A Comparative Study of the Structural Dynamics of Four Terminal Uridylyl Transferases
Abstract
:1. Introduction
2. Materials and Methods
2.1. Molecular Dynamics Simulations
2.1.1. TbRET1
2.1.2. TbRET2
2.2. Ensemble-Averaged Electrostatics
2.2.1. TbRET1
2.2.2. TbRET2
2.2.3. TbMEAT1
2.2.4. TbTUT4
2.3. Clustering
3. Results
3.1. POVME Analysis
3.2. Ensemble-Averaged Electrostatics
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Berriman, M.; Ghedin, E.; Hertz-Fowler, C.; Blandin, G.; Renauld, H.; Bartholomeu, D.C.; Lennard, N.J.; Caler, E.; Hamlin, N.E.; Haas, B.; et al. The genome of the african trypanosome trypanosoma brucei. Science 2005, 309, 416–422. [Google Scholar] [CrossRef] [PubMed]
- Demir, O.; Amaro, R.E. Elements of nucleotide specificity in the trypanosoma brucei mitochondrial RNA editing enzyme RET2. J. Chem. Inf. Model. 2012, 52, 1308–1318. [Google Scholar] [CrossRef] [PubMed]
- Buchanan-Kilbey, G.; Djumpah, J.; Papadopoulou, M.V.; Bloomer, W.; Hu, L.; Wilkinson, S.R.; Ashworth, R. Evaluating the developmental toxicity of trypanocidal nitroaromatic compounds on zebrafish. Acta Trop. 2013, 128, 701–705. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, A.; Coote, M.L.; Barakat, K. Molecular dynamics-driven drug discovery: Leaping forward with confidence. Drug Discov. Today 2017, 22, 249–269. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Ernst, N.L.; Turley, S.; Stuart, K.D.; Hol, W.G. Structural basis for UTP specificity of RNA editing tutases from trypanosoma brucei. EMBO J 2005, 24, 4007–4017. [Google Scholar] [CrossRef] [PubMed]
- Aphasizhev, R.; Aphasizheva, I. Uridine insertion/deletion editing in trypanosomes: A playground for RNA-guided information transfer. Wiley Interdiscip. Rev. RNA 2011, 2, 669–685. [Google Scholar] [CrossRef] [PubMed]
- Aphasizhev, R.; Aphasizheva, I. Mitochondrial RNA editing in trypanosomes: Small rnas in control. Biochimie 2014, 100, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Stagno, J.; Aphasizheva, I.; Rosengarth, A.; Luecke, H.; Aphasizhev, R. UTP-bound and apo structures of a minimal RNA uridylyltransferase. J. Mol. Biol. 2007, 366, 882–899. [Google Scholar] [CrossRef] [PubMed]
- Alfonzo, J.D.; Thiemann, O.; Simpson, L. The mechanism of u insertion/deletion RNA editing in kinetoplastid mitochondria. Nucleic Acids Res. 1997, 25, 3751–3759. [Google Scholar] [CrossRef] [PubMed]
- Kuchta, K.; Knizewski, L.; Wyrwicz, L.S.; Rychlewski, L.; Ginalski, K. Comprehensive classification of nucleotidyltransferase fold proteins: Identification of novel families and their representatives in human. Nucleic Acids Res. 2009, 37, 7701–7714. [Google Scholar] [CrossRef] [PubMed]
