On Controls in Ancient Microbiome Studies, and Microbial Resilience in Ancient Samples
Abstract
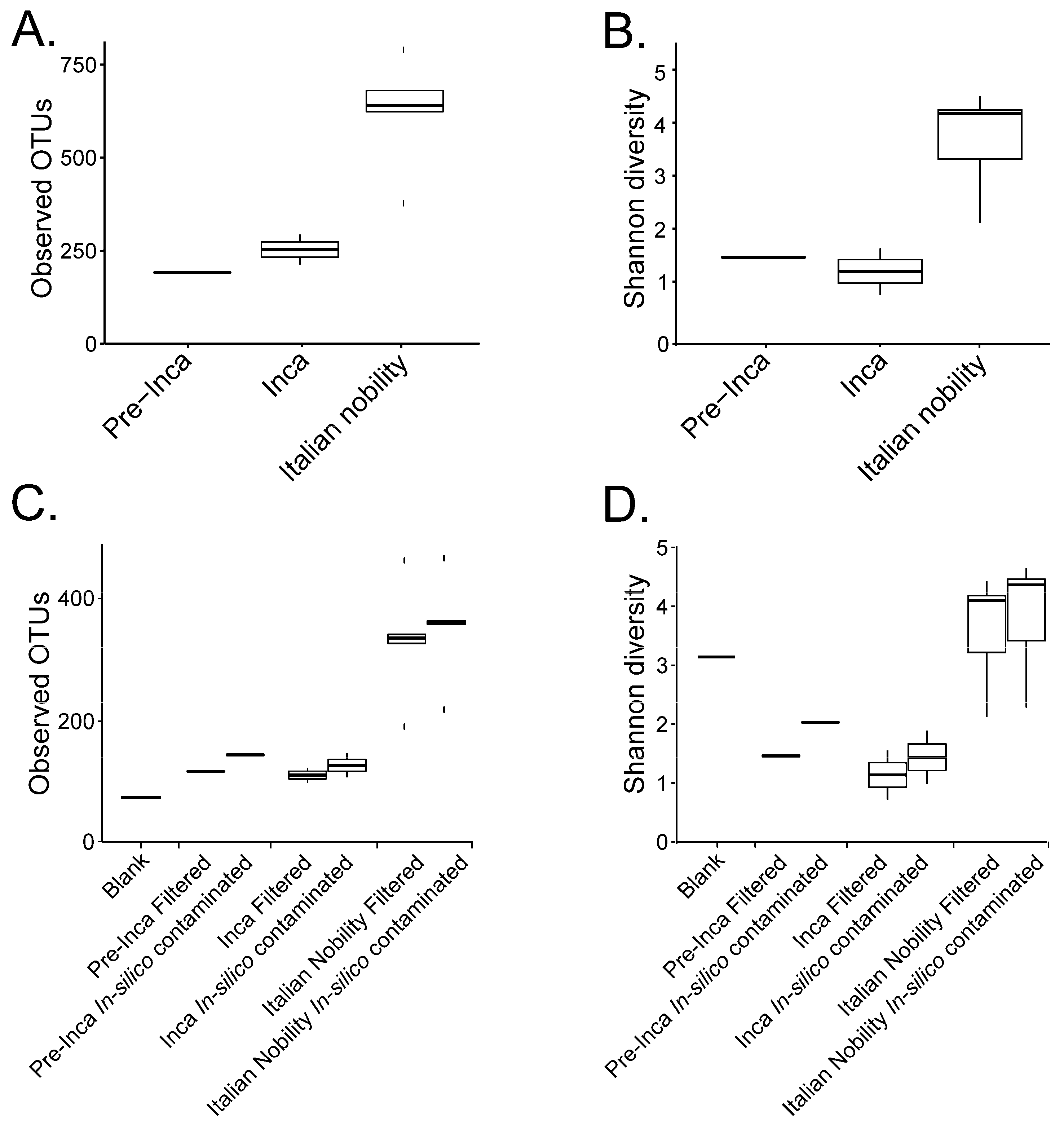
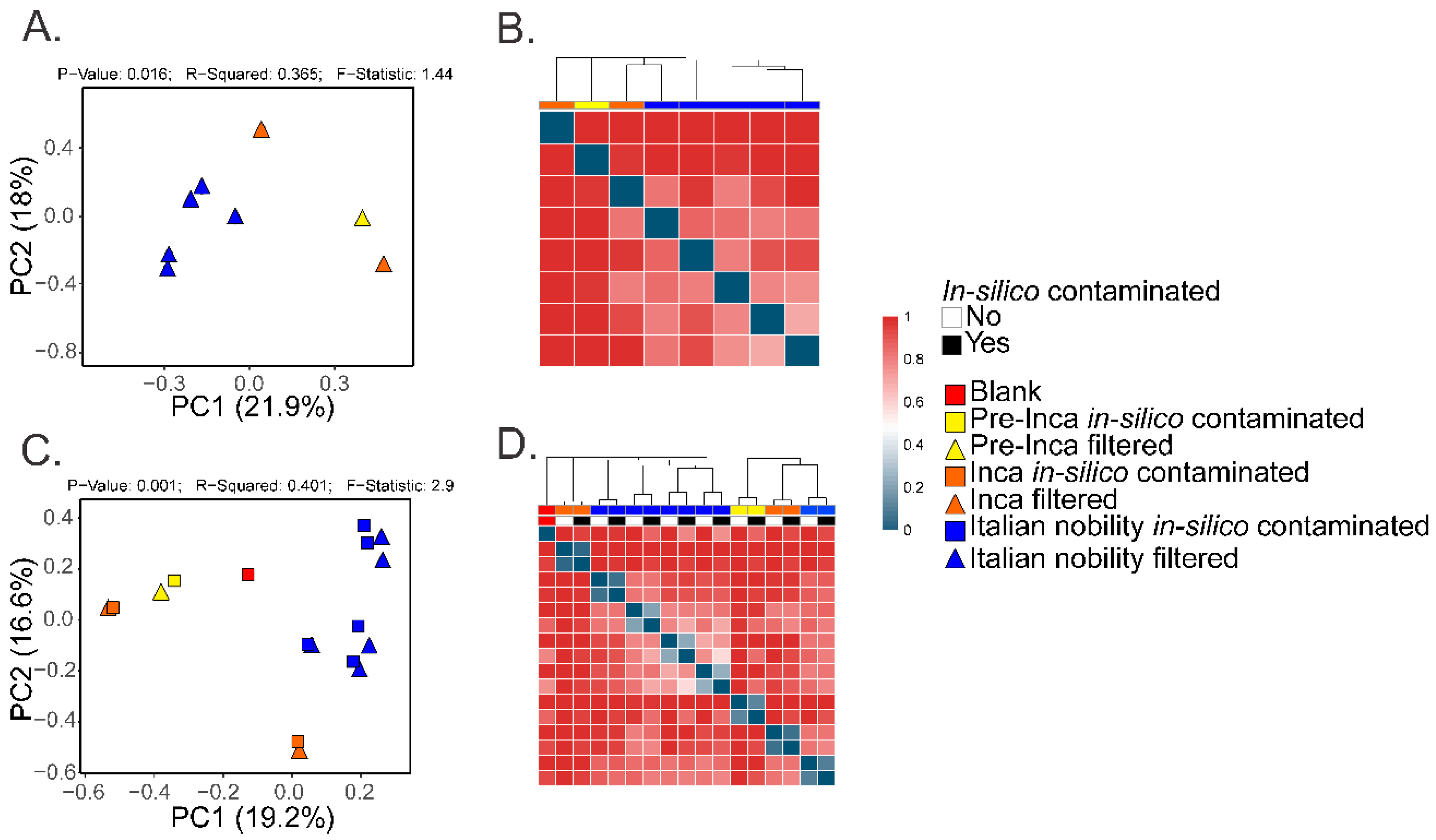
:1. Controls in Ancient Microbiome Studies
2. Considerations about Microbial Resilience in Ancient Samples
Author Contributions
Funding
Conflicts of Interest
References
- Eisenhofer, R.; Weyrich, L.S. Proper authentication of ancient DNA is still essential. Genes 2018, 9, 122. [Google Scholar] [CrossRef] [PubMed]
- Santiago-Rodriguez, T.M.; Fornaciari, G.; Luciani, S.; Toranzos, G.A.; Marota, I.; Giuffra, V.; Cano, R.J. Gut microbiome and putative resistome of inca and italian nobility mummies. Genes 2017, 8, 310. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Caporaso, J.G. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Velsko, I.M.; Frantz, L.A.F.; Herbig, A.; Larson, G.; Warinner, C. Selection of appropriate metagenome taxonomic classifiers for ancient microbiome research. bioRxiv 2018, 260042. [Google Scholar] [CrossRef] [Green Version]
- Menzel, P.; Ng, K.L.; Krogh, A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat. Commun. 2016, 7, 11257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Auria, G.; Peris-Bondia, F.; Džunková, M.; Mira, A.; Collado, M.C.; Latorre, A.; Moya, A. Active and secreted IgA-coated bacterial fractions from the human gut reveal an under-represented microbiota core. Sci. Rep. 2013, 3, 3515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adler, C.J.; Dobney, K.; Weyrich, L.S.; Kaidonis, J.; Walker, A.W.; Haak, W.; Bradshaw, C.J.A.; Townsend, G.; Sołtysiak, A.; Alt, K.W.; et al. Sequencing ancient calcified dental plaque shows changes in oral microbiota with dietary shifts of the Neolithic and Industrial revolutions. Nat. Genet. 