Study of mcr-1 Gene-Mediated Colistin Resistance in Enterobacteriaceae Isolated from Humans and Animals in Different Countries
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains
2.2. Conjugation and Plasmid Analysis
2.3. Whole Genome Analysis
3. Results
3.1. Strain Characteristics
3.2. Location of the mcr-1 Gene
3.3. Genome Analysis
Mcr-1 Database
3.4. Sequenced Plasmids
3.5. Genetic Integration of mcr-1 Gene
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Grundmann, H.; Glasner, C.; Albiger, B.; Aanensen, D.M.; Tomlinson, C.T.; Andrasević, A.T.; Cantón, R.; Carmeli, Y.; Friedrich, A.W.; Giske, C.G.; et al. Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): A prospective, multinational study. Lancet Infect. Dis. 2017, 17, 153–163. [Google Scholar] [CrossRef]
- Biswas, S.; Brunel, J.-M.; Dubus, J.-C.; Reynaud-Gaubert, M.; Rolain, J.-M. Colistin: An update on the antibiotic of the 21st century. Expert Rev. Anti. Infect. Ther. 2012, 10, 917–934. [Google Scholar] [CrossRef] [PubMed]
- Stein, A.; Raoult, D. Colistin: An Antimicrobial for the 21st Century? Clin. Infect. Dis. 2002, 35, 901–902. [Google Scholar] [CrossRef] [PubMed]
- Jeannot, K.; Bolard, A.; Plésiat, P. Resistance to polymyxins in Gram-negative organisms. Int. J. Antimicrob. Agents 2017, 49, 526–535. [Google Scholar] [CrossRef] [PubMed]
- Baron, S.; Hadjadj, L.; Rolain, J.M.; Olaitan, A.O. Molecular mechanisms of polymyxin resistance: Knowns and unknowns. Int. J. Antimicrob. Agents 2016. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Yang, Y.-Q.; Li, Y.-X.; Song, T.; Yang, Y.-X.; Jiang, W.; Zhang, A.-Y.; Guo, X.-Y.; Liu, B.-H.; Wang, Y.-X.; Lei, C.-W.; et al. Colistin Resistance Gene mcr-1 and Its Variant in Escherichia coli Isolates from Chickens in China. Antimicrob. Agents Chemother. 2017, 61, e01204-16. [Google Scholar] [CrossRef] [PubMed]
- Xavier, B.B.; Lammens, C.; Ruhal, R.; Kumar-Singh, S.; Butaye, P.; Goossens, H.; Malhotra-Kumar, S. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Eurosurveillance 2016, 21, 30280. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Li, H.; Shen, Y.; Liu, Z.; Wang, S.; Shen, Z.; Zhang, R.; Walsh, T.R.; Shen, J.; Wang, Y. Novel Plasmid-Mediated Colistin Resistance Gene mcr-3 in Escherichia coli. mBio 2017, 8, e00543-17. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Villa, L.; Feudi, C.; Curcio, L.; Orsini, S.; Luppi, A.; Pezzotti, G.; Magistrali, C.F. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill. 2017, 22, 30589. [Google Scholar] [CrossRef] [PubMed]
- Borowiak, M.; Fischer, J.; Hammerl, J.A.; Hendriksen, R.S.; Szabo, I.; Malorny, B. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J. Antimicrob. Chemother. 2017. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Fang, L.-X.; Wu, Z.; Deng, H.; Yang, R.-S.; Li, X.-P.; Li, S.-M.; Liao, X.-P.; Feng, Y.; Liu, Y.-H. Genetic Analysis of the IncX4 Plasmids: Implications for a Unique Pattern in the mcr-1 Acquisition. Sci. Rep. 2017, 7, 424. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Sun, J.; Li, J.; Ding, Y.; Li, X.-P.; Lin, J.; Hassan, B.; Feng, Y. Expanding landscapes of the diversified mcr-1-bearing plasmid reservoirs. Microbiome 2017, 5, 70. [Google Scholar] [CrossRef] [PubMed]
- Tegetmeyer, H.E.; Jones, S.C.P.; Langford, P.R.; Baltes, N. ISApl1, a novel insertion element of Actinobacillus pleuropneumoniae, prevents ApxIV-based serological detection of serotype 7 strain AP76. Vet. Microbiol. 2008, 128, 342–353. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Kieffer, N.; Nordmann, P. In-vitro study of IS Apl1 -mediated mobilization of the colistin resistance gene mcr-1. Antimicrob. Agents Chemother. 2017. [Google Scholar] [CrossRef] [PubMed]
- Snesrud, E.; He, S.; Chandler, M.; Dekker, J.P.; Hickman, A.B.; McGann, P.; Dyda, F. A Model for Transposition of the Colistin Resistance Gene mcr-1 by IS Apl1. Antimicrob. Agents Chemother. 2016, 60, 6973–6976. [Google Scholar] [CrossRef] [PubMed]
- Berrazeg, M.; Hadjadj, L.; Ayad, A.; Drissi, M.; Rolain, J.M. First detected human case in Algeria of mcr-1 plasmid-mediated colistin resistance in a 2011 Escherichia coli isolate. Antimicrob. Agents Chemother. 2016, 60, 6996–6997. [Google Scholar] [CrossRef] [PubMed]
- Olaitan, A.O.; Chabou, S.; Okdah, L.; Morand, S.; Rolain, J.-M. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect. Dis. 2016, 16, 147. [Google Scholar] [CrossRef]
- Rolain, J.-M.; Kempf, M.; Leangapichart, T.; Chabou, S.; Olaitan, A.O.; Le Page, S.; Morand, S.; Raoult, D. Plasmid-Mediated mcr-1 Gene in Colistin-Resistant Clinical Isolates of Klebsiella pneumoniae in France and Laos. Antimicrob. Agents Chemother. 2016, 60, 6994–6995. [Google Scholar] [CrossRef] [PubMed]
- Leangapichart, T.; Gautret, P.; Brouqui, P.; Memish, Z.A.; Raoult, D.; Rolain, J.-M. Correction for Leangapichart et al., Acquisition of mcr-1 Plasmid-Mediated Colistin Resistance in Escherichia coli and Klebsiella pneumoniae during Hajj 2013 and 2014. Antimicrob. Agents Chemother. 2016, 60, 7537. [Google Scholar] [CrossRef] [PubMed]
- Olaitan, A.O.; Thongmalayvong, B.; Akkhavong, K.; Somphavong, S.; Paboriboune, P.; Khounsy, S.; Morand, S.; Rolain, J.-M. Clonal transmission of a colistin-resistant Escherichia coli from a domesticated pig to a human in Laos: Table 1. J. Antimicrob. Chemother. 2015, 47, dkv252. [Google Scholar] [CrossRef] [PubMed]
- Olaitan, A.O.; Diene, S.M.; Kempf, M.; Berrazeg, M.; Bakour, S.; Gupta, S.K.; Thongmalayvong, B.; Akkhavong, K.; Somphavong, S.; Paboriboune, P.; et al. Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: An epidemiological and molecular study. Int. J. Antimicrob. Agents 2014, 44, 500–507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- EUCAST. European Committee on Antimicrobial Susceptibility Testing Breakpoint tables for interpretation of MICs and zone diameters European Committee on Antimicrobial Susceptibility Testing Breakpoint tables for interpretation of MICs and zone diameters. Available online: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_50_Breakpoint_Table_01.pdf (accessed on 4 January 2017).