- Aphasizheva, I.; Aphasizhev, R. Ret1-catalyzed uridylylation shapes the mitochondrial transcriptome in trypanosoma brucei. Mol. Cell. Biol. 2010, 30, 1555–1567. [Google Scholar] [CrossRef] [PubMed]
- Rajappa, L.T.; Suematsu, T.; Munoz-Tello, P.; Long, M.; Qian, Y.; Demir, Ö.; Cheng, K.J.; Stagno, J.R.; Luecke, H.; Costello, C.E.; et al. RNA editing tutase 1: Structural foundation of substrate recognition, complex interactions and drug targeting. Nucleic Acids Res. 2016, 44, 10862–10878. [Google Scholar] [CrossRef] [PubMed]
- Stagno, J.; Aphasizheva, I.; Bruystens, J.; Luecke, H.; Aphasizhev, R. Structure of the mitochondrial editosome-like complex associated tutase 1 reveals divergent mechanisms of UTP selection and domain organization. J. Mol. Biol. 2010, 399, 464–475. [Google Scholar] [CrossRef] [PubMed]
- Dror, R.O.; Dirks, R.M.; Grossman, J.P.; Xu, H.; Shaw, D.E. Biomolecular simulation: A computational microscope for molecular biology. Annu. Rev. Biophys. 2012, 41, 429–452. [Google Scholar] [CrossRef] [PubMed]
