Genetic Analysis of the Major Capsid Protein of the Archaeal Fusellovirus SSV1: Mutational Flexibility and Conformational Change
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Cell Growth
2.2. Purification of Template DNA for Long Inverse PCR from E. coli
2.3. Long Inverse PCR
2.4. Complementation in cis of SSV1 Mutants
2.5. Transformation of Sulfolobus
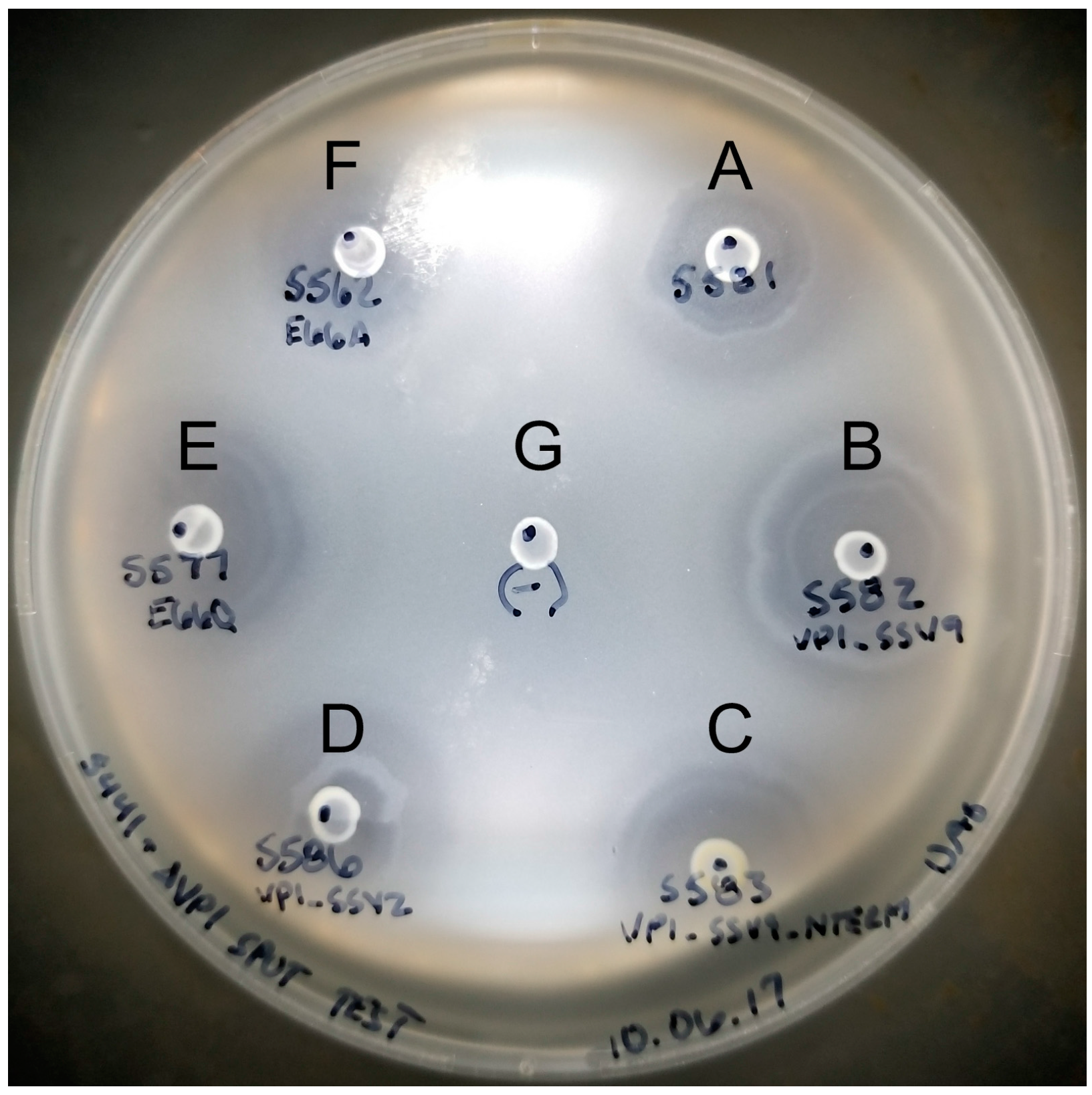
2.6. Halo Assay to Check for Infectious Virus Production
2.7. Confirmation of Infectious SSV DNA
2.8. Transmission Electron Microscopy
2.9. Particle Analysis
3. Results
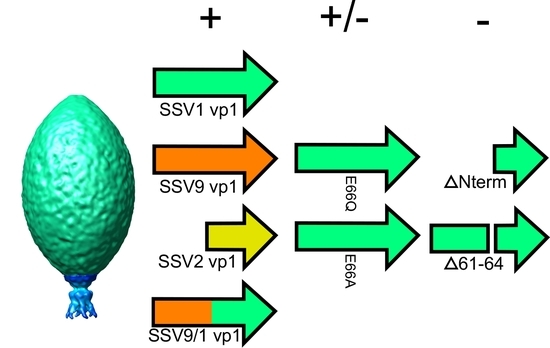
3.1. The N-Terminus of the SSV1 Major Capsid Protein VP1 Is Essential for Infectivity
3.2. Complementation in cis of SSV1 Deletion Mutants
3.3. Changing the Conserved Glutamate in the Major Capsid Gene VP1 Allows Infectious Virus but Generates Many Abnormal Virions
4. Discussion
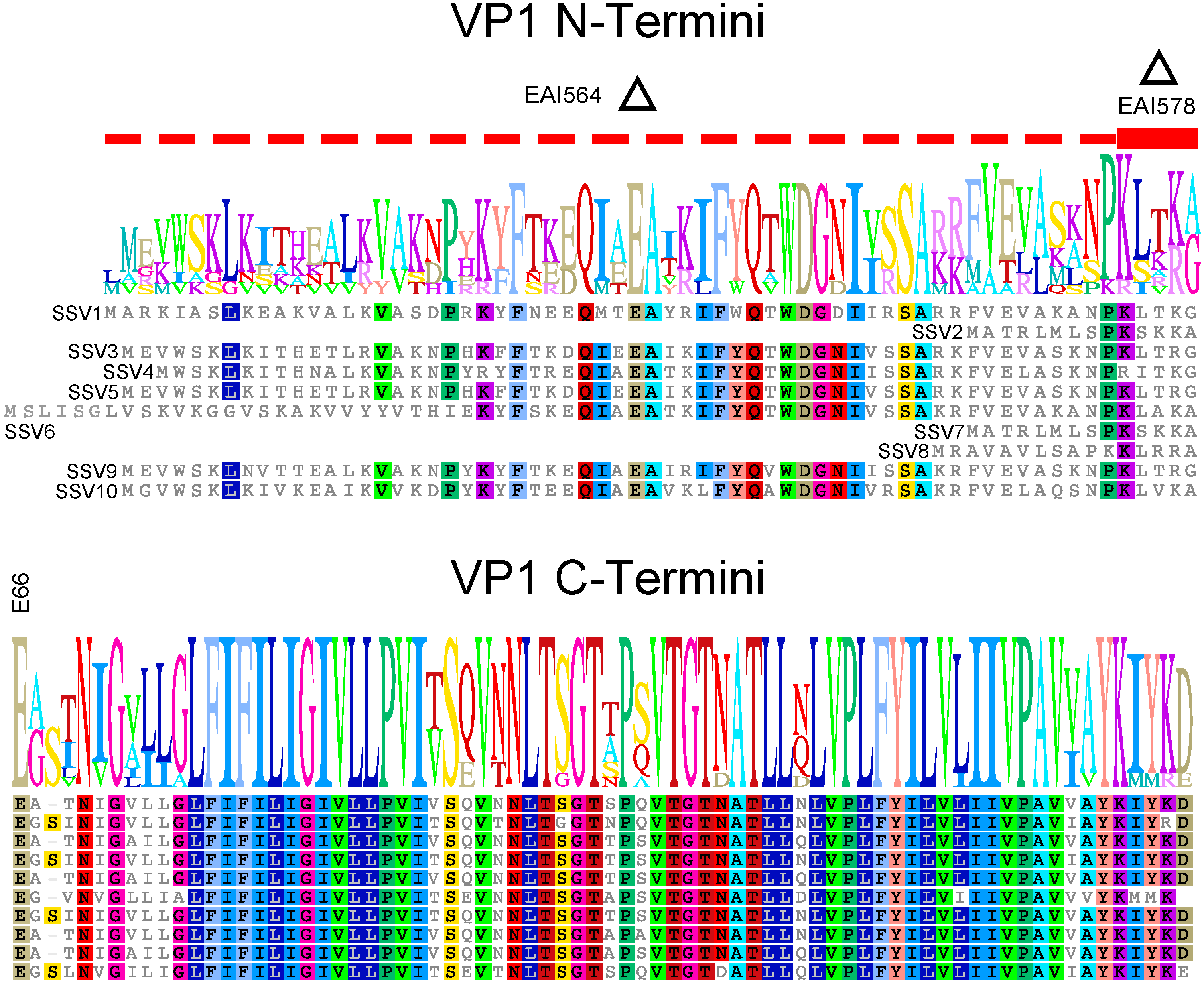
4.1. The Translational Start of the VP1 Protein Is Not Clear
4.2. The N-Terminus of SSV1 VP1 Is Essential, But the Cleavage Site Is Not
4.3. Virions Containing Point Mutations in VP1 Have Unusual Morphology
4.4. The SSV1 VP1 N-Terminus Can Be Functionally Replaced with the SSV9 VP1 N-Terminus
4.5. Proteolysis and Assembly of SSV1
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Krupovic, M.; Quemin, E.R.J.; Bamford, D.H.; Forterre, P.; Prangishvili, D. Unification of the globally distributed spindle-shaped viruses of the Archaea. J. Virol. 2014, 88, 2354–2358. [Google Scholar] [CrossRef] [PubMed]
- Prangishvili, D. The wonderful world of archaeal viruses. Annu. Rev. Microbiol. 2013, 67, 565–585. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.; Yeats, S.; Janekovic, D.; Reiter, W.D.; Aicher, W.; Zillig, W. SAV-1, A Temperate UV-Inducible DNA Virus-like Particle from the Archaebacterium Sulfolobus acidocaldarius Isolate B-12. EMBO J. 1984, 3, 2165–2168. [Google Scholar] [PubMed]
- Yeats, S.; McWilliam, P.; Zillig, W. A plasmid in the archaebacterium Sulfolobus acidocaldarius. EMBO J. 1982, 1, 1035–1038. [Google Scholar] [PubMed]
- Palm, P.; Schleper, C.; Grampp, B.; Yeats, S.; McWilliam, P.; Reiter, W.D.; Zillig, W. Complete nucleotide sequence of the virus SSV1 of the archaebacterium Sulfolobus shibatae. Virology 1991, 185, 242–250. [Google Scholar] [CrossRef]