- Chabou, S.; Leangapichart, T.; Okdah, L.; Le Page, S.; Hadjadj, L.; Rolain, J.M. Real-time quantitative PCR assay with Taqman probe for rapid detection of MCR-1 plasmid-mediated colistin resistance. New Microbes New Infect. 2016, 13, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Escherichia coli_Enterobase. Available online: http://mlst.warwick.ac.uk/mlst/dbs/Ecoli (accessed on 20 January 2017).
- Klebsiella sequence typing. Available online: http://bigsdb.web.pasteur.fr/klebsiella (accessed on 20 January 2017).
- Sennati, S.; Di Pilato, V.; Riccobono, E.; Di Maggio, T.; Villagran, A.L.; Pallecchi, L.; Bartoloni, A.; Rossolini, G.M.; Giani, T. Citrobacter braakii carrying plasmid-borne mcr-1 colistin resistance gene from ready-to-eat food from a market in the Chaco region of Bolivia. J. Antimicrob. Chemother. 2017, 72, 2127–2129. [Google Scholar] [CrossRef] [PubMed]
- Bachiri, T.; Bakour, S.; Lalaoui, R.; Belkebla, N.; Allouache, M.; Rolain, J.M.; Touati, A. Occurrence of Carbapenemase-Producing Enterobacteriaceae Isolates in the Wildlife: First Report of OXA-48 in Wild Boars in Algeria. Microb. Drug Resist. 2017. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Bertini, A.; Villa, L.; Falbo, V.; Hopkins, K.L.; Threlfall, E.J. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 2005, 63, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chavda, K.D.; Al Laham, N.; Melano, R.G.; Jacobs, M.R.; Bonomo, R.A.; Kreiswirth, B.N. Complete Nucleotide Sequence of a blaKPC-Harboring IncI2 Plasmid and Its Dissemination in New Jersey and New York Hospitals. Antimicrob. Agents Chemother. 2013, 57, 5019–5025. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.-T.; Song, F.-J.; Zou, M.; Hao, Z.-H.; Shan, H. Emergence of Colistin Resistance Gene mcr-1 in Cronobacter sakazakii Producing NDM-9 and in Escherichia coli from the Same Animal. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.-M. ARG-ANNOT, a New Bioinformatic Tool To Discover Antibiotic Resistance Genes in Bacterial Genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; Garcia-Fernandez, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Moller Aarestrup, F.; Hasman, H. In Silico Detection and Typing of Plasmids using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rossello-Mora, R.; Oliver Glackner, F.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Arantes, A.S.; Stothard, P. Comparing thousands of circular genomes using the CGView Comparison Tool. BMC Genom. 2012, 13, 202. [Google Scholar] [CrossRef] [PubMed]
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef] [PubMed]
- Meinersmann, R.J.; Ladely, S.R.; Plumblee, J.R.; Hall, M.C.; Simpson, S.A.; Ballard, L.L.; Scheffler, B.E.; Genzlinger, L.L.; Cook, K.L. Complete Genome Sequence of a Colistin Resistance mcr-1 Gene-Bearing Escherichia coli Strain from the United States. Genome Announc. 2016. [Google Scholar] [CrossRef]
- Fernandes, M.R.; McCulloch, J.A.; Vianello, M.A.; Moura, Q.; Pérez-Chaparro, P.J.; Esposito, F.; Sartori, L.; Dropa, M.; Matté, M.H.; Lira, D.P.A.; et al. First Report of the Globally Disseminated IncX4 Plasmid Carrying the mcr-1 Gene in a Colistin-Resistant Escherichia coli Sequence Type 101 Isolate from a Human Infection in Brazil. Antimicrob. Agents Chemother. 2016, 60, 6415–6417. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.Y.; Ang, G.Y.; Chong, T.-M.; Chin, P.S.; Ngeow, Y.F.; Yin, W.-F.; Chan, K.-G. Complete genome sequencing revealed novel genetic contexts of the mcr-1 gene in Escherichia coli strains. J. Antimicrob. Chemother. 2017, 72, 1253–1255. [Google Scholar] [CrossRef] [PubMed]
- Meinersmann, R.J.; Ladely, S.R.; Bono, J.L.; Plumblee, J.R.; Hall, M.C.; Genzlinger, L.L.; Cook, K.L. Complete Genome Sequence of a Colistin Resistance Gene (mcr-1)-Bearing Isolate of Escherichia coli from the United States. Genome Announc. 2016, 4, e01283-16. [Google Scholar] [CrossRef] [PubMed]
- Sellera, F.P.; Fernandes, M.R.; Carvalho, M.N.; Nascimiento, C.L.; Dutra, G.P.; McCulloch, J.A.; Pérez-Chaparro, P.J.; Sartori, L.; Lincopan, N. Identification of mcr-1 in Escherichia coli isolates from footpad lesions in South American penguin. (Unpublished).
- Bai, L.; Wang, J.; Hurley, D.; Yu, Z.; Wang, L.; Chen, Q.; Li, J.; Li, F.; Fanning, S. Characterisation of two large plasmids containing ESBL genes; a novel disrupted mcr-1 gene and a lysogenized phage P-1 like sequence, cultured from a human atypical enteropathogenic Escherichia coli (aEPEC) recovered in China. Unpublished.