2013, 45, 450–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rascovan, N.; Huynh, H.; Chouin, G.; Adekola, K.; Georges-Zimmermann, P.; Signoli, M.; Desfosses, Y.; Aboudharam, G.; Drancourt, M.; Desnues, C. Tracing back ancient oral microbiomes and oral pathogens using dental pulps from ancient teeth. npj Biofilms Microbiomes 2016, 2, 6. [Google Scholar] [CrossRef] [PubMed]
- Hallmaier-Wacker, L.K.; Lueert, S.; Roos, C.; Knauf, S. The impact of storage buffer, DNA extraction method, and polymerase on microbial analysis. Sci. Rep. 2018, 8, 6292. [Google Scholar] [CrossRef] [PubMed]
- Edmonds, K.; Williams, L. The Role of the Negative Control in Microbiome Analyses. FASEB J. 2017, 31, 940–943. [Google Scholar]
- Kim, D.; Hofstaedter, C.E.; Zhao, C.; Mattei, L.; Tanes, C.; Clarke, E.; Lauder, A.; Sherrill-Mix, S.; Chehoud, C.; Kelsen, J.; et al. Optimizing methods and dodging pitfalls in microbiome research. Microbiome 2017, 5, 52. [Google Scholar] [CrossRef] [PubMed]
- Plaza-Díaz, J.; Fernández-Caballero, J.Á.; Chueca, N.; García, F.; Gómez-Llorente, C.; Sáez-Lara, M.J.; Fontana, L.; Gil, Á. Pyrosequencing analysis reveals changes in intestinal microbiota of healthy adults who received a daily dose of immunomodulatory probiotic strains. Nutrients 2015, 7, 3999–4015. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Santiago, J.; Gianella, S.; Massanella, M.; Spina, C.A.; Karris, M.Y.; Var, S.R.; Patel, D.; Jordan, P.S.; Young, J.A.; Little, S.J.; et al. Gut Lactobacillales are associated with higher CD4 and less microbial translocation during HIV infection. AIDS 2013, 27, 1921–1931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, S.; Mohammed, M.; Ghosh, T.; Kanungo, S.; Nair, G.; Mande, S.S. Metagenome of the gut of a malnourished child. Gut Pathog. 2011, 3, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peris-Bondia, F.; Latorre, A.; Artacho, A.; Moya, A.; D’Auria, G. The active human gut microbiota differs from the total microbiota. PLoS ONE 2011, 6, e22448. [Google Scholar] [CrossRef] [PubMed]
- Sommer, M.O.A.; Church, G.M.; Dantas, G. The human microbiome harbors a diverse reservoir of antibiotic resistance genes. Virulence 2010, 1, 299–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allali, I.; Delgado, S.; Marron, P.I.; Astudillo, A.; Yeh, J.J.; Ghazal, H.; Amzazi, S.; Keku, T.; Azcarate-Peril, M.A. Gut microbiome compositional and functional differences between tumor and non-tumor adjacent tissues from cohorts from the US and Spain. Gut Microbes 2015, 6, 161–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Ji, F.; Guo, J.; Shi, D.; Fang, D.; Li, L. Dysbiosis of small intestinal microbiota in liver cirrhosis and its association with etiology. Sci. Rep. 2016, 6, 34055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bashiardes, S.; Shapiro, H.; Rozin, S.; Shibolet, O.; Elinav, E. Non-alcoholic fatty liver and the gut microbiota. Mol. Metab. 2016, 5, 782–794. [Google Scholar] [CrossRef] [PubMed]