- Nadal, M.; Mirambeau, G.; Forterre, P.; Reiter, W.D.; Duguet, M. Positively supercoiled DNA in a virus-like particle of an archaebacterium. Nature 1986, 321, 256–258. [Google Scholar] [CrossRef]
- Schleper, C.; Kubo, K.; Zillig, W. The particle SSV1 from the extremely thermophilic archaeon Sulfolobus is a virus-Demonstration of infectivity and of transfection with viral-DNA. Proc. Natl. Acad. Sci. USA 1992, 89, 7645–7649. [Google Scholar] [CrossRef] [PubMed]
- Ceballos, R.M.; Marceau, C.D.; Marceau, J.O.; Morris, S.; Clore, A.J.; Stedman, K.M. Differential virus host-ranges of the Fuselloviridae of hyperthermophilic Archaea: Implications for evolution in extreme environments. Front. Microbiol. 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Muskhelishvili, G.; Palm, P.; Zillig, W. SSV1-encoded site-specific recombination system in Sulfolobus shibatae. Mol. Gen. Genet. 1993, 237, 334–342. [Google Scholar] [PubMed]
- Quemin, E.R.J.; Chlanda, P.; Sachse, M.; Forterre, P.; Prangishvili, D.; Krupovic, M. Eukaryotic-like virus budding in Archaea. mBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Reiter, W.D.; Palm, P.; Henschen, A.; Lottspeich, F.; Zillig, W.; Grampp, B. Identification and characterization of the genes encoding 3-structural proteins of the Sulfolobus virus-like particle SSV1. Mol. Gen. Genet. 1987, 206, 144–153. [Google Scholar] [CrossRef]
- Quemin, E.R.J.; Pietila, M.K.; Oksanen, H.M.; Forterre, P.; Rijpstra, W.I.C.; Schouten, S.; Bamford, D.H.; Prangishvili, D.; Krupovic, M. Sulfolobus spindle-shaped Virus 1 contains glycosylated capsid proteins, a cellular chromatin protein, and host-derived lipids. J. Virol. 2015, 89, 11681–11691. [Google Scholar] [CrossRef] [PubMed]
- Stedman, K.M.; DeYoung, M.; Saha, M.; Sherman, M.B.; Morais, M.C. Structural insights into the architecture of the hyperthermophilic Fusellovirus SSV1. Virology 2015, 474, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Perilla, J.R.; Yufenyuy, E.L.; Meng, X.; Chen, B.; Ning, J.; Ahn, J.; Gronenborn, A.M.; Schulten, K.; Aiken, C.; et al. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature 2013, 497, 643–646. [Google Scholar] [CrossRef] [PubMed]