- Snesrud, E.; Ong, A.C.; Corey, B.; Kwak, Y.I.; Clifford, R.; Gleeson, T.; Wood, S.; Whitman, T.J.; Lesho, E.P.; Hinkle, M.; McGann, P. Analysis of Serial Isolates of mcr-1 -Positive Escherichia coli Reveals a Highly Active IS Apl1 Transposon. Antimicrob. Agents Chemother. 2017, 61, e00056-17. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, R.L.; Batra, D.; Rowe, L.; Loparev, V.N.; Stripling, D.; Garcia-Toledo, L.; Knipe, K.; Juieng, P.; Sheth, M.; Martin, H.; et al. High-Quality Genome Sequence of an Escherichia coli O157 Strain Carrying an mcr-1 Resistance Gene Isolated from a Patient in the United States. Genome Announc. 2017, 5, e01725-16. [Google Scholar] [CrossRef] [PubMed]
- Malhotra-Kumar, S.; Xavier, B.B.; Das, A.J.; Lammens, C.; Butaye, P.; Goossens, H. Colistin resistance gene mcr-1 harboured on a multidrug resistant plasmid. Lancet. Infect. Dis. 2016, 16, 283–284. [Google Scholar] [CrossRef]
- Yang, R.-S.J.; Sun, J.; Fang, L.-X. Escherichia coli strain CQ02-121 plasmid pCQ02-121, complete sequence. Unpublished.
- Brauer, A.; Telling, K.; Laht, M.; Kalmus, P.; Lutsar, I.; Remm, M.; Kisand, V.; Tenson, T. Plasmid with Colistin Resistance Gene mcr-1 in Extended-Spectrum-β-Lactamase-Producing Escherichia coli Strains Isolated from Pig Slurry in Estonia. Antimicrob. Agents Chemother. 2016, 60, 6933–6936. [Google Scholar] [CrossRef] [PubMed]
- Sonnevend, Á.; Ghazawi, A.; Alqahtani, M.; Shibl, A.; Jamal, W.; Hashmey, R.; Pal, T. Plasmid-mediated colistin resistance in Escherichia coli from the Arabian Peninsula. Int. J. Infect. Dis. 2016, 50, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Yang, Y.; Miao, M.; Chavda, K.D.; Mediavilla, J.R.; Xie, X.; Feng, P.; Tang, Y.-W.; Kreiswirth, B.N.; Chen, L.; Du, H. Complete Sequences of mcr-1 -Harboring Plasmids from Extended-Spectrum-β-Lactamase- and Carbapenemase-Producing Enterobacteriaceae. Antimicrob. Agents Chemother. 2016, 60, 4351–4354. [Google Scholar] [CrossRef] [PubMed]
- Perreten, V.; Strauss, C.; Collaud, A.; Gerber, D. Colistin Resistance Gene mcr-1 in Avian-Pathogenic Escherichia coli in South Africa. Antimicrob. Agents Chemother. 2016, 60, 4414–4415. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Zong, Z. Kluyvera ascorbata carrying the mcr-1 colistin resistance gene from hospital sewage. Antimicrob. Agents Chemother. 2016. [Google Scholar] [CrossRef]
- Yang, Y.-Q.; Zhang, A.-Y.; Ma, S.-Z.; Kong, L.-H.; Li, Y.-X.; Liu, J.-X.; Davis, M.A.; Guo, X.-Y.; Liu, B.-H.; Lei, C.-W.; Wang, H.-N. Co-occurrence of mcr-1 and ESBL on a single plasmid in Salmonella enterica. J. Antimicrob. Chemother. 2016, 71, 2336–2338. [Google Scholar] [CrossRef] [PubMed]
- Xavier, B.B.; Lammens, C.; Butaye, P.; Goossens, H.; Malhotra-Kumar, S. Complete sequence of an IncFII plasmid harbouring the colistin resistance gene mcr-1 isolated from Belgian pig farms. J. Antimicrob. Chemother. 2016, 71, 2342–2344. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Kieffer, N.; Brink, A.; Coetze, J.; Jayol, A.; Nordmann, P. Genetic Features of MCR-1-Producing Colistin-Resistant Escherichia coli Isolates in South Africa. Antimicrob. Agents Chemother. 2016, 60, 4394–4397. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Li, X.-P.; Yang, R.-S.; Fang, L.-X.; Huo, W.; Li, S.-M.; Jiang, P.; Liao, X.-P.; Liu, Y.-H. Complete Nucleotide Sequence of an IncI2 Plasmid Coharboring blaCTX-M-55 and mcr-1. Antimicrob. Agents Chemother. 2016, 60, 5014–5017. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Xie, M.; Zhang, J.; Yang, Z.; Liu, L.; Liu, X.; Zheng, Z.; Chan, E.W.-C.; Chen, S. Genetic characterization of mcr-1 -bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J. Antimicrob. Chemother. 2017, 72, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Zurfluh, K.; Klumpp, J.; Nüesch-Inderbinen, M.; Stephan, R. Full-Length Nucleotide Sequences of mcr-1 -Harboring Plasmids Isolated from Extended-Spectrum-β-Lactamase-Producing Escherichia coli Isolates of Different Origins. Antimicrob. Agents Chemother. 2016, 60, 5589–5591. [Google Scholar] [CrossRef] [PubMed]
- Di Pilato, V.; Arena, F.; Tascini, C.; Cannatelli, A.; Henrici De Angelis, L.; Fortunato, S.; Giani, T.; Menichetti, F.; Rossolini, G.M. mcr-1.2, a New mcr Variant Carried on a Transferable Plasmid from a Colistin-Resistant KPC Carbapenemase-Producing Klebsiella pneumoniae Strain of Sequence Type 512. Antimicrob. Agents Chemother. 2016, 60, 5612–5615. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.H.; Yi, L.X.; Wang, J. Distribution of mcr-1 in Salmonella enterica. Unpublished.