- Klepeis, J.L.; Lindorff-Larsen, K.; Dror, R.O.; Shaw, D.E. Long-timescale molecular dynamics simulations of protein structure and function. Curr. Opin. Struct. Biol. 2009, 19, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Adcock, S.A.; McCammon, J.A. Molecular dynamics: Survey of methods for simulating the activity of proteins. Chem. Rev. 2006, 106, 1589–1615. [Google Scholar] [CrossRef] [PubMed]
- Demir, O.; Labaied, M.; Merritt, C.; Stuart, K.; Amaro, R.E. Computer-aided discovery of trypanosoma brucei RNA-editing terminal uridylyl transferase 2 inhibitors. Chem. Biol. Drug Des. 2014, 84, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. Vmd: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Votapka, L.W.; Czapla, L.; Zhenirovskyy, M.; Amaro, R.E. Delensembleelec: Computing ensemble-averaged electrostatics using delphi. Commun. Comput. Phys. 2013, 13, 256–268. [Google Scholar] [CrossRef]
- Durrant, J.D.; Votapka, L.; Sorensen, J.; Amaro, R.E. Povme 2.0: An enhanced tool for determining pocket shape and volume characteristics. J. Chem. Theory Comput. 2014, 10, 5047–5056. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.R.; Sorensen, J.; Hensley, N.; Wong, C.; Zhu, C.; Amaro, R.E. Povme 3.0: Software for mapping binding pocket flexibility. 2017; Submitted. [Google Scholar]