- Iverson, E.A.; Goodman, D.A.; Gorchels, M.E.; Stedman, K.M. Extreme Mutation Tolerance: Nearly Half of the Archaeal Fusellovirus Sulfolobus Spindle-Shaped Virus 1 Genes Are Not Required for Virus Function, Including the Minor Capsid Protein Gene vp3. J. Virol. 2017, 91, e02406-16. [Google Scholar] [CrossRef] [PubMed]
- Yost, S.A.; Marcotrigiano, J. Viral precursor polyproteins: Keys of regulation from replication to maturation. Curr. Opin. Virol. 2013, 3, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Bertram, S.; Glowacka, I.; Steffen, I.; Kühl, A.; Pöhlmann, S. Novel insights into proteolytic cleavage of influenza virus hemagglutinin. Rev. Med. Virol. 2010, 20, 298–310. [Google Scholar] [CrossRef] [PubMed]
- Clore, A.J.; Stedman, K.M. The SSV1 viral integrase is not essential. Virology 2007, 361, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Iverson, E.; Stedman, K. A genetic study of SSV1 the prototypical fusellovirus. Front. Microbiol. 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Zillig, W.; Kletzin, A.; Schleper, C.; Holz, I.; Janekovic, D.; Hain, J.; Lanzendorfer, M.; Kristjansson, J.K. Screening for Sulfolobales, their Plasmids and their Viruses in Icelandic Solfataras. Syst. Appl. Microbiol. 1994, 16, 609–628. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2012; ISBN 978-1-936113-41-5. [Google Scholar]
- Bertani, G. Lysogeny at Mid-Twentieth Century: P1, P2, and Other Experimental Systems. J. Bacteriol. 2004, 186, 595–600. [Google Scholar] [CrossRef] [PubMed]
- Birnboim, H.C.; Doly, J. Rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979, 7, 1513–1523. [Google Scholar] [CrossRef] [PubMed]
- Arnold, H.P.; She, Q.; Phan, H.; Stedman, K.; Prangishvili, D.; Holz, I.; Kristjansson, J.K.; Garrett, R.; Zillig, W. The genetic element pSSVx of the extremely thermophilic crenarchaeon Sulfolobus is a hybrid between a plasmid and a virus. Mol. Microbiol. 1999, 34, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Wiedenheft, B.; Stedman, K.; Roberto, F.; Willits, D.; Gleske, A.K.; Zoeller, L.; Snyder, J.; Douglas, T.; Young, M. Comparative genomic analysis of hyperthermophilic archaeal Fuselloviridae viruses. J. Virol. 2004, 78, 1954–1961. [Google Scholar] [CrossRef] [PubMed]