- McGann, P.; Snesrud, E.; Maybank, R.; Corey, B.; Ong, A.C.; Clifford, R.; Hinkle, M.; Whitman, T.; Lesho, E.; Schaecher, K.E. Escherichia coli Harboring mcr-1 and blaCTX-M on a Novel IncF Plasmid: First Report of mcr-1 in the United States. Antimicrob. Agents Chemother. 2016, 60, 4420–4421. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Feng, Y.; Lü, X.; McNally, A.; Zong, Z. IncP Plasmid Carrying Colistin Resistance Gene mcr-1 in Klebsiella pneumoniae from Hospital Sewage. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Mediavilla, J.R.; Patrawalla, A.; Chen, L.; Chavda, K.D.; Mathema, B.; Vinnard, C.; Dever, L.L.; Kreiswirth, B.N. Colistin- and Carbapenem-Resistant Escherichia coli Harboring mcr-1 and blaNDM-5, Causing a Complicated Urinary Tract Infection in a Patient from the United States: TABLE 1. mBio 2016, 7, e01191-16. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Xie, M.; Lv, J.; Wai-Chi Chan, E.; Chen, S. Complete genetic analysis of plasmids carrying mcr-1 and other resistance genes in an Escherichia coli isolate of animal origin. J. Antimicrob. Chemother. 2016, 72. [Google Scholar] [CrossRef]
- Malhotra-Kumar, S.; Xavier, B.B.; Das, A.J.; Lammens, C.; Hoang, H.T.T.; Pham, N.T.; Goossens, H. Colistin-resistant Escherichia coli harbouring mcr-1 isolated from food animals in Hanoi, Vietnam. Lancet Infect. Dis. 2016, 16, 286–287. [Google Scholar] [CrossRef]
- Kong, L.-H.; Lei, C.-W.; Ma, S.-Z.; Jiang, W.; Liu, B.-H.; Wang, Y.-X.; Guan, R.; Men, S.; Yuan, Q.-W.; Cheng, G.-Y.; et al. Various Sequence Types of Escherichia coli Isolates Coharboring blaNDM-5 and mcr-1 Genes from a Commercial Swine Farm in China. Antimicrob. Agents Chemother. 2017, 61, e02167-16. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Xie, M.; Chen, K.; Dong, N.; Zhang, R.; Chen, S. Characterization of a hybrid plasmid harboring a composite transposon encoding mcr-1. (Unpublished).
- Gao, R.; Hu, Y.; Li, Z.; Sun, J.; Wang, Q.; Lin, J.; Ye, H.; Liu, F.; Srinivas, S.; Li, D.; et al. Dissemination and Mechanism for the MCR-1 Colistin Resistance. PLOS Pathog. 2016, 12, e1005957. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Feng, Y.; Liu, F.; Jiang, H.; Qu, Z.; Lei, M.; Wang, J.; Zhang, B.; Hu, Y.; Ding, J.; Zhu, B. A Phage-Like IncY Plasmid Carrying the mcr-1 Gene in Escherichia coli from a Pig Farm in China. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Pham Thanh, D.; Thanh Tuyen, H.; Nguyen Thi Nguyen, T.; Chung The, H.; Wick, R.R.; Thwaites, G.E.; Baker, S.; Holt, K.E. Inducible colistin resistance via a disrupted plasmid-borne mcr-1 gene in a 2008 Vietnamese Shigella sonnei isolate. J. Antimicrob. Chemother. 2016, 71, 2314–2317. [Google Scholar] [CrossRef] [PubMed]
- Donà, V.; Bernasconi, O.J.; Pires, J.; Collaud, A.; Overesch, G.; Ramette, A.; Perreten, V.; Endimiani, A. Heterogeneous Genetic Location of mcr-1 in Colistin-Resistant Escherichia coli Isolated from Humans and Retail Chicken Meat in Switzerland: Emergence of mcr-1 -Carrying IncK2 Plasmids. Antimicrob. Agents Chemother. 2017. [Google Scholar] [CrossRef] [PubMed]
