Replication Termination: Containing Fork Fusion-Mediated Pathologies in Escherichia coli
Abstract
:1. Introduction
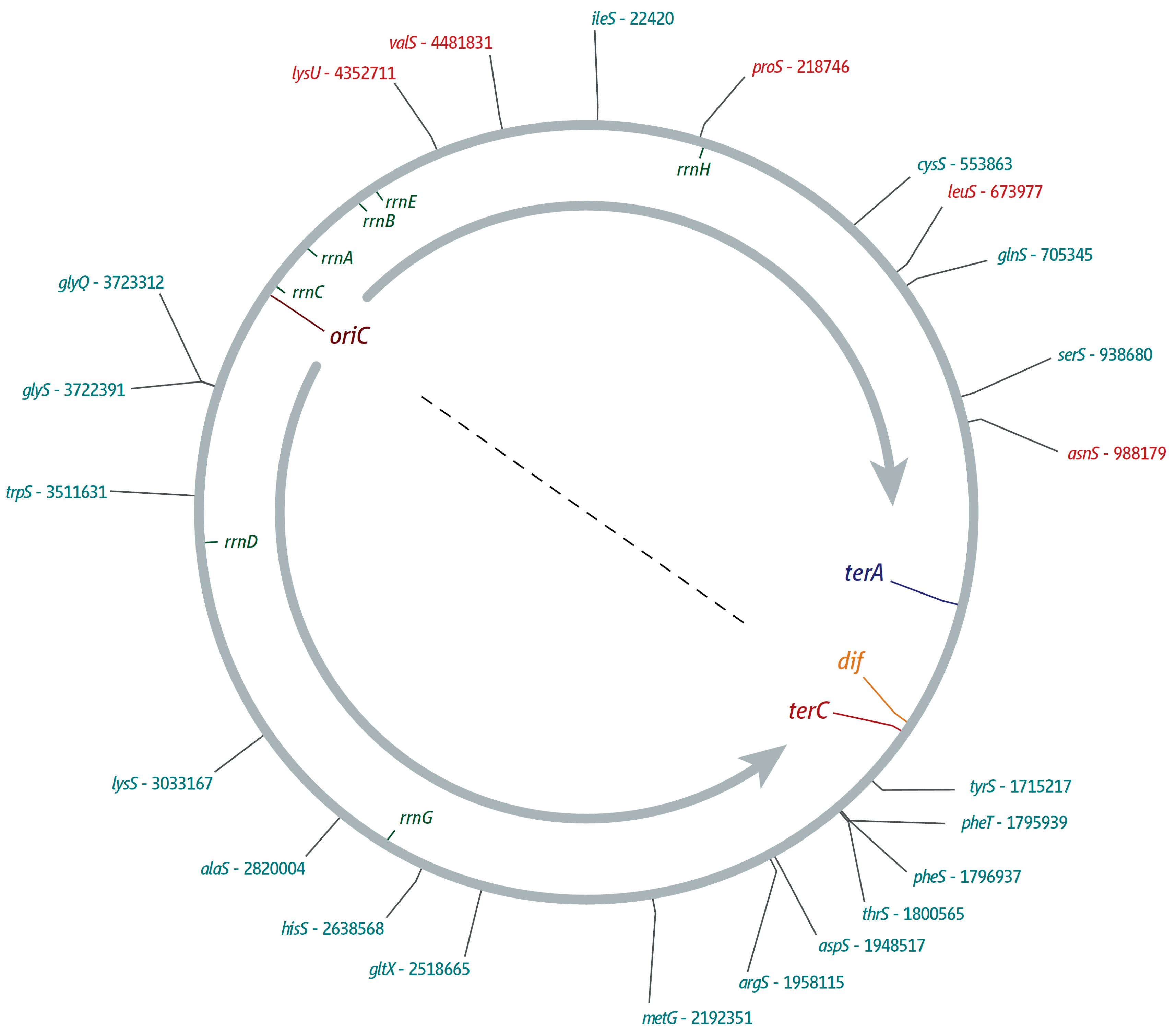
2. ter/Tus Complexes Block Replication Forks with High Efficiency
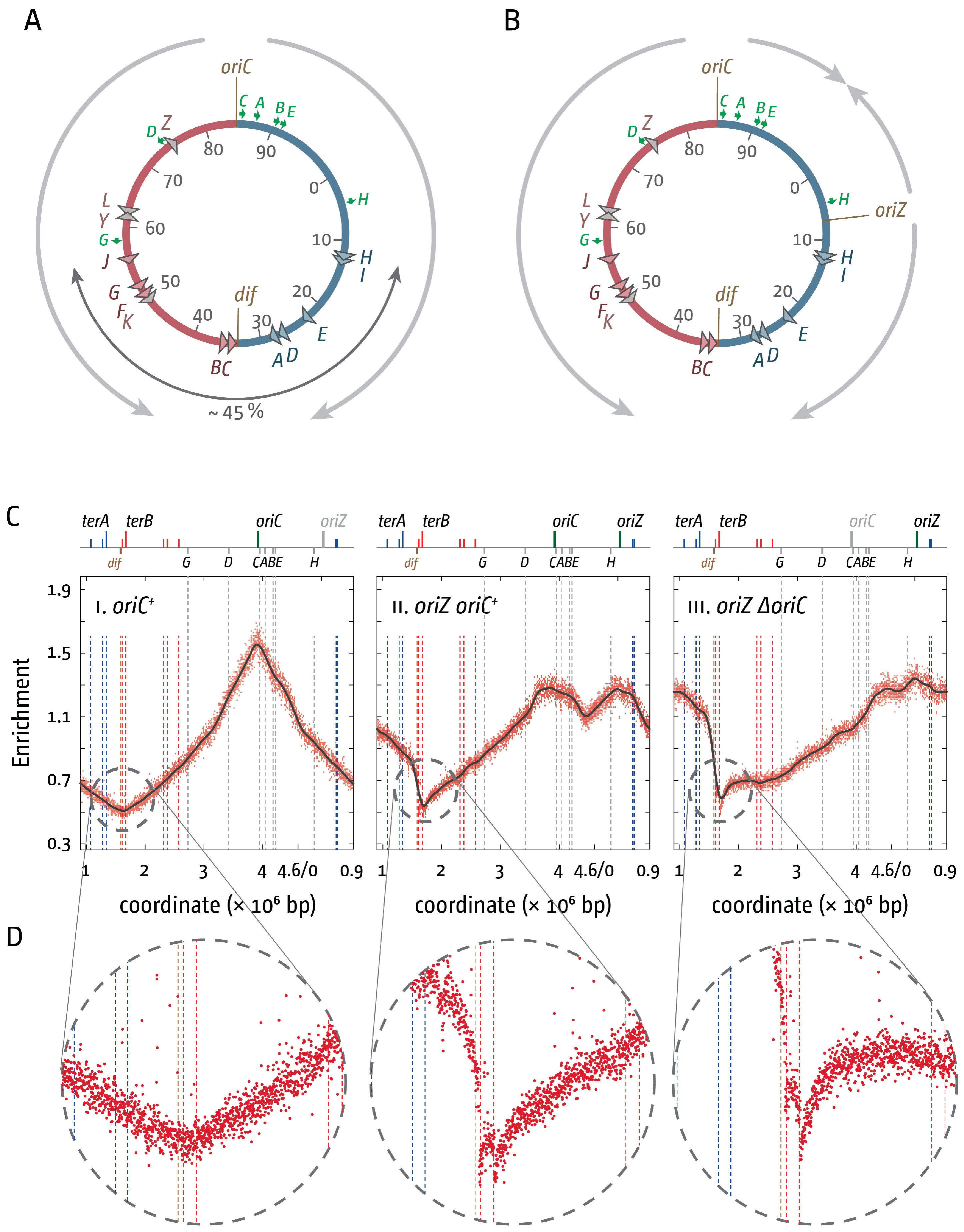
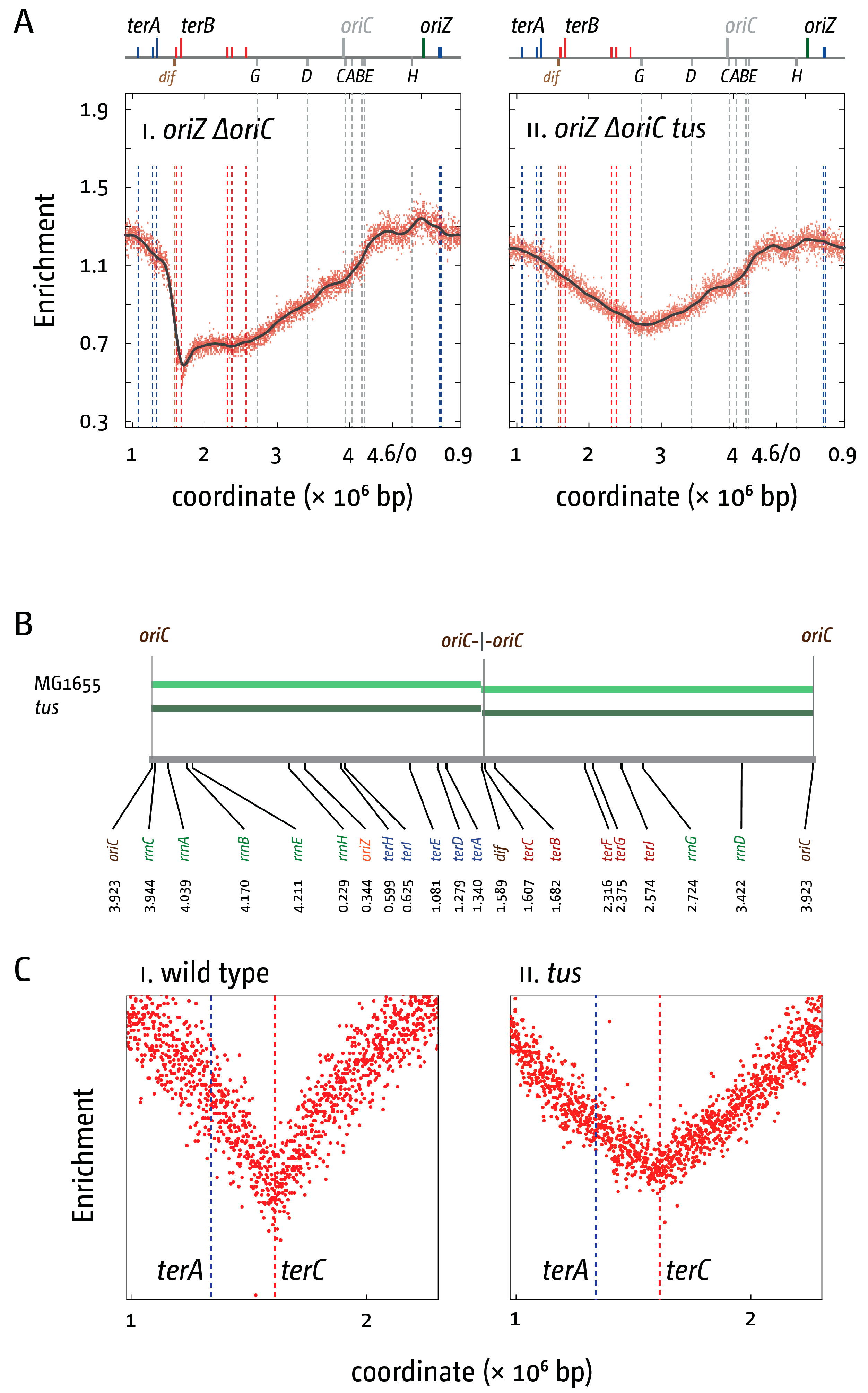
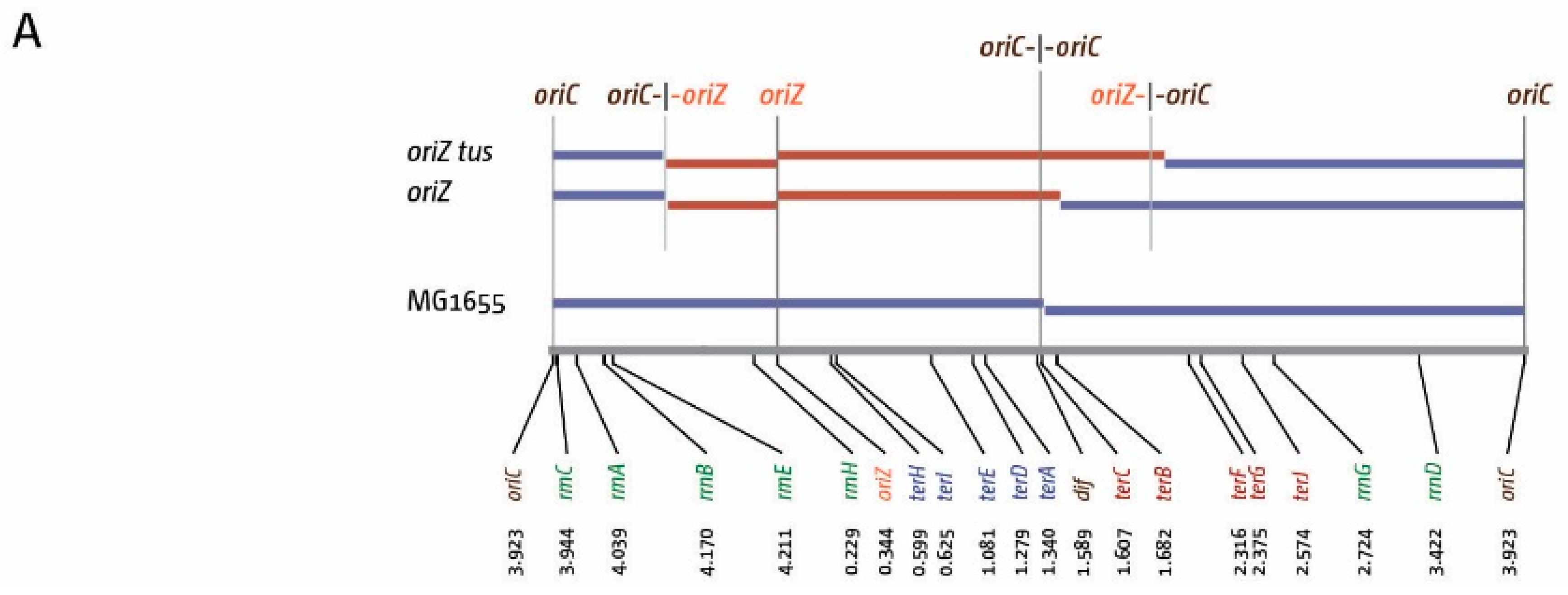
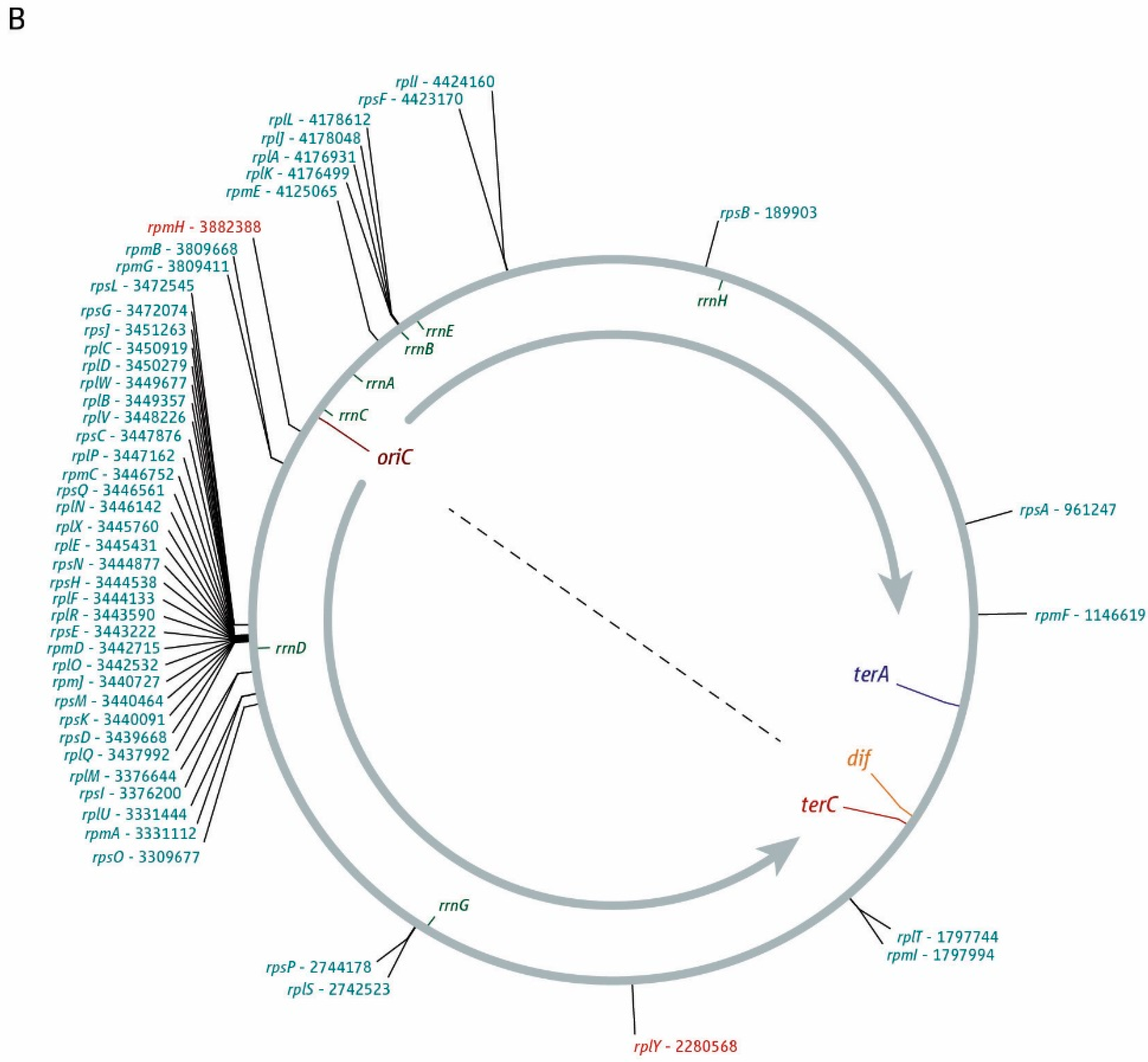
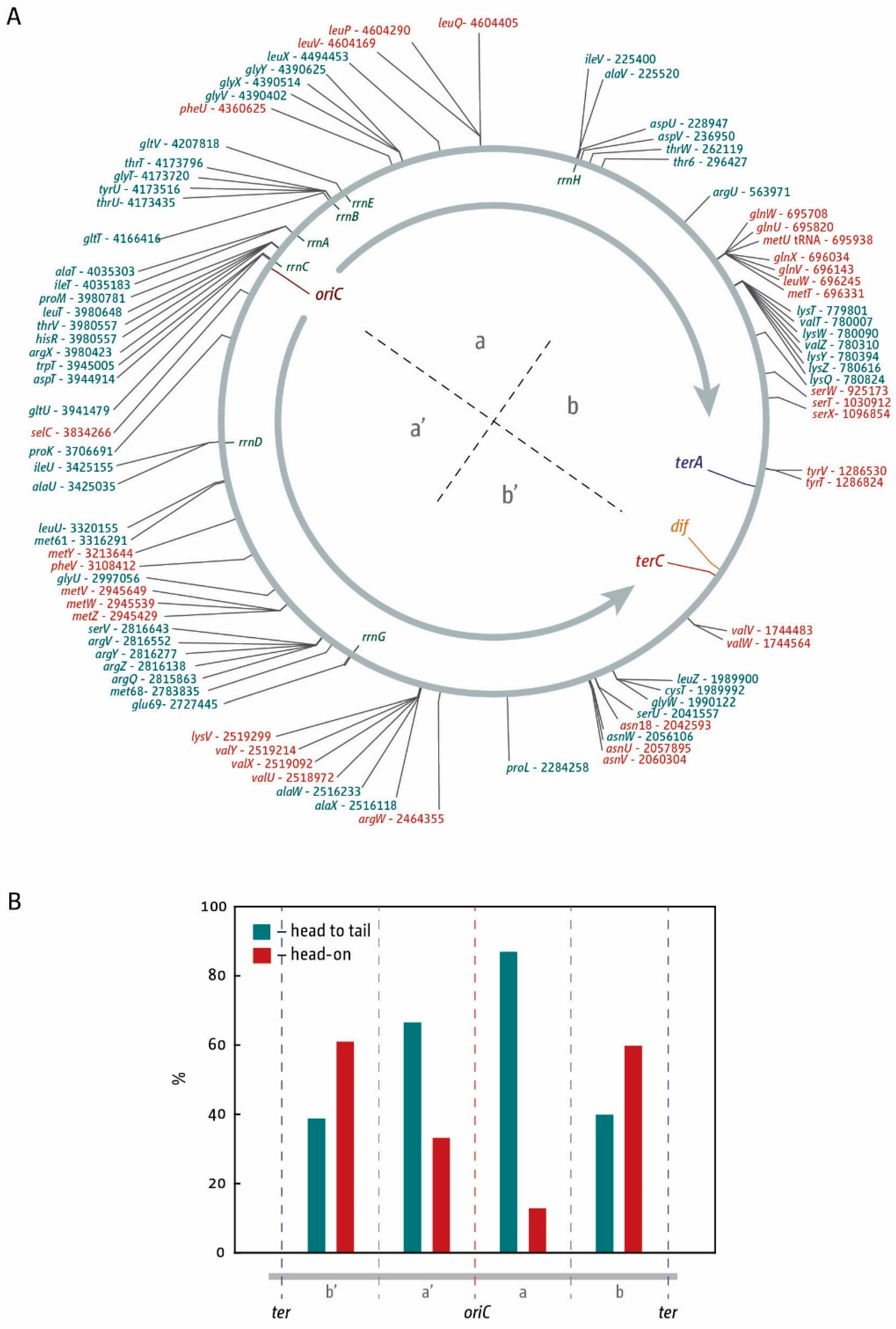
3. The Location of Fork Fusion Events
4. Coordinating Replication and Transcription
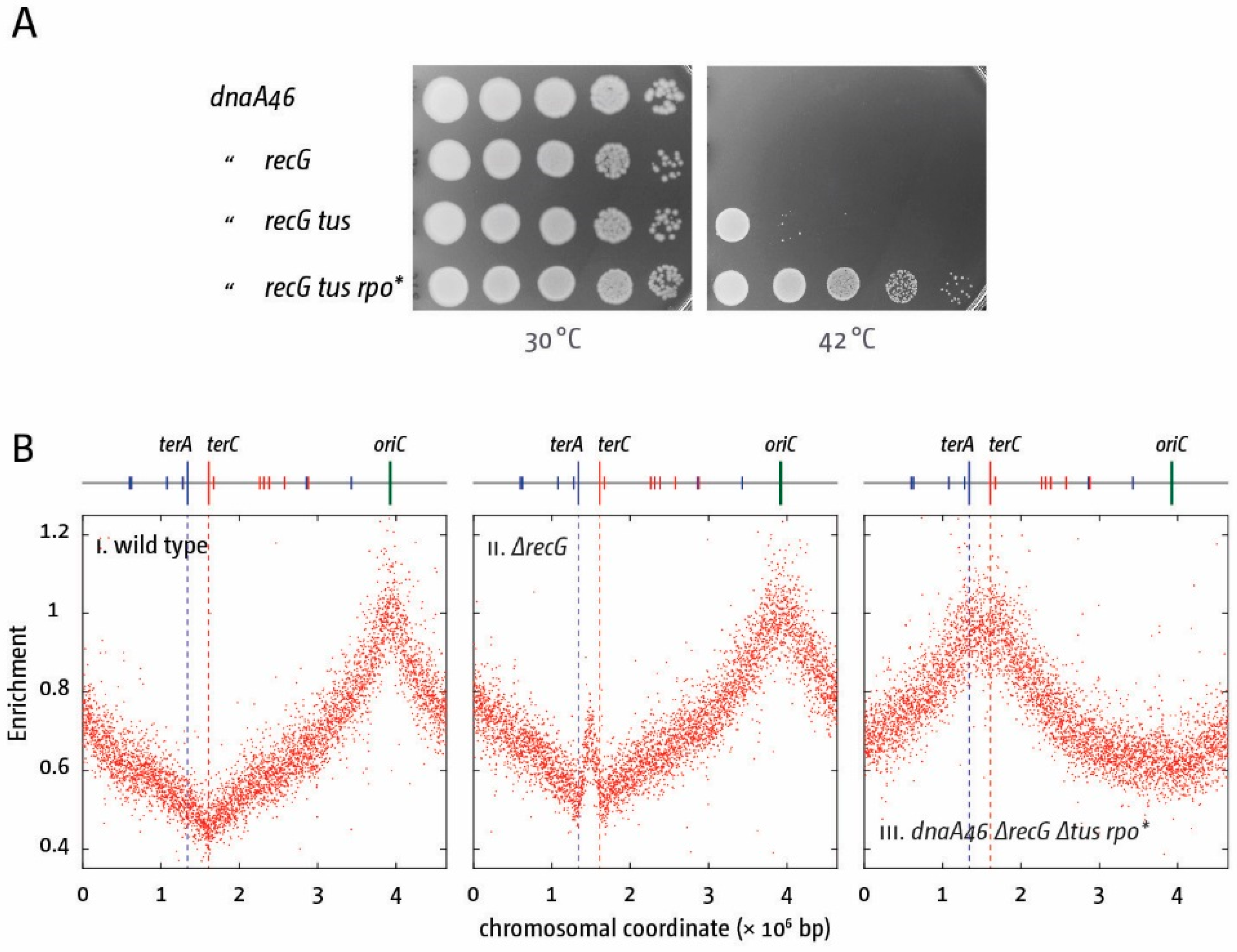
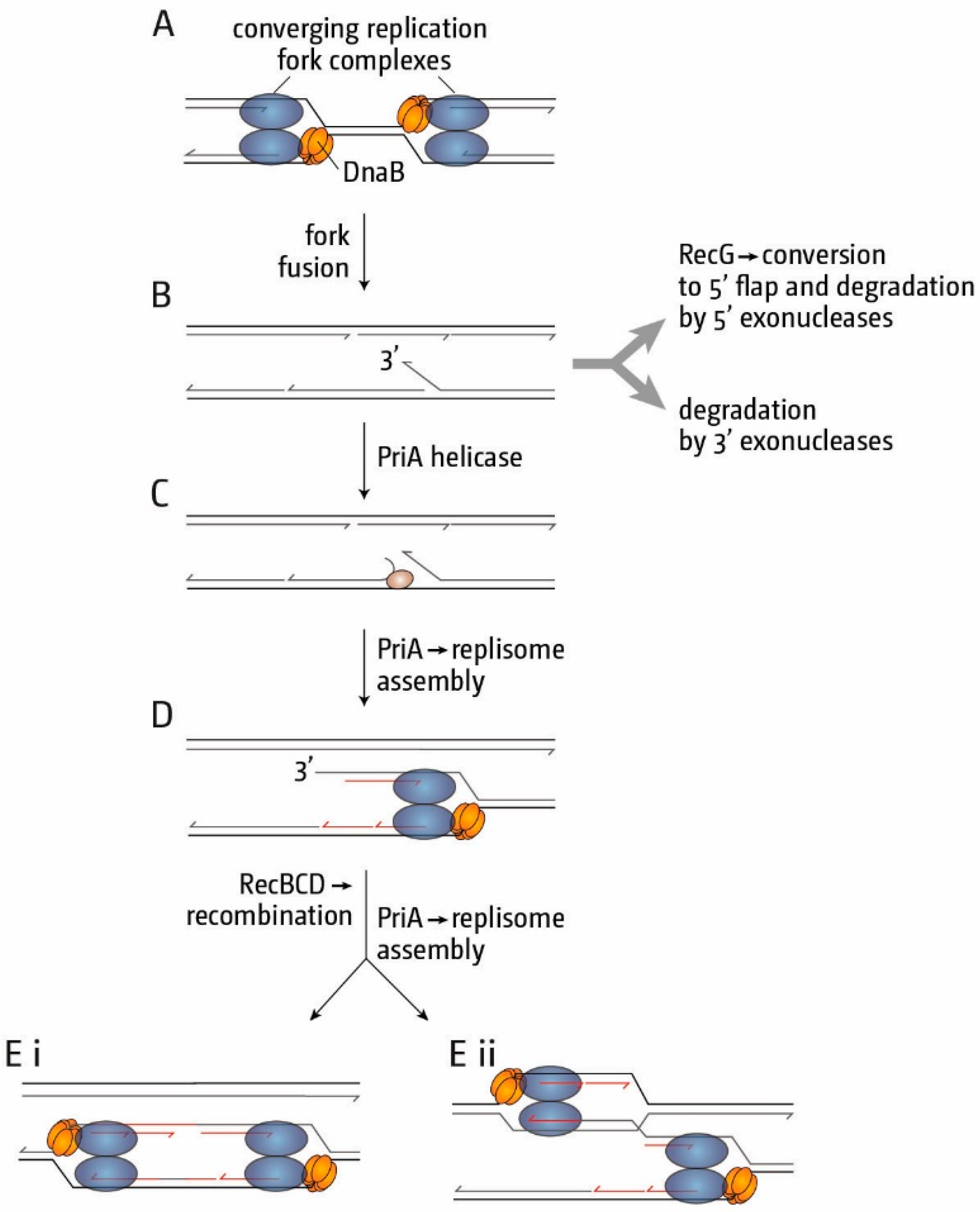
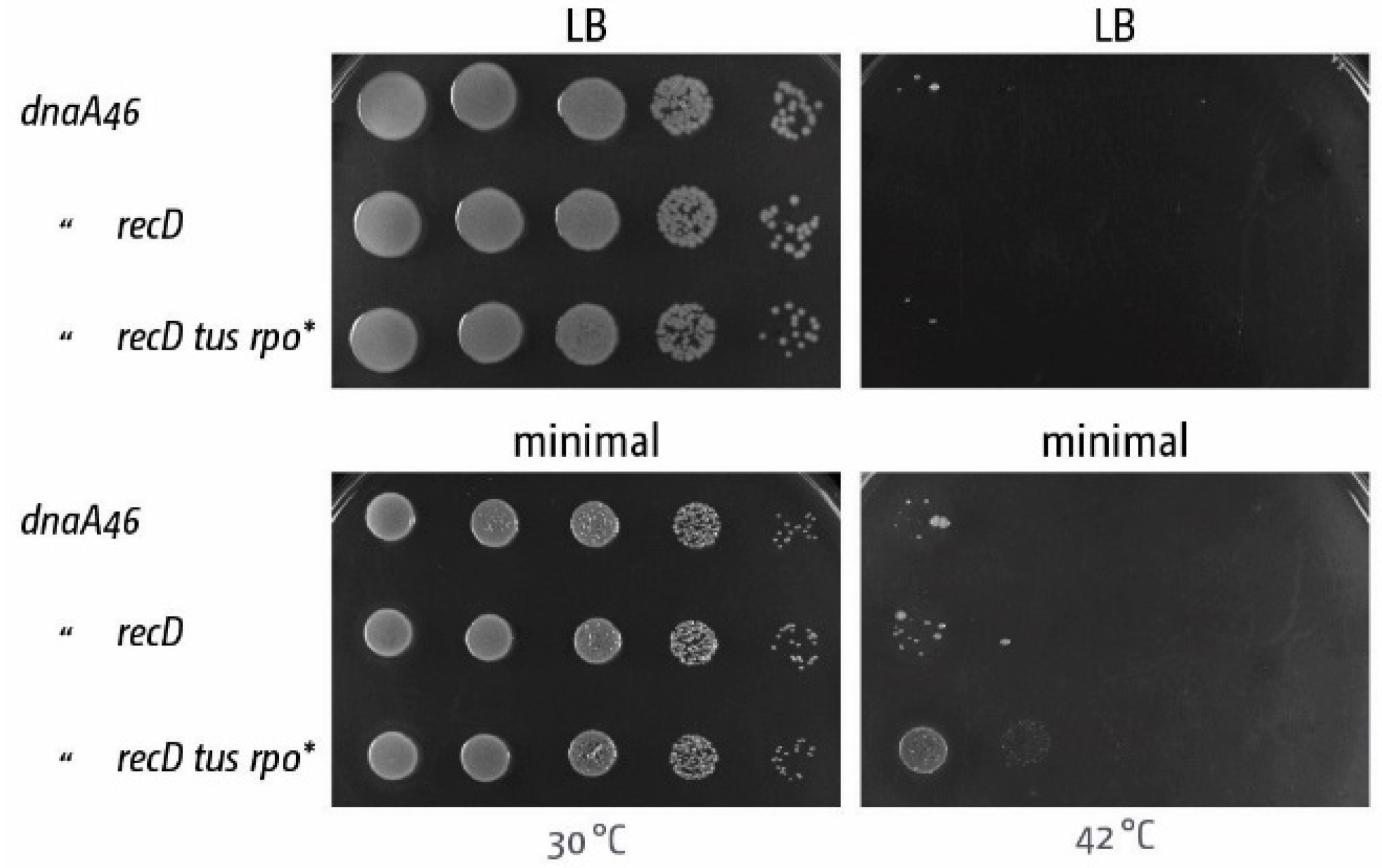
5. The Replication Fork Trap Contains Over-Replication
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fang, L.; Davey, M.J.; O’Donnell, M. Replisome assembly at oriC, the replication origin of E. coli, reveals an explanation for initiation sites outside an origin. Mol. Cell 1999, 4, 541–553. [Google Scholar] [CrossRef]