- Stedman, K.M.; She, Q.X.; Phan, H.; Arnold, H.P.; Holz, I.; Garrett, R.A.; Zillig, W. Relationships between fuselloviruses infecting the extremely thermophilic archaeon Sulfolobus: SSV1 and SSV2. Res. Microbiol. 2003, 154, 295–302. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Belinky, F.; Rogozin, I.B.; Koonin, E.V. Selection on start codons in prokaryotes and potential compensatory nucleotide substitutions. Sci. Rep. 2017, 7, 12422. [Google Scholar] [CrossRef] [PubMed]
- Condò, I.; Ciammaruconi, A.; Benelli, D.; Ruggero, D.; Londei, P. Cis-acting signals controlling translational initiation in the thermophilic archaeon Sulfolobus solfataricus. Mol. Microbiol. 1999, 34, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Torarinsson, E.; Klenk, H.-P.; Garrett, R.A. Divergent transcriptional and translational signals in Archaea. Environ. Microbiol. 2005, 7, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, W.G.; Semler, B.L. Expression of virus-encoded proteinases: Functional and structural similarities with cellular enzymes. Microbiol. Rev. 1993, 57, 781–822. [Google Scholar] [PubMed]
- Guagliardi, A.; Cerchia, L.; Rossi, M. An intracellular protease of the crenarchaeon Sulfolobus solfataricus, which has sequence similarity to eukaryotic peptidases of the CD clan. Biochem. J. 2002, 368, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Tang, J. Purification, characterization, and gene cloning of thermopsin, a thermostable acid protease from Sulfolobus acidocaldarius. J. Biol. Chem. 1990, 265, 1490–1495. [Google Scholar] [PubMed]

| Plasmid | Description | Infectivity | Reference |
|---|---|---|---|
| pAJC97 | SSV1 shuttle vector (TOPO PCR Blunt II inserted into ORF A e178 at bp 3173) | + | [18] |
| EAI283 | SSV1::Tn5 mutant, EZ-Tn5 inserted at bp 3572 (ORF e178) | + | [15] |
| EAI427 | pAJC97 background with E66Q mutation in VP1 | + | This Work |
| EAI500 | pAJC97 background with E66A mutation in VP1 | + | This Work |
| EAI551 | EAI283 background with SSV1-VP1 complementation | + | This work |
| EAI566 | EAI283 background with SSV2-VP1 complementation | + | This Work |
| EAI553 | EAI283 background with SSV9-VP1 complementation | + | This Work |