- Seiffert, S.N.; Carattoli, A.; Schwendener, S.; Collaud, A.; Endimiani, A.; Perreten, V. Plasmids Carrying blaCMY -2/4 in Escherichia coli from Poultry, Poultry Meat, and Humans Belong to a Novel IncK Subgroup Designated IncK2. Front. Microbiol. 2017, 8, 407. [Google Scholar] [CrossRef] [PubMed]
- Zurfluh, K.; Nüesch-Inderbinen, M.; Klumpp, J.; Poirel, L.; Nordmann, P.; Stephan, R. Key features of mcr-1-bearing plasmids from Escherichia coli isolated from humans and food. Antimicrob. Resist. Infect. Control 2017, 6, 91. [Google Scholar] [CrossRef] [PubMed]
- Szabó, M.; Kiss, J.; Kótány, G.; Olasz, F. Importance of illegitimate recombination and transposition in IS30-associated excision events. Plasmid 1999, 42, 192–209. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, B.S. Homing endonucleases: Structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 2001, 29, 3757–3774. [Google Scholar] [CrossRef] [PubMed]
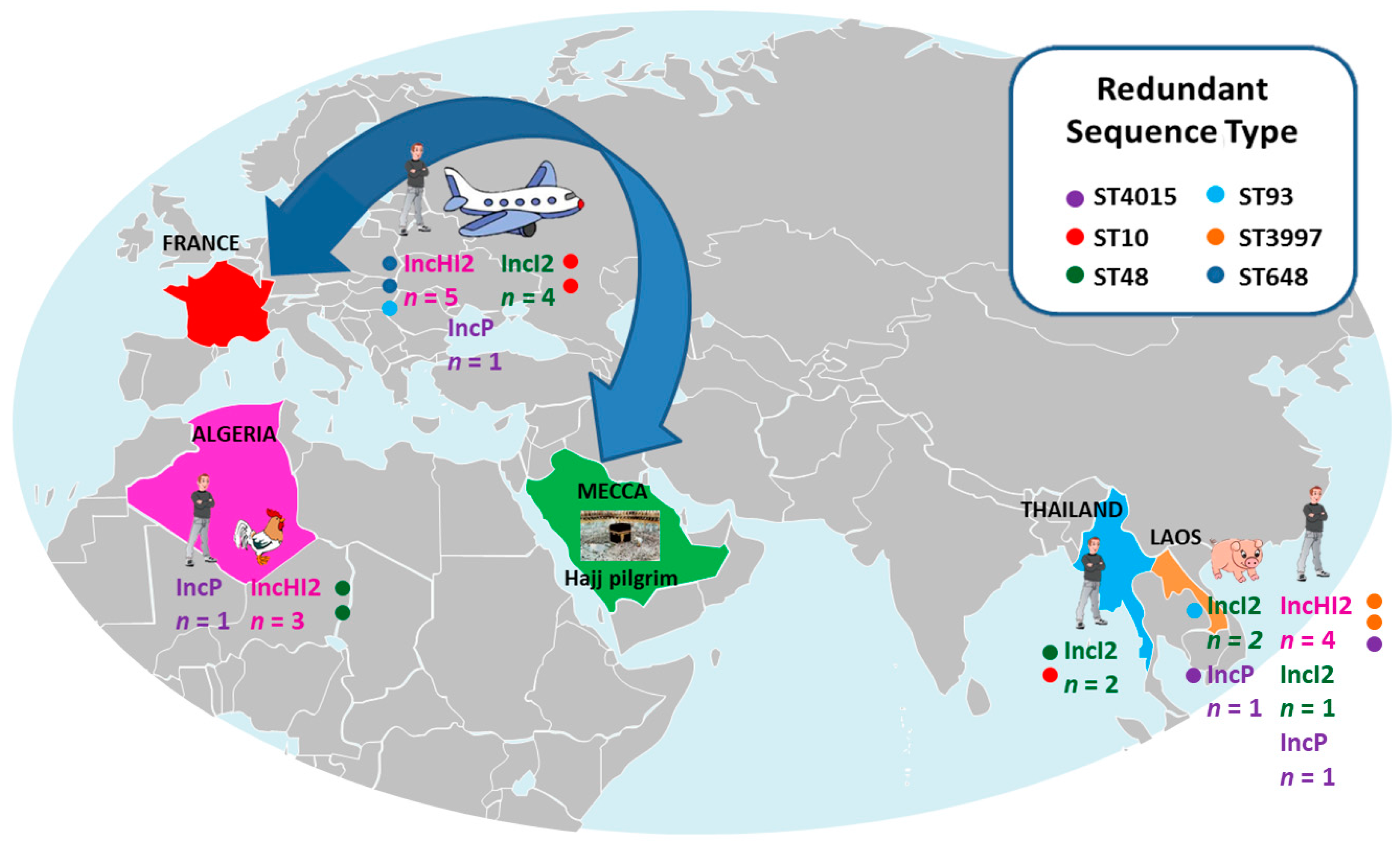
| Strains mcr-1 | Country | Origin | Other Known Colistin Mechanisms | MIC Colistin | ST | Plasmid Stability | Conjugation | Transformation | Plasmid Typing | ISApl1 |
|---|---|---|---|---|---|---|---|---|---|---|
| Escherichia coli LH1 | Laos | Human | 6 | 4015 | + | - | - | IncHI2 | downstream | |
| E. coli LH30 | Laos | Human | 6 | 4012 | −/− (0.25) | + | ND | IncHI2 | down+upstream | |
| E. coli LH57 | Laos | Human | PhoQ mut (E375K) | 8 | 3997 | + | + | ND | IncHI2 | down *+upstream * |
| E. coli LH121 | Laos | Human | 16 | 4013 | + | + | ND | IncP | / | |
| E. coli LH140 | Laos | Human | PhoQ mut (E375K) | 12 | 3997 | + | + | ND | IncHI2 | downstream |
| E. coli LH257 | Laos | Human | 12 | 4014 | + | + | ND | IncI2 | down+upstream | |
| E. coli P10 | Laos | Pig | 6 | 4015 | + | - | + | IncP | / | |
| E. coli P6 | Laos | Pig | 6 | 4704 | + | + | ND | IncI2 | / | |
| E. coli P17 | Laos | Pig | 4 | 93 | + | - | + | IncI2 | downstream | |
| E. coli TH214 | Thailand | Human | 6 | 10 | + | + | ND | IncI2 | downstream | |
| E. coli TH99 | Thailand | Human | 4 | 48 | + | + | ND | IncI2 | / | |
| E. coli SE65 | Algeria | Human | 4 | 405 | + | + | ND | IncP | downstream | |
| E. coli 235 | Algeria | Chicken | 4 | 5758 | + | + | ND | IncHI2 | downstream | |
| E. coli SA9 | Algeria | Chicken | 3 | 48 | + | + | ND | IncHI2 | downstream | |
| E. coli SE3 | Algeria | Chicken | 3 | 48 | + | + | ND | IncHI2 | downstream | |
| E. coli 1R | Traveler | Human | 4 | 453 | + | - | + | IncP | downstream | |
| E. coli 6R | Traveler | Human | 4 | 648 | + | + | - | IncHI2 | downstream | |
| E. coli 44A | Traveler | Human | 4 | 93 | + | + | ND | IncHI2 | / | |
| E. coli 85R | Traveler | Human | 4 | 656 | + | + | ND | IncI2 | down+upstream | |