- Pomerantz, R.T.; O’Donnell, M. Replisome mechanics: Insights into a twin DNA polymerase machine. Trends Microbiol. 2007, 15, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Breier, A.M.; Weier, H.-U.G.; Cozzarelli, N.R. Independence of replisomes in Escherichia coli chromosomal replication. Proc. Natl. Acad. Sci. USA 2005, 102, 3942–3947. [Google Scholar] [CrossRef] [PubMed]
- Bastia, D.; Zaman, S. Mechanism and physiological significance of programmed replication termination. Semin. Cell Dev. Biol. 2014, 30, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Duggin, I.G.; Wake, R.G.; Bell, S.D.; Hill, T.M. The replication fork trap and termination of chromosome replication. Mol. Microbiol. 2008, 70, 1323–1333. [Google Scholar] [CrossRef] [PubMed]
- Neylon, C.; Kralicek, A.V.; Hill, T.M.; Dixon, N.E. Replication termination in Escherichia coli: Structure and antihelicase activity of the Tus-Ter complex. Microbiol. Mol. Biol. Rev. MMBR 2005, 69, 501–526. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Lamothe, R.; Nicolas, E.; Sherratt, D.J. Chromosome replication and segregation in bacteria. Annu. Rev. Genet. 2012, 46, 121–143. [Google Scholar] [CrossRef] [PubMed]
- Barre, F.X.; Søballe, B.; Michel, B.; Aroyo, M.; Robertson, M.; Sherratt, D. Circles: The replication-recombination-chromosome segregation connection. Proc. Natl. Acad. Sci. USA 2001, 98, 8189–8195. [Google Scholar] [CrossRef] [PubMed]
- Crozat, E.; Grainge, I. FtsK DNA translocase: The fast motor that knows where it’s going. Chembiochem Eur. J. Chem. Biol. 2010, 11, 2232–2243. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, D.; Taylor, T.; Smith, S.L.; Dimude, J.U.; Upton, A.L.; Mehrjouy, M.M.; Skovgaard, O.; Sherratt, D.J.; Retkute, R.; Rudolph, C.J. Shaping the landscape of the Escherichia coli chromosome: Replication-transcription encounters in cells with an ectopic replication origin. Nucleic Acids Res. 2015, 43, 7865–7877. [Google Scholar] [CrossRef] [PubMed]
- Iismaa, T.P.; Wake, R.G. The normal replication terminus of the Bacillus subtilis chromosome, terC, is dispensable for vegetative growth and sporulation. J. Mol. Biol. 1987, 195, 299–310. [Google Scholar] [CrossRef]
- Roecklein, B.; Pelletier, A.; Kuempel, P. The tus gene of Escherichia coli: Autoregulation, analysis of flanking sequences and identification of a complementary system in Salmonella typhimurium. Res. Microbiol. 1991, 142, 169–175. [Google Scholar] [CrossRef]
- Dimude, J.U.; Stockum, A.; Midgley-Smith, S.L.; Upton, A.L.; Foster, H.A.; Khan, A.; Saunders, N.J.; Retkute, R.; Rudolph, C.J. The consequences of replicating in the wrong orientation: Bacterial chromosome duplication without an active replication origin. mBio 2015, 6, e01294-15. [Google Scholar] [CrossRef] [PubMed]
- Rudolph, C.J.; Upton, A.L.; Harris, L.; Lloyd, R.G. Pathological replication in cells lacking RecG DNA translocase. Mol. Microbiol. 2009, 73, 352–366. [Google Scholar] [CrossRef] [PubMed]
- Rudolph, C.J.; Upton, A.L.; Lloyd, R.G. Replication fork collisions cause pathological chromosomal amplification in cells lacking RecG DNA translocase. Mol. Microbiol. 2009, 74, 940–955. [Google Scholar] [CrossRef] [PubMed]
- Rudolph, C.J.; Upton, A.L.; Briggs, G.S.; Lloyd, R.G. Is RecG a general guardian of the bacterial genome? DNA Repair 2010, 9, 210–223. [Google Scholar] [CrossRef] [PubMed]
