC transcription factor JUNGBRUNNEN1 enhances drought tolerance in tomato. Plant Biotechnol. J. 2018, 16, 354–366. [Google Scholar] [CrossRef]
- Fang, L.; Su, L.; Sun, X.; Li, X.; Sun, M.; Karungo, S.K.; Fang, S.; Chu, J.; Li, S.; Xin, H. Expression of Vitis amurensis NAC26 in Arabidopsis enhances drought tolerance by modulating jasmonic acid synthesis. J. Exp. Bot. 2016, 67, 2829–2845. [Google Scholar] [CrossRef] [PubMed]
- Mitsuda, N.; Iwase, A.; Yamamoto, H.; Yoshida, M.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 2007, 19, 270. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, M.; Nagahage, I.S.P.; Ohtani, M.; Ishikawa, T.; Uchimiya, H.; Kawai-Yamada, M.; Demura, T. Arabidopsis NAC domain proteins VND-INTERACTING1 and ANAC103 interact with multiple NAC domain proteins. Plant Biotechnol. J. 2015, 32, 119–123. [Google Scholar] [CrossRef]
- Huh, S.U.; Lee, S.-B.; Kim, H.H.; Paek, K.-H. ATAF2, a NAC transcription factor, binds to the promoter and regulates NIT2 gene expression involved in auxin biosynthesis. Mol. Cells 2012, 34, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Willemsen, V.; Bauch, M.; Bennett, T.; Campilho, A.; Wolkenfelt, H.; Xu, J.; Haseloff, J.; Scheres, B. The NAC domain transcription factors FEZ and SOMBRERO control the orientation of cell division plane in Arabidopsis root stem cells. Dev. Cell 2008, 15, 913–922. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Qi, G.; Kong, Y.; Kong, D.; Gao, Q.; Zhou, G. Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biol. 2010, 10, 145. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Song, X.; Duan, W.; Huang, Z.; Liu, G.; Li, Y.; Hou, X. Genome-wide analysis and expression patterns of NAC transcription factor family under different developmental stages and abiotic stresses in chinese cabbage. Plant Mol. Biol. Rep. 2014, 32, 1041–1056. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Manimekalai, R.; Sharoni, A.M.; Satoh, K.; Kondoh, H.; Ooka, H.; Kikuchi, S. Genome-wide analysis of NAC transcription factor family in rice. Gene 2010, 465, 30–44. [Google Scholar] [CrossRef] [PubMed]
- Hussey, S.G.; Mizrachi, E.; Spokevicius, A.V.; Bossinger, G.; Berger, D.K.; Myburg, A.A. SND2, a NAC transcription factor gene, regulates genes involved in secondary cell wall development in Arabidopsis fibres and increases fibre cell area in Eucalyptus. BMC Plant Biol. 2011, 11, 173. [Google Scholar] [CrossRef]
- Yan, J.; Tong, T.; Li, X.; Chen, Q.; Dai, M.; Niu, F.; Yang, M.; Deyholos, M.K.; Yang, B.; Jiang, Y.-Q. A novel NAC-type transcription factor, NAC87, from oilseed rape modulates reactive oxygen species accumulation and cell death. Plant Cell Physiol. 2018, 59, 290–303. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Wang, X.; Guo, H.; Cheng, Y.; Hou, C.; Chen, J.-G.; Wang, S. NTL8 regulates trichome formation in Arabidopsis by directly activating R3 MYB genes TRY and TCL1. Plant Physiol. 2017, 174, 2363–2375. [Google Scholar] [CrossRef] [PubMed]
- Duan, M.; Zhang, R.; Zhu, F.; Zhang, Z.; Gou, L.; Wen, J.; Dong, J.; Wang, T. A lipid-anchored NAC transcription factor is translocated into the nucleus and activates glyoxalase I expression during drought stress. Plant Cell 2017, 29, 1748. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Dai, M.; Yao, J.; Xiao, B.; Li, X.; Zhang, Q.; Xiong, L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. USA 2006, 103, 12987. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.K.; Hagedorn, P.H.; De Torres-Zabala, M.; Grant, M.R.; Rung, J.H.; Collinge, D.B.; Lyngkjaer, M.F. Transcriptional regulation by an NAC (NAM–ATAF1,2–CUC2) transcription factor attenuates ABA signaling for efficient basal defence towards Blumeria graminis f. sp. hordei in Arabidopsis. Plant J. 2008, 56, 867–880. [Google Scholar] [CrossRef] [PubMed]