| E. coli 95R | Traveler | Human | 4 | 10 | + | + | ND | IncI2 | downstream | |
| E. coli 96R | Traveler | Human | 4 | 10 | + | + | ND | IncI2 | downstream | |
| E. coli 117R | Traveler | Human | 4 | 648 | + | + | ND | IncHI2 | downstream | |
| E. coli 1RC4 | Traveler | Human | 4 | 155 | + | + | ND | IncHI2 | down+upstream | |
| E. coli 134R | Traveler | Human | 3 | 602 | + | + | ND | IncI2 | downstream | |
| E. coli 143R | Traveler | Human | 3 | 1300 | + | + | ND | IncHI2 | downstream | |
| Klebsiella pneumoniae 119R | Traveler | Human | 3 | 788 | + | + | ND | IncI2 | downstream | |
| K. pneumoniae LH131 | Laos | Human | MgrB (stop) | 32 | 1319 | −/+ (32) | - | - | ND | / |
| K. pneumoniae LH17 | Laos | Human | PmrB mut (T157P) | 12 | 37 | −/+ (12) | - | - | ND | / |
| K. pneumoniae LH61 | Laos | Human | MgrB (substitution) | 16 | 491 | −/+ (16) | + | ND | IncI2 | / |
| K. pneumoniae LH92 | Laos | Human | 12 | 39 | −/− (12) | - | - | ND | / | |
| K. pneumoniae FHM128 | France | Human | 4 | 1310 | + | - | - | ND | downstream | |
| K. pneumoniae FHA60 | France | Human | 8 | 1307 | + | - | - | ND | downstream |
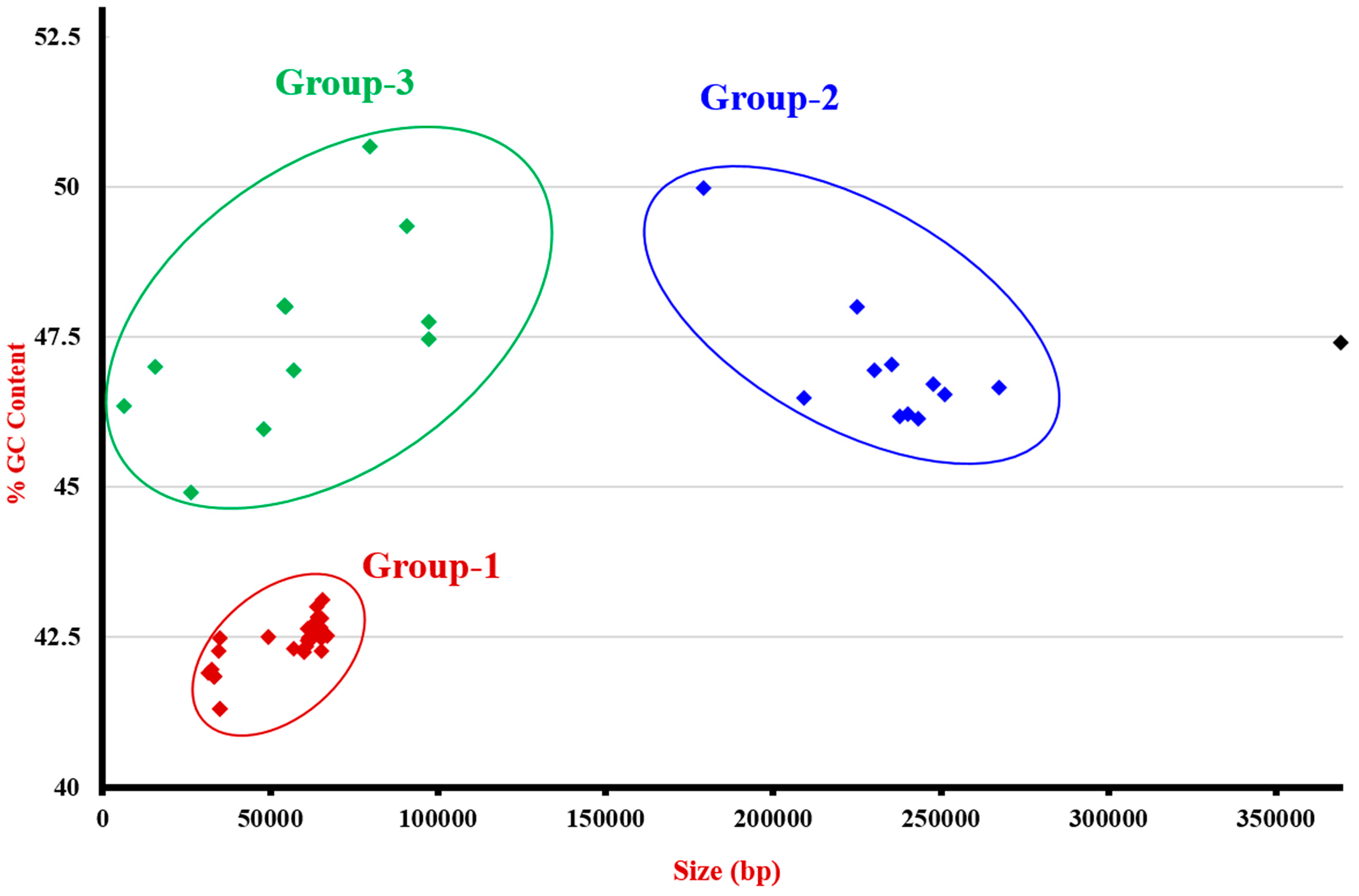
| Genbank Accession Number | Type of Plasmid | GC% | Size (bp) | Strain mcr-1 | Country | Origin | ISApl1 Presence in Plasmid | Reference |
|---|---|---|---|---|---|---|---|---|
| CP015913.1 | IncI2 | 43.11 | 65,888 | Escherichia coli | USA | Animal | Not present | [39] |
| CP015977.1 | IncX4 | 41.85 | 33,304 | E. coli | Brazil | Human | Not present | [40] |
| CP016183.1 | IncHI1B | 46.93 | 230,278 | E. coli | Malaysia | Animal | Downstream mcr-1 | [41] |
| CP016184.1 | IncHI1B | 47.04 | 235,403 | E. coli | Malaysia | Animal | Downstream mcr-1 + 1 other copy | [41] |
| CP016185.1 | IncI2 | 42.48 | 61,735 | E. coli | Malaysia | Animal | Not present | [41] |
| CP016186.1 | IncI2 | 42.25 | 60,218 | E. coli | Malaysia | Environment | Not present | [41] |
| CP016187.1 | IncI2 | 42.35 | 60,950 | E. coli | Malaysia | Animal | Not present | [41] |
| CP016405.1 | IncI2 | 42.65 | 63,329 | E. coli | USA | Animal | Not present | [42] |
| CP016550.1 | IncX4 | 42.5 | 49,695 | E. coli | Netherlands | Human | Not present | [39] |
| CP017246.1 | IncX4 | 42.48 | 34,992 | E. coli | Brazil | Animal | Not present | [43] |
| CP017632.1 | IncN | 47.4 | 369,298 | E. coli | China | Human | Tn6330 + 2 other copies | [44] |
| CP018106.1 | IncI2 | 42.82 | 64,467 | E. coli | Germany | Human | Downstream mcr-1 | [45] |
| CP018112.1 | IncI2 | 42.82 | 64,467 | E. coli | USA | Human | Downstream mcr-1 | [45] |
| CP018118.1 | IncI2 | 42.82 | 64,467 | E. coli | USA | Human | Downstream mcr-1 | [45] |
| CP018124.1 | IncI2 | 42.8 | 65,539 | E. coli | USA | Human | Downstream mcr-1 + 1 other copy | [45] |
| CP018773.1 | IncX4 | 41.84 | 33,305 | E. coli | USA | Human | Not present | [46] |
| KP347127.1 | IncI2 | 43 | 64,015 | E. coli | China | Animal | Downstream mcr-1 | [6] |