| EAI557 | EAI283 background with N-terminus of SSV9-VP1 complementation | + | This Work |
| EAI237 | pAJC97 background with N-terminus of VP1 deleted | − | This Work |
| EAI564 | EAI283 background with N-terminus of VP1 deleted | − | This Work |
| EAI578 | EAI283 background with residues 61–65 deleted from VP1 | − | This Work |
| Product | Forward Primer | Reverse Primer |
|---|---|---|
| SSV1 VP1 deletion | TGA GGG ATG GAA ATC AGT TTA AAG | CAA ACT CCT TAG GAG TCT CAT CC |
| SSV1 VP1 N-terminus deletion | ATG GAA GCA ACC AAC ATA GG (61) and GAA GCA ACC AAC ATA GG (54) | CAA ACT CCT TAG GAG TCT CAT CC |
| SSV1 VP1 aa61–65 deletion | GAA GCA ACC AAC ATA GGC | GGG GTT TGC CTT TGC TAC |
| SSV1 VP1 point mutant (E66A) A | GCA GCA ACC AAC ATA GGC | ACC TTT TGT GAG CTT GGG G |
| SSV1 VP1 point mutant (E66Q) B | CAA GCA ACC AAC ATA GGC | ACC TTT TGT GAG CTT GGG G |
| SSV1 VP1 C | GCcAGAAAGATAGCCTCAC | ACCTTTTGTGAGCTTGGG |
| SSV9 VP1 | GAAGTTTGGTCAAAGTTAAACG | ATCTTTGTAGATTTTATACG |
| SSV2 VP1 | GCCACCAGACTAATGCTAAGC | GTCACGATATATCTTATACGCTATGAC |
| SSV9 VP1 N-terminus | GAAGTTTGGTCAAAGTTAAACG | ACCCCTAGTAAGTTTGGG |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iverson, E.A.; Goodman, D.A.; Gorchels, M.E.; Stedman, K.M. Genetic Analysis of the Major Capsid Protein of the Archaeal Fusellovirus SSV1: Mutational Flexibility and Conformational Change. Genes 2017, 8, 373. https://doi.org/10.3390/genes8120373
Iverson EA, Goodman DA, Gorchels ME, Stedman KM. Genetic Analysis of the Major Capsid Protein of the Archaeal Fusellovirus SSV1: Mutational Flexibility and Conformational Change. Genes. 2017; 8(12):373. https://doi.org/10.3390/genes8120373
Chicago/Turabian StyleIverson, Eric A., David A. Goodman, Madeline E. Gorchels, and Kenneth M. Stedman. 2017. "Genetic Analysis of the Major Capsid Protein of the Archaeal Fusellovirus SSV1: Mutational Flexibility and Conformational Change" Genes 8, no. 12: 373. https://doi.org/10.3390/genes8120373
APA StyleIverson, E. A., Goodman, D. A., Gorchels, M. E., & Stedman, K. M. (2017). Genetic Analysis of the Major Capsid Protein of the Archaeal Fusellovirus SSV1: Mutational Flexibility and Conformational Change. Genes, 8(12), 373. https://doi.org/10.3390/genes8120373