- Stearns, J.C.; Lynch, M.D.J.; Senadheera, D.B.; Tenenbaum, H.C.; Goldberg, M.B.; Cvitkovitch, D.G.; Croitoru, K.; Moreno-Hagelsieb, G.; Neufeld, J.D. Bacterial biogeography of the human digestive tract. Sci. Rep. 2011, 1, 170. [Google Scholar] [CrossRef] [PubMed]
- Rajilić-Stojanović, M.; de Vos, W.M. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol. Rev. 2014, 38, 996–1047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bull, M.J.; Plummer, N.T. Part 1: The Human Gut Microbiome in Health and Disease. Integr. Med. 2014, 13, 17–22. [Google Scholar]
- Struzycka, I. The oral microbiome in dental caries. Pol. J. Microbiol. 2014, 63, 127–135. [Google Scholar] [PubMed]
- The Human Microbiome Project Consortium; Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; FitzGerald, M.G.; et al. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [Green Version]
- Hartman, A.L.; Lough, D.M.; Barupal, D.K.; Fiehn, O.; Fishbein, T.; Zasloff, M.; Eisen, J.A. Human gut microbiome adopts an alternative state following small bowel transplantation. Proc. Natl. Acad. Sci. USA 2009, 106, 17187–17192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuczynski, J.; Stombaugh, J.; Walters, W.A.; González, A.; Caporaso, J.G.; Knight, R. Using QIIME to analyze 16s rRNA gene sequences from microbial communities. Curr. Protoc. Microbiol. 2012, 27. [Google Scholar] [CrossRef]
- MacKelprang, R.; Burkert, A.; Haw, M.; Mahendrarajah, T.; Conaway, C.H.; Douglas, T.A.; Waldrop, M.P. Microbial survival strategies in ancient permafrost: Insights from metagenomics. ISME J. 2017, 11, 2305–2318. [Google Scholar] [CrossRef] [PubMed]