| KU341381.1 | IncHI2 | 46.53 | 251,493 | E. coli | China | Animal | Downstream mcr-1 | [6] |
| KU353730.1 | IncFII | 50.67 | 79,798 | E. coli | Belgium | Animal | Not present | [47] |
| KU647721.2 | IncX4 | 45.95 | 48,350 | E. coli | Unknown | Animal | Not present | [48] |
| KU743383.1 | IncX4 | 41.85 | 33,311 | E. coli | Estonia | Animal | Not present | [49] |
| KU743384.1 | IncHI2 | 46.21 | 240,367 | E. coli | Saudi Arabia | Human | Tn6330 + 1 other copy | [50] |
| KU761326.1 | IncI2 | 42.65 | 64,964 | E. coli | China | Human | Not present | [51] |
| KU761327.1 | IncX4 | 41.84 | 33,287 | Klebsiella pneumoniae | China | Human | Not present | [51] |
| KU870627.1 | IncI2 | 42.46 | 62,219 | E. coli | South Africa | Animal | Downstream mcr-1 | [52] |
| KU922754.1 | IncI2 | 42.3 | 57,059 | Kluyvera ascorbata | China | Environment | Not present | [53] |
| KU934209.1 | IncI2 | 42.26 | 65,419 | Salmonella enterica | China | Animal | Downstream mcr-1 | [54] |
| KU994859.1 | IncFII | 49.34 | 91,041 | E. coli | Belgium | Animal | Downstream mcr-1 + 1 other partial copy | [55] |
| KX013538.1 | IncI2 | 42.43 | 61,228 | E. coli | United Arab Emirates | Human | Downstream mcr-1 | [50] |
| KX013539.1 | IncI2 | 42.69 | 62,661 | E. coli | Bahrain | Human | Not present | [50] |
| KX013540.1 | IncI2 | 42.49 | 64,942 | E. coli | Bahrain | Human | Not present | [50] |
| KX032519.1 | IncI2 | 42.63 | 61,177 | E. coli | South Africa | Human | Downstream mcr-1 | [56] |
| KX032520.1 | IncX4 | 41.9 | 31,808 | E. coli | South Africa | Human | Not present | [56] |
| KX034083.1 | IncI2 | 42.51 | 67,134 | E. coli | China | Animal | Downstream mcr-1 | [57] |
| KX084392.1 | IncX4 | 41.85 | 33,298 | E. coli | China | Animal | Not present | [58] |
| KX084393.1 | IncI2 | 42.64 | 63,656 | E. coli | China | Animal | Not present | [58] |
| KX084394.1 | IncHI2 | 46.13 | 243,572 | E. coli | China | Animal | Tn6330 | [58] |
| KX129782.1 | IncHI2 | 46.7 | 247,885 | E. coli | Italy | Food | Tn6330 | [59] |
| KX129783.1 | IncX4 | 42.27 | 34,640 | E. coli | Switzerland | Environment | Not present | [59] |
| KX129784.1 | IncHI1B | 46.48 | 209,401 | E. coli | Thailand | Food | Not present | [59] |
| KX236309.1 | IncX4 | 41.85 | 33,303 | K. pneumoniae | Italy | Human | Not present | [60] |
| KX254341.1 | IncHI2 | 46.64 | 267,486 | E. coli | China | Animal | Not present | [58] |
| KX254342.1 | IncI2 | 42.64 | 63,656 | E. coli | China | Animal | Downstream mcr-1 | [58] |
| KX254343.1 | IncX4 | 41.84 | 33,307 | E. coli | China | Animal | Not present | [58] |
| KX257480 | IncFII | 48.01 | 54,502 | S. enterica | Unknown | Animal | Incomplete downstream mcr-1 | [61] |
| KX257481 | IncFII | 47.99 | 54,670 | S. enterica | Unknown | Animal | Incomplete downstream mcr-1 | [61] |
| KX257482 | IncFII | 48.02 | 54,494 | S. enterica | Unknown | Animal | Not present | [61] |
| KX276657.1 | IncN | 47.99 | 225,069 | E. coli | USA | Human | Downstream mcr-1 + 1 other copy | [62] |