- Kazemi-Shahandashti, S.-S.; Maali-Amiri, R. Global insights of protein responses to cold stress in plants: Signaling, defence, and degradation. J. Plant Physiol. 2018, 226, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Ding, Y.; Yang, S. Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Zhu, J.; Zhu, J.-K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Angelcheva, L.; Mishra, Y.; Antti, H.; Kjellsen, T.D.; Funk, C.; Strimbeck, R.G.; Schröder, W.P. Metabolomic analysis of extreme freezing tolerance in Siberian spruce (Picea obovata). New Phytol. 2014, 204, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ye, K.; Shi, Y.; Cheng, J.; Zhang, X.; Yang, S. BZR1 positively regulates freezing tolerance via CBF-dependent and CBF-independent pathways in Arabidopsis. Mol. Plant 2017, 10, 545–559. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.-M.; Zhao, Q.; Zhao, L.-L.; Qiao, Y.; Xie, X.-B.; Li, H.-F.; Yao, Y.-X.; You, C.-X.; Hao, Y.-J. The cold-induced basic helix-loop-helix transcription factor gene MdCIbHLH1 encodes an ICE-like protein in apple. BMC Plant Biol. 2012, 12, 22. [Google Scholar] [CrossRef]
- Lee, C.-M.; Thomashow, M.F. Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2012, 109, 15054–15059. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.-H.; Hong, X.; Agarwal, M.; Zhu, J.-K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003, 17, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.-L.; Wang, S.-B.; Chen, R.-G.; Chen, B.-H.; Du, X.-H.; Yin, Y.-X.; Gong, Z.-H.; Zhang, Y.-Y. Characterization and expression profile of CaNAC2 pepper gene. Front. Plant Sci. 2015, 6, 755. [Google Scholar] [CrossRef] [PubMed]
- Shan, W.; Kuang, J.F.; Lu, W.J.; Chen, J.Y. Banana fruit NAC transcription factor MaNAC1 is a direct target of MaICE1 and involved in cold stress through interacting with MaCBF1. Plant Cell Environ. 2014, 37, 2116–2127. [Google Scholar] [CrossRef] [PubMed]
- An, J.-P.; Li, R.; Qu, F.-J.; You, C.-X.; Wang, X.-F.; Hao, Y.-J. An apple NAC transcription factor negatively regulates cold tolerance via CBF-dependent pathway. J. Plant Physiol. 2018, 221, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-L.; Yang, X.; Hu, Y.-X.; Yu, X.-D.; Li, Q.-L. A novel NAC transcription factor from Suaeda liaotungensis K. enhanced transgenic Arabidopsis drought, salt, and cold stress tolerance. Plant Cell Rep. 2014, 33, 767–778. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Hong, Y.; Zhang, H.; Li, D.; Song, F. Rice NAC transcription factor ONAC095 plays opposite roles in drought and cold stress tolerance. BMC Plant Biol. 2016, 16, 203. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Yang, W.; Zhang, Q.; Cheng, T.; Pan, H.; Xu, Z.; Zhang, J.; Chen, C. Genome-wide characterization and linkage mapping of simple sequence repeats in mei (Prunus mume Sieb. et Zucc.). PLoS ONE 2013, 8, e59562. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, K.; Nakamura, S.; Matsumoto, T.; Ohta, T.; Ogawa, K.; Tamura, H.; Matsuda, H.; Yoshikawa, M. Medicinal flowers. XXXVIII. Structures of acylated sucroses and inhibitory effects of constituents on aldose reducatase from the flower buds of Prunus mume. Chem. Pharm. Bull. 2013, 61, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Y. China Mei Flower (Prunus mume) Cultivars in Colour; China Forestry Publishing House: Beijing, China, 2017. [Google Scholar]
- Zhai, P.; Zhang, X.; Wan, H.; Pan, X. Trends in total precipitation and frequency of daily precipitation extremes over China. J. Clim. 2005, 18, 1096–1108. [Google Scholar] [CrossRef]
- Chen, R.; Zhang, Q.; Chen, J. Studies on breeding hardy cultivars of mei flower (Prunus mume) for gardens in Beijing. Acta Hortic. 2008, 769, 305–311. [Google Scholar] [CrossRef]
- Stamatakis, A. Raxml version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. Mafft multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H. Mcscanx: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Blanc, G.; Wolfe, K.H. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 2004, 16, 1667–1678. [Google Scholar] [CrossRef] [PubMed]