- Rudolph, C.J.; Mahdi, A.A.; Upton, A.L.; Lloyd, R.G. RecG protein and single-strand DNA exonucleases avoid cell lethality associated with PriA helicase activity in Escherichia coli. Genetics 2010, 186, 473–492. [Google Scholar] [CrossRef] [PubMed]
- Rudolph, C.J.; Upton, A.L.; Stockum, A.; Nieduszynski, C.A.; Lloyd, R.G. Avoiding chromosome pathology when replication forks collide. Nature 2013, 500, 608–611. [Google Scholar] [CrossRef] [PubMed]
- Kuempel, P.L.; Duerr, S.A.; Seeley, N.R. Terminus region of the chromosome in Escherichia coli inhibits replication forks. Proc. Natl. Acad. Sci. USA 1977, 74, 3927–3931. [Google Scholar] [CrossRef] [PubMed]
- Kuempel, P.L.; Duerr, S.A.; Maglothin, P.D. Chromosome replication in an Escherichia coli dnaA mutant integratively suppressed by prophage P2. J. Bacteriol. 1978, 134, 902–912. [Google Scholar] [PubMed]
- Louarn, J.; Patte, J.; Louarn, J.M. Evidence for a fixed termination site of chromosome replication in Escherichia coli K12. J. Mol. Biol. 1977, 115, 295–314. [Google Scholar] [CrossRef]