- Roberts, E.; Eargle, J.; Wright, D.; Luthey-Schulten, Z. Multiseq: Unifying sequence and structure data for evolutionary analysis. BMC Bioinform. 2006, 7, 382. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Grove, L.E.; Hall, D.R.; Bohnuud, T.; Mottarella, S.E.; Luo, L.; Xia, B.; Beglov, D.; Vajda, S. The ftmap family of web servers for determining and characterizing ligand-binding hot spots of proteins. Nat. Protoc. 2015, 10, 733–755. [Google Scholar] [CrossRef] [PubMed]
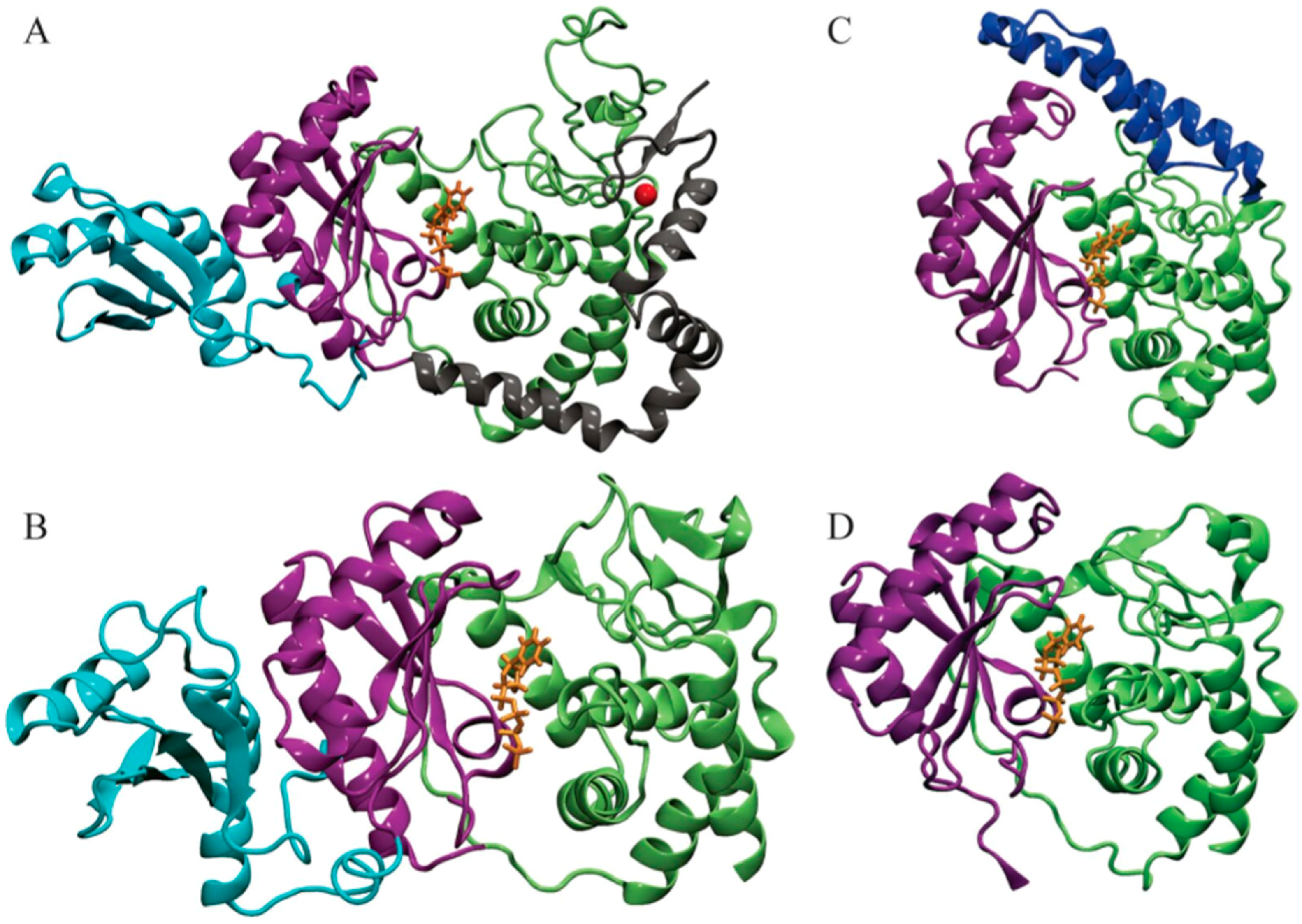
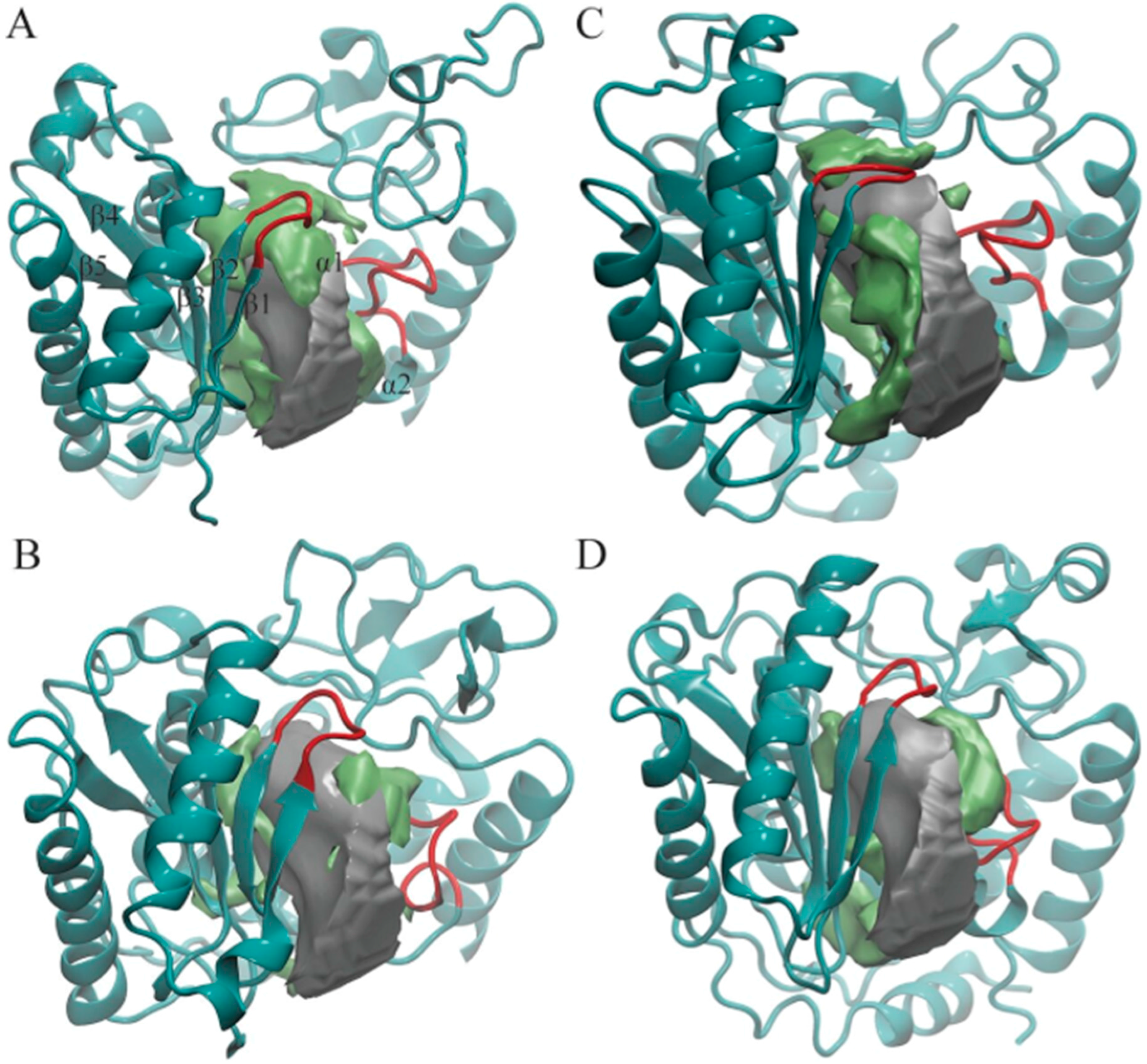
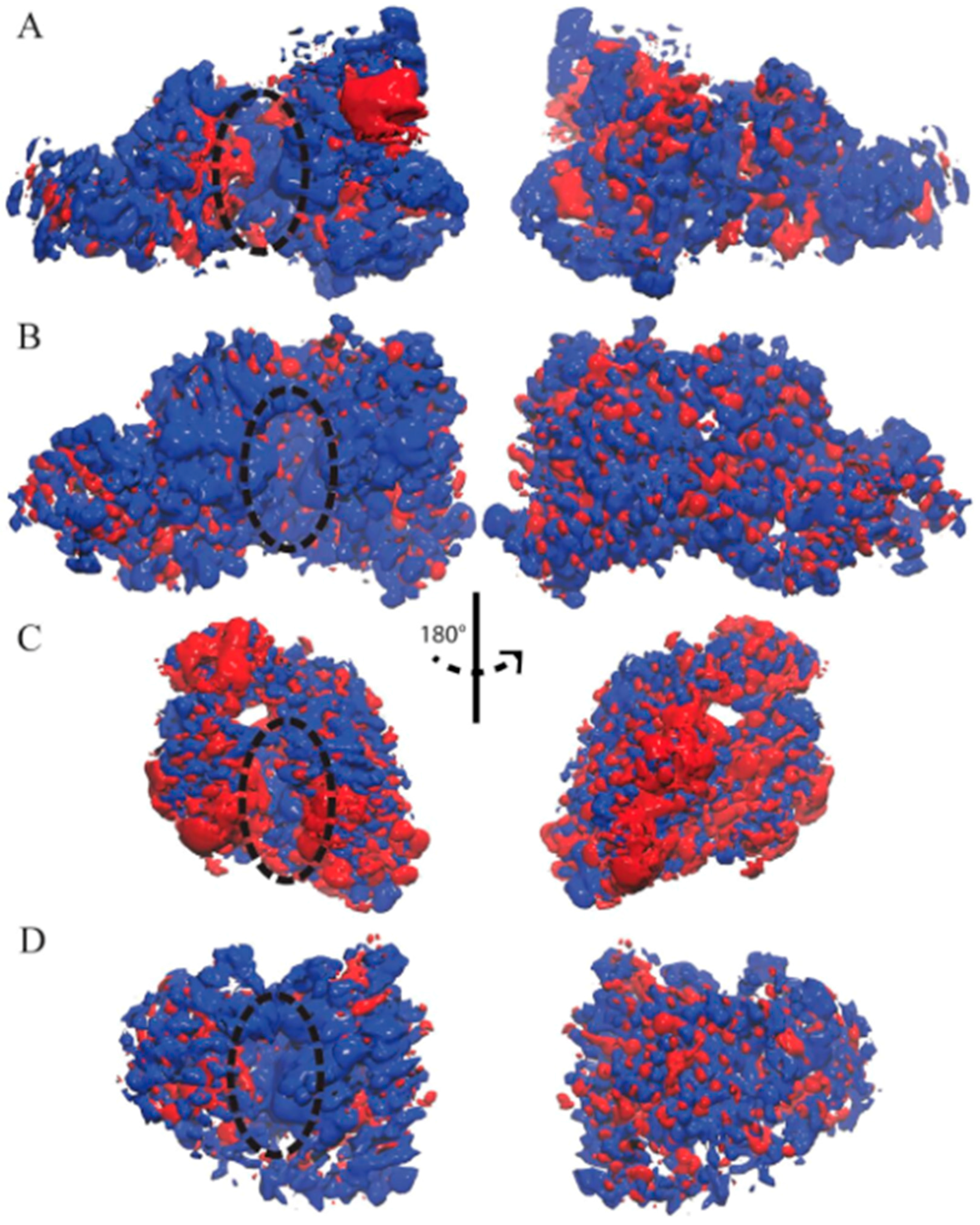
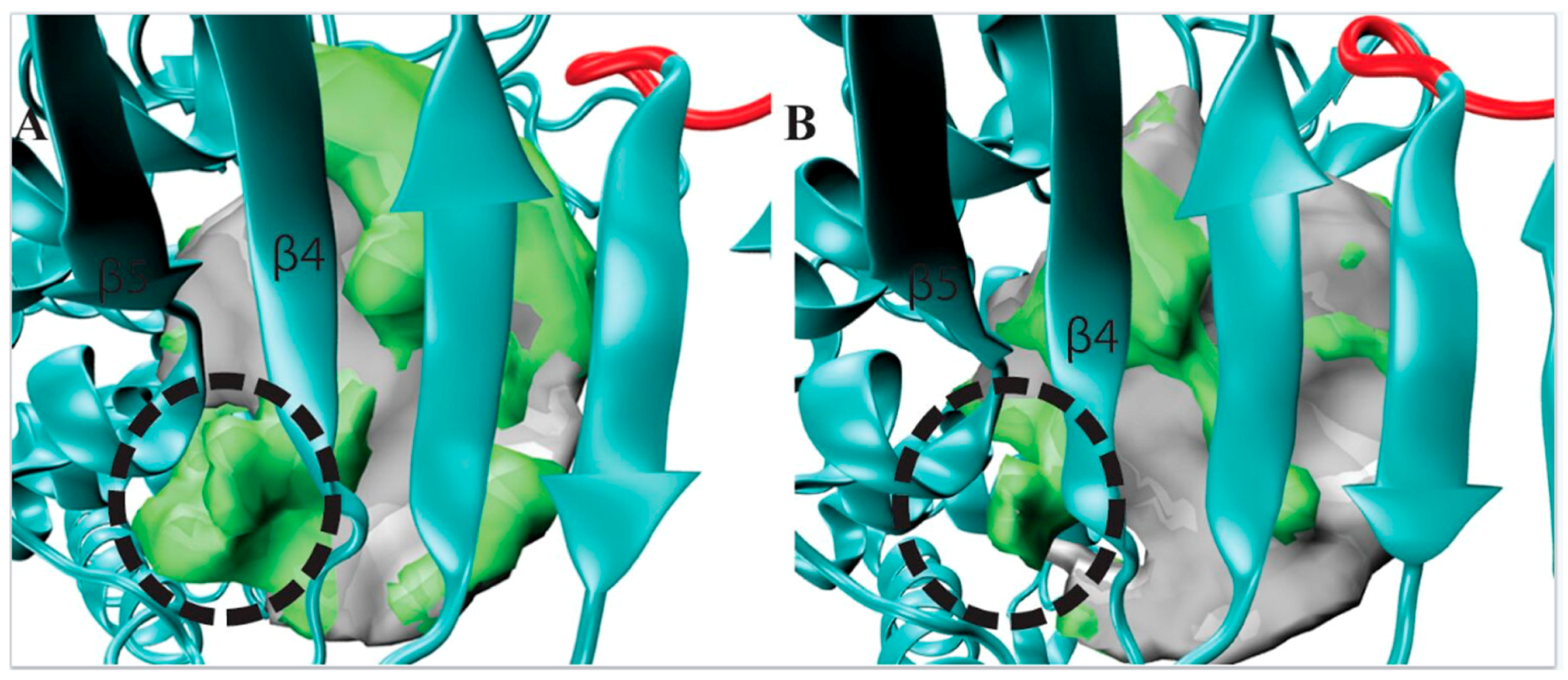
| Enzyme | In Complex With | Domains | Substrates | Ref. |
|---|---|---|---|---|
| RET1 | Mitochondrial 3’ processome (MPsome) | C2H2 Zn Finger, CTD, NTD, RRM | UTP, high affinity for ssRNA | [12] |
| RET2 | RNA Editing Core Complex (Editosome) | Middle Domain, CTD, NTD | UTP, ssRNA, dsRNA (needs gRNAs) | [5] |
| MEAT1 | Resembles editosome | Bridge Domain, CTD, NTD | Exclusively U Specific, ssRNA, dsRNA | [13] |
| TUT4 | Unknown | NTD, CTD (minimal TUTase) | Prefers UTP and exclusively single-stranded RNA as a primer | [8] |
| Enzyme | Average Volume/Å3 | Standard Deviation | Number of Frames |
|---|---|---|---|
| TbRET1 | 2434 | 194 | 1500 |
| TbRET2 | 2061 | 145 | 1000 |
| TbMEAT1 | 1831 | 247 | 1000 |