| KX377410.1 | IncFIB | 46.94 | 57,278 | K. pneumoniae | China | Environment | Downstream mcr-1 + 1 other copy | [63] |
| KX447768.1 | IncX4 | 41.84 | 33,395 | E. coli | USA | Human | Not present | [64] |
| KX505142.1 | IncI2 | 42.64 | 65,203 | Cronobacter sakazakii | China | Animal | Downstream mcr-1 | [31] |
| KX518745.1 | IncY | 47.46 | 97,559 | E. coli | China | Animal | Tn6630 | [65] |
| KX528699.1 | IncFIB | 46.99 | 15,998 | E. coli | Vietnam | Animal | Downstream mcr-1 + 1 other copy | [66] |
| KX570748.1 | IncX4 | 41.96 | 32,751 | E. coli | China | Animal | Not present | [67] |
| KX772391.1 | IncFIB | 49.97 | 179,444 | E. coli | China | Human | Tn6330 | [68] |
| KX772777.1 | IncX4 | 41.84 | 33,309 | E. coli | China | Human | Not present | [69] |
| KX772778.1 | IncI2 | 42.47 | 65,375 | E. coli | China | Human | Not present | [69] |
| KX880944.1 | IncY | 47.75 | 97,386 | E. coli | China | Animal | Tn6630 | [70] |
| LT174530 | IncI2 | 42.5 | 61,826 | Shigella sonnei | Vietnam | Human | Downstream mcr-1 | [71] |
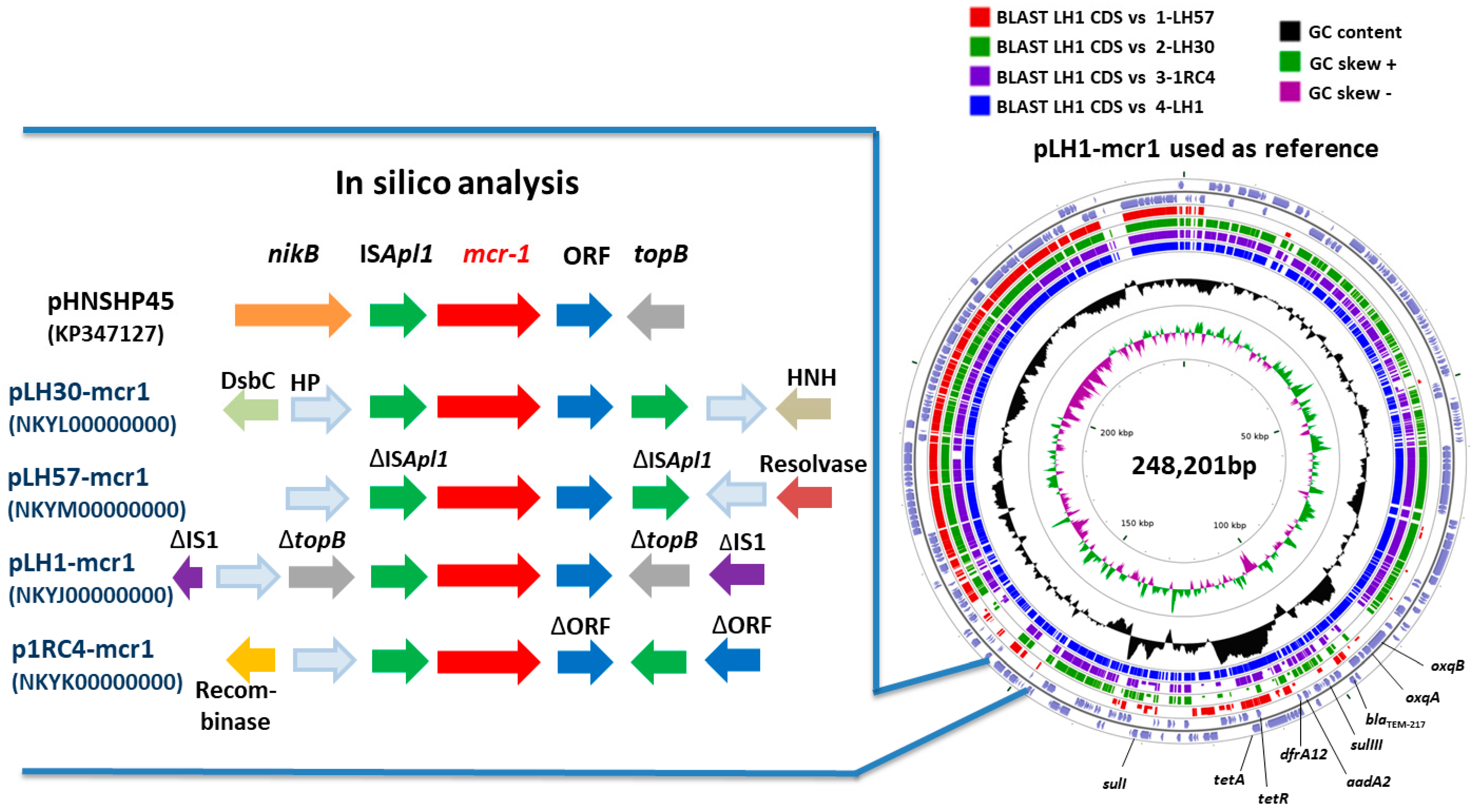
| mcr-1 Plasmid | Type of Plasmid | pMLST | GC% | Size (bp) | Resistance Genes | ISApl1 Presence in Plasmid | Genbank Accession Number |
|---|---|---|---|---|---|---|---|
| pLH30-mcr1 | IncHI2 | ST 3 | 45.91 | 223,898 | mcr-1, blaTEM-217, cmlA1, floR, aph3-Ia, aadA2, sulIII, dfrA17, mefB | 2 sequences: 1 downstream and 1 upstream mcr-1 gene | NKYL00000000 |
| pLH57-mcr1 | IncHI2 | Unknown ST | 48.00 | 218,800 | mcr-1, blaTEM-217, tetA, tetR, strA, strB, aph3’’-Ib, aph6-Id, sulII, dfrA14 | 2 truncated sequences: 1 downstream and 1 upstream mcr-1 gene | NKYM00000000 |
| pLH1-mcr1 | IncHI2 | ST 3 | 46.38 | 248,201 | mcr-1, blaTEM-217, tetA, tetR, aadA2, oqxA, oqxBgb, sulI, sulIII, dfrA12 | 2 sequences: 1 downstream mcr-1 gene and 1 inside traE gene | NKYJ00000000 |
| p1RC4-mcr1 | IncHI2 | ST 4 | 46.22 | 239,098 | mcr-1, blaTEM-217, tetA, tetR, floR, aph3-Ia, aph3-Ib, aadA1, strA, sulIII, dfrA14 | 2 sequences: 1 downstream and 1 upstream mcr-1 gene | NKYK00000000 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hadjadj, L.; Riziki, T.; Zhu, Y.; Li, J.; Diene, S.M.; Rolain, J.-M. Study of mcr-1 Gene-Mediated Colistin Resistance in Enterobacteriaceae Isolated from Humans and Animals in Different Countries. Genes 2017, 8, 394. https://doi.org/10.3390/genes8120394
Hadjadj L, Riziki T, Zhu Y, Li J, Diene SM, Rolain J-M. Study of mcr-1 Gene-Mediated Colistin Resistance in Enterobacteriaceae Isolated from Humans and Animals in Different Countries. Genes. 2017; 8(12):394. https://doi.org/10.3390/genes8120394
Chicago/Turabian StyleHadjadj, Linda, Toilhata Riziki, Yan Zhu, Jian Li, Seydina M. Diene, and Jean-Marc Rolain. 2017. "Study of mcr-1 Gene-Mediated Colistin Resistance in Enterobacteriaceae Isolated from Humans and Animals in Different Countries" Genes 8, no. 12: 394. https://doi.org/10.3390/genes8120394