- Jónsson, H.; Ginolhac, A.; Schubert, M. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 2013, 29, 1682–1684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kistler, L.; Ware, R.; Smith, O.; Collins, M.; Allaby, R.G. A new model for ancient DNA decay based on paleogenomic meta-analysis. Nucleic Acids Res. 2017, 45, 6310–6320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santiago-Rodriguez, T.M.; Fornaciari, G.; Luciani, S.; Dowd, S.E.; Toranzos, G.A.; Marota, I.; Cano, R.J. Natural mummification of the human gut preserves bacteriophage DNA. FEMS Microbiol. Lett. 2015, 363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
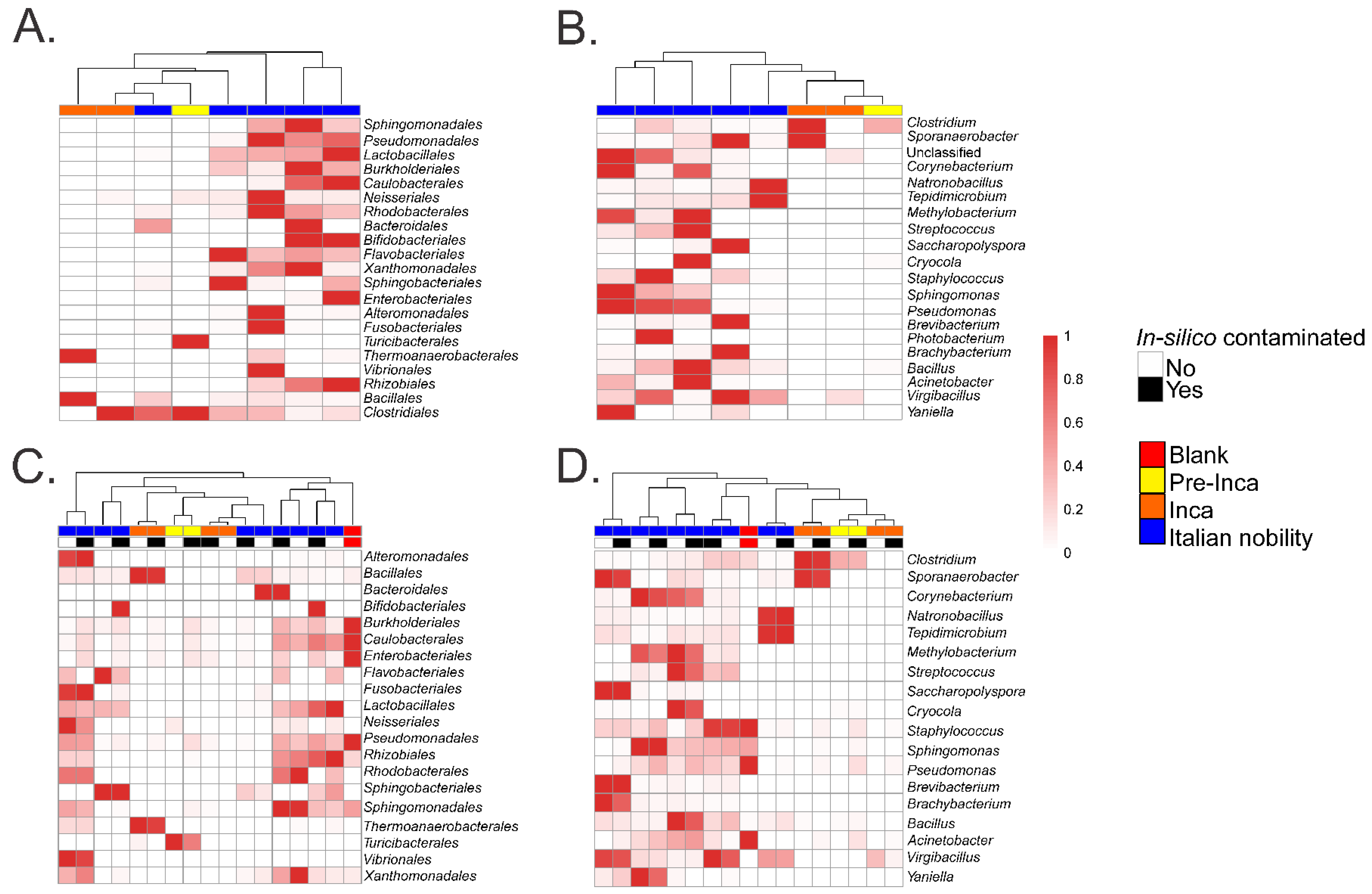
| Order (16S Data) | Mummy | Presence in Human Gut References | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| FI3 | FI9 | FI12 | NASD3 | NASD14 | NASD22 | NASD27 | NASD29 | Blank | ||
| Sphingomonadales | <0.01 | 0.20 | 0.02 | 35.14 | 14.934 | 0.51 | 8.74 | 0.22 | 3.01 | [6,12] |
| Pseudomonadales | <0.01 | 0.15 | <0.01 | 6.35 | 6.66 | 0.54 | 7.27 | 0.17 | 20.42 | [6,13,14] |
| Lactobacillales | <0.01 | 0.12 | 0.01 | 2.87 | 2.76 | 3.65 | 5.98 | 0.34 | 0.10 | [12,13,15] |
| Burkholderiales | <0.01 | <0.01 | 0.00 | 2.52 | 0.67 | 0.90 | 1.32 | 0.08 | 15.43 | [12,16] |
| Caulobacteriales | <0.01 | 0.01 | 0.00 | 1.08 | 0.24 | 0.04 | 1.69 | 0.04 | 6.90 | [17] |
| Neisseriales | <0.01 | 0.02 | <0.01 | <0.01 | 0.08 | <0.01 | <0.01 | <0.01 | 0.00 | [13,18] |
| Rhodobacterales | <0.01 | 0.00 | 0.00 | 0.02 | 0.05 | 0.01 | 0.01 | <0.01 | 0.05 | [12,19] |