| TbTUT4 | 1768 | 316 | 1000 |
| Cluster | Enzyme | Average Volume/Å3 | Standard Deviation |
|---|---|---|---|
| 1 | TbRET1 | 2508 | 147 |
| 2 | TbRET2 | 2073 | 134 |
| 3 | TbMEAT1 | 2071 | 143 |
| 4 | TbTUT4 | 1750 | 83 |
| 5 | TbRET1 | 2195 | 116 |
| 6 | TbTUT4 | 2175 | 132 |
| 7 | TbMEAT1 | 1661 | 90 |
| 8 | TbMEAT1 | 1810 | 102 |
| 9 | TbTUT4 | 1506 | 90 |
| 10 | TbMEAT1 | 1535 | 91 |
| 11 | TbTUT4 | 1191 | 65 |
| 12 | TbTUT4 | 1393 | 97 |
| 13 | TbRET2 | 1772 | 95 |
| 14 | TbTUT4 | 1610 | 75 |
| 15 | TbMEAT1 | 1318 | 52 |
| 16 | TbTUT4 | 1749 | 80 |
| 17 | TbMEAT1 | 1711 | 48 |
| 18 | TbRET1 | 1848 | 4 |
| 19 | TbTUT4 | 1620 | 0 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, K.J.; Demir, Ö.; Amaro, R.E. A Comparative Study of the Structural Dynamics of Four Terminal Uridylyl Transferases. Genes 2017, 8, 166. https://doi.org/10.3390/genes8060166
Cheng KJ, Demir Ö, Amaro RE. A Comparative Study of the Structural Dynamics of Four Terminal Uridylyl Transferases. Genes. 2017; 8(6):166. https://doi.org/10.3390/genes8060166
Chicago/Turabian StyleCheng, Kevin J., Özlem Demir, and Rommie E. Amaro. 2017. "A Comparative Study of the Structural Dynamics of Four Terminal Uridylyl Transferases" Genes 8, no. 6: 166. https://doi.org/10.3390/genes8060166