- Koch, M.A.; Haubold, B.; Mitchell-Olds, T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and Related Genera (Brassicaceae). Mol. Biol. Evol. 2000, 17, 1483–1498. [Google Scholar] [CrossRef] [PubMed]
- Abdul Kayum, M.; Nath, U.; Park, J.-I.; Biswas, M.; Choi, E.; Song, J.-Y.; Kim, H.-T.; Nou, I.-S. Genome-wide identification, characterization, and expression profiling of glutathione S-transferase (GST) family in pumpkin reveals likely role in cold-stress tolerance. Genes 2018, 9, 84. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Chen, W.; Sun, L.; Zhao, F.; Huang, B.; Yang, W.; Tao, Y.; Wang, J.; Yuan, Z.; Fan, G. The genome of Prunus mume. Nat. Commun. 2012, 3, 1318. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhuo, X.; Zhao, K.; Zheng, T.; Han, Y.; Yuan, C.; Zhang, Q. Transcriptome profiles reveal the crucial roles of hormone and sugar in the bud dormancy of Prunus mume. Sci. Rep. 2018, 8, 5090. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Sun, L.; Zhou, Y.; Yang, W.; Cheng, T.; Wang, J.; Zhang, Q. Identification and expression analysis of the SQUAMOSA promoter-binding protein (SBP)-box gene family in Prunus mume. Mol. Genet. Genom. 2015, 290, 1701–1715. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta CT) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Sharma, V.; Pal, A.K.; Acharya, V.; Ahuja, P.S. Genome-wide organization and expression profiling of the NAC transcription factor family in potato (Solanum tuberosum L.). DNA Res. 2013, 20, 403–423. [Google Scholar] [CrossRef] [PubMed]
- The Arabidopsis Genome, Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis. Nature 2000, 408, 796. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Gu, Z.; Wang, H.; Nekrutenko, A. Evolutionary analyses of the human genome. Nature 2001, 409, 847–849. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, M.; Goué, N.; Igarashi, H.; Ohtani, M.; Nakano, Y.; Mortimer, J.C.; Nishikubo, N.; Kubo, M.; Katayama, Y.; Kakegawa, K.; et al. Vascular-related NAC-domain6 and vascular-related NAC-domain7 effectively induce transdifferentiation into xylem vessel elements under control of an induction system. Plant Physiol. 2010, 153, 906. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Lee, C.; Ye, Z.-H. Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol. Plant 2010, 3, 1087–1103. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, M.; Ohtani, M.; Mitsuda, N.; Kubo, M.; Ohmetakagi, M.; Fukuda, H.; Demura, T. VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 2010, 22, 1249. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. Micrornas: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Wang, T.; Pan, H.; Wang, J.; Yang, W.; Cheng, T.; Zhang, Q. Identification and profiling of novel and conserved microRNAs during the flower opening process in Prunus mume via deep sequencing. Mol. Genet. Genom. 2014, 289, 169–183. [Google Scholar] [CrossRef] [PubMed]
- Kadier, Y.; Zu, Y.-Y.; Dai, Q.-M.; Song, G.; Lin, S.-W.; Sun, Q.-P.; Pan, J.-B.; Lu, M. Genome-wide identification, classification and expression analysis of NAC family of genes in sorghum [Sorghum bicolor (L.) Moench]. Plant Growth Regul. 2017, 83, 301–312. [Google Scholar] [CrossRef]
- Balazadeh, S.; Kwasniewski, M.; Caldana, C.; Mehrnia, M.; Zanor, M.I.; Xue, G.-P.; Mueller-Roeber, B. ORS1, an H2O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol. Plant 2011, 4, 346–360. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, G.L.; Marques, C.S.; Costa, M.D.; Reis, P.A.; Alves, M.S.; Carvalho, C.M.; Fietto, L.G.; Fontes, E.P. Complete inventory of soybean NAC transcription factors: Sequence conservation and expression analysis uncover their distinct roles in stress response. Gene 2009, 444, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Force, A.; Lynch, M.; Pickett, F.B.; Amores, A.; Yan, Y.-L.; Postlethwait, J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 1999, 151, 1531–1545. [Google Scholar] [PubMed]