| Xanthomonadales | <0.01 | 0.00 | 0.00 | 0.15 | 0.11 | 0.02 | 0.02 | 0.01 | 0.05 | [13,20] |
| Sphingobacteriales | <0.01 | 0.00 | 0.00 | 0.00 | <0.01 | 0.19 | 0.05 | 0.02 | 0.03 | [12,21] |
| Enterobacteriales | <0.01 | 0.00 | <0.01 | 0.10 | 0.01 | <0.01 | 0.25 | 0.00 | 7.01 | [12,13,21] |
| Rhizobiales | 0.04 | 0.47 | 0.12 | 18.9 | 8.93 | 0.39 | 44.94 | 0.65 | 5.36 | [12,13] |
| Bacillales | 96.82 | 0.36 | 0.01 | 7.30 | 29.66 | 14.81 | 2.91 | 23.83 | 9.91 | [13,15,21] |
| Clostridiales | 0.04 | 97.65 | 99.6 | 4.10 | 19.79 | 34.00 | 9.49 | 73.93 | 17.34 | [12,13] |
| Sample | Counts Prior Filtering | Counts after Filtering |
|---|---|---|
| Blank | 3824 | NA |
| FI3 | 78,903 | 78,881 |
| FI9 | 24,956 | 24,495 |
| FI12 | 56,971 | 56,846 |
| NASD3 | 32,086 | 14,768 |
| NASD14 | 44,255 | 24,740 |
| NASD22 | 60,266 | 55,727 |
| NASD27 | 24,633 | 13,392 |
| NASD29 | 101,517 | 100,028 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santiago-Rodriguez, T.M.; Toranzos, G.A. On Controls in Ancient Microbiome Studies, and Microbial Resilience in Ancient Samples. Genes 2018, 9, 471. https://doi.org/10.3390/genes9100471
Santiago-Rodriguez TM, Toranzos GA. On Controls in Ancient Microbiome Studies, and Microbial Resilience in Ancient Samples. Genes. 2018; 9(10):471. https://doi.org/10.3390/genes9100471
Chicago/Turabian StyleSantiago-Rodriguez, Tasha M., and Gary A. Toranzos. 2018. "On Controls in Ancient Microbiome Studies, and Microbial Resilience in Ancient Samples" Genes 9, no. 10: 471. https://doi.org/10.3390/genes9100471