- Lan, T.; Yang, Z.-L.; Yang, X.; Liu, Y.-J.; Wang, X.-R.; Zeng, Q.-Y. Extensive functional diversification of the Populus glutathione S-transferase supergene family. Plant Cell 2009, 21, 3749–3766. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, O.; Nahal, H.; Foong, J.; Provart, N.J.; Campbell, M.M. Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 2009, 149, 981–993. [Google Scholar] [CrossRef] [PubMed]
- Strimbeck, G.R.; Schaberg, P.G.; Fossdal, C.G.; Schröder, W.P.; Kjellsen, T.D. Extreme low temperature tolerance in woody plants. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Kim, M.J.; Park, J.Y.; Kim, S.Y.; Jeon, J.; Lee, Y.H.; Kim, J.; Park, C.M. Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J. 2010, 61, 661–671. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Park, M.-J.; Seo, P.J.; Song, J.-S.; Kim, H.-J.; Park, C.-M. Controlled nuclear import of the transcription factor NTL6 reveals a cytoplasmic role of SnRk2.8 in the drought-stress response. Biochem. J. 2012, 448, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Park, C.-M. A membrane-bound NAC transcription factor as an integrator of biotic and abiotic stress signals. Plant Signal. Behav. 2010, 5, 481–483. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.; Ivanova, A.; Duncan, O.; Law, S.R.; Van Aken, O.; De Clercq, I.; Wang, Y.; Carrie, C.; Xu, L.; Kmiec, B. A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 2013, 25, 3450–3471. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.-H.; Jeon, H.S.; Kim, H.G.; Park, O.K. An Arabidopsis NAC transcription factor NAC4 promotes pathogen-induced cell death under negative regulation by microRNA164. New Phytol. 2017, 214, 343–360. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.-S.; Xie, Q.; Fei, J.-F.; Chua, N.-H. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 2005, 17, 1376–1386. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Xie, K.; Xiong, L. Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice. J. Exp. Bot. 2014, 65, 2119–2135. [Google Scholar] [CrossRef] [PubMed]
- Vialette-Guiraud, A.C.M.; Adam, H.; Finet, C.; Jasinski, S.; Jouannic, S.; Scutt, C.P. Insights from ANA-grade angiosperms into the early evolution of CUP-SHAPED COTYLEDON genes. Ann. Bot. 2011, 107, 1511–1519. [Google Scholar] [CrossRef] [PubMed]
- Mallory, A.C.; Dugas, D.V.; Bartel, D.P.; Bartel, B. MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr. Biol. 2004, 14, 1035–1046. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, M.; Haynor, D.; Johnson, J.M. Protein interaction networks. Expert Rev. Proteom. 2004, 1, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Deng, Z.; Lai, J.; Zhang, Y.; Yang, C.; Yin, B.; Zhao, Q.; Zhang, L.; Li, Y.; Yang, C.; et al. Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 2009, 19, 1279. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.; Nardmann, J.; Werr, W. Plant development revolves around axes. Trends Plant Sci. 2008, 13, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Sanghera, G.S.; Wani, S.H.; Hussain, W.; Singh, N.B. Engineering cold stress tolerance in crop plants. Curr. Genom. 2011, 12, 30–43. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.; Kim, J. Cold stress signaling networks in Arabidopsis. J. Plant Biol. 2013, 56, 69–76. [Google Scholar] [CrossRef]
- Thomashow, M.F. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Li, X.; Wang, Z.; Sun, Z.; Zhu, X.; Liu, S.; Song, F.; Liu, F.; Wang, Y. Cold priming induced tolerance to subsequent low temperature stress is enhanced by melatonin application during recovery in wheat. Molecules 2018, 23. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Brestic, M.; Tan, D.-X.; Zivcak, M.; Zhu, X.; Liu, S.; Song, F.; Reiter, R.J.; Liu, F. Melatonin alleviates low PS I-limited carbon assimilation under elevated CO2 and enhances the cold tolerance of offspring in chlorophyll b-deficient mutant wheat. J. Pineal Res. 2018, 64, e12453. [Google Scholar] [CrossRef] [PubMed]
- Mauch-Mani, B.; Flors, V. The ATAF1 transcription factor: At the convergence point of ABA-dependent plant defense against biotic and abiotic stresses. Cell Res. 2009, 19, 1322. [Google Scholar] [CrossRef] [PubMed][Green Version]
| Duplicated Gene Pairs | Ks | Ka | Ka/Ks | Type of Duplication | Type of Selection | Divergence Time (Mya) | ||
|---|---|---|---|---|---|---|---|---|
| PmNAC003 | vs. | PmNAC004 | 0.253 | 0.072 | 0.283 | Tandem | Purifying | 8.43 |
| PmNAC011 | vs. | PmNAC012 | 0.240 | 0.189 | 0.787 | Tandem | Purifying | 8.01 |
| PmNAC021 | vs. | PmNAC022 | 0.574 | 0.656 | 1.144 | Tandem | Purifying | 19.12 |
| PmNAC036 | vs. | PmNAC037 | 0.076 | 0.033 | 0.438 | Tandem | Purifying | 2.54 |
| PmNAC037 | vs. | PmNAC038 | 1.102 | 1.670 | 1.515 | Tandem | Purifying | 36.73 |
| PmNAC038 | vs. | PmNAC039 | 1.050 | 1.659 | 1.580 | Tandem | Positive | 35.00 |
| PmNAC044 | vs. | PmNAC045 | 1.452 | 2.173 | 1.497 | Tandem | Positive | 48.39 |
| PmNAC045 | vs. | PmNAC046 | 0.093 | 0.025 | 0.270 | Tandem | Purifying | 3.09 |
| PmNAC058 | vs. | PmNAC059 | 1.743 | 2.088 | 1.198 | Tandem | Positive | 58.11 |
| PmNAC061 | vs. | PmNAC062 | 1.220 | 1.101 | 0.903 | Tandem | Purifying | 40.65 |
| PmNAC062 | vs. | PmNAC063 | 1.033 | 1.221 | 1.183 | Tandem | Positive | 34.42 |
| PmNAC074 | vs. | PmNAC075 | 0.034 | 0.036 | 1.035 | Tandem | Positive | 1.15 |
| PmNAC075 | vs. | PmNAC076 | 0.441 | 0.166 | 0.377 | Tandem | Purifying | 14.69 |
| PmNAC080 | vs. | PmNAC081 | 1.188 | 2.878 | 2.424 | Tandem | Positive | 39.58 |
| PmNAC088 | vs. | PmNAC089 | 1.454 | 1.926 | 1.324 | Tandem | Positive | 48.47 |
| PmNAC005 | vs. | PmNAC112 | 1.550 | 0.484 | 0.312 | Segmental | Purifying | 51.66 |
| PmNAC011 | vs. | PmNAC093 | 1.301 | 0.443 | 0.340 | Segmental | Purifying | 43.37 |
| PmNAC024 | vs. | PmNAC055 | 1.341 | 0.336 | 0.250 | Segmental | Purifying | 44.71 |
| PmNAC025 | vs. | PmNAC082 | 1.352 | 0.276 | 0.204 | Segmental | Purifying | 45.05 |
| PmNAC056 | vs. | PmNAC083 | 1.462 | 0.263 | 0.180 | Segmental | Purifying | 48.72 |
| PmNAC057 | vs. | PmNAC082 | 2.613 | 0.566 | 0.217 | Segmental | Purifying | 87.11 |
| PmNAC060 | vs. | PmNAC081 | 2.246 | 0.360 | 0.160 | Segmental | Purifying | 74.86 |
| PmNAC064 | vs. | PmNAC080 | 1.466 | 0.245 | 0.167 | Segmental | Purifying | 48.87 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhuo, X.; Zheng, T.; Zhang, Z.; Zhang, Y.; Jiang, L.; Ahmad, S.; Sun, L.; Wang, J.; Cheng, T.; Zhang, Q. Genome-Wide Analysis of the NAC Transcription Factor Gene Family Reveals Differential Expression Patterns and Cold-Stress Responses in the Woody Plant Prunus mume. Genes 2018, 9, 494. https://doi.org/10.3390/genes9100494
Zhuo X, Zheng T, Zhang Z, Zhang Y, Jiang L, Ahmad S, Sun L, Wang J, Cheng T, Zhang Q. Genome-Wide Analysis of the NAC Transcription Factor Gene Family Reveals Differential Expression Patterns and Cold-Stress Responses in the Woody Plant Prunus mume. Genes. 2018; 9(10):494. https://doi.org/10.3390/genes9100494
Chicago/Turabian StyleZhuo, Xiaokang, Tangchun Zheng, Zhiyong Zhang, Yichi Zhang, Liangbao Jiang, Sagheer Ahmad, Lidan Sun, Jia Wang, Tangren Cheng, and Qixiang Zhang. 2018. "Genome-Wide Analysis of the NAC Transcription Factor Gene Family Reveals Differential Expression Patterns and Cold-Stress Responses in the Woody Plant Prunus mume" Genes 9, no. 10: 494. https://doi.org/10.3390/genes9100494
APA StyleZhuo, X., Zheng, T., Zhang, Z., Zhang, Y., Jiang, L., Ahmad, S., Sun, L., Wang, J., Cheng, T., & Zhang, Q. (2018). Genome-Wide Analysis of the NAC Transcription Factor Gene Family Reveals Differential Expression Patterns and Cold-Stress Responses in the Woody Plant Prunus mume. Genes, 9(10), 494. https://doi.org/10.3390/genes9100494