- Hill, T.M.; Henson, J.M.; Kuempel, P.L. The terminus region of the Escherichia coli chromosome contains two separate loci that exhibit polar inhibition of replication. Proc. Natl. Acad. Sci. USA 1987, 84, 1754–1758. [Google Scholar] [CrossRef] [PubMed]
- De Massy, B.; Béjar, S.; Louarn, J.; Louarn, J.M.; Bouché, J.P. Inhibition of replication forks exiting the terminus region of the Escherichia coli chromosome occurs at two loci separated by 5 min. Proc. Natl. Acad. Sci. USA 1987, 84, 1759–1763. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, A.J.; Hill, T.M.; Kuempel, P.L. Location of sites that inhibit progression of replication forks in the terminus region of Escherichia coli. J. Bacteriol. 1988, 170, 4293–4298. [Google Scholar] [PubMed]
- Hill, T.M.; Kopp, B.J.; Kuempel, P.L. Termination of DNA replication in Escherichia coli requires a trans-acting factor. J. Bacteriol. 1988, 170, 662–668. [Google Scholar] [PubMed]
- Hidaka, M.; Akiyama, M.; Horiuchi, T. A consensus sequence of three DNA replication terminus sites on the E. coli chromosome is highly homologous to the terR sites of the R6K plasmid. Cell 1988, 55, 467–475. [Google Scholar] [CrossRef]
- Hidaka, M.; Kobayashi, T.; Horiuchi, T. A newly identified DNA replication terminus site, TerE, on the Escherichia coli chromosome. J. Bacteriol. 1991, 173, 391–393. [Google Scholar] [PubMed]
- Hill, T.M.; Pelletier, A.J.; Tecklenburg, M.L.; Kuempel, P.L. Identification of the DNA sequence from the E. coli terminus region that halts replication forks. Cell 1988, 55, 459–466. [Google Scholar] [CrossRef]
- Sharma, B.; Hill, T.M. TerF, the sixth identified replication arrest site in Escherichia coli, is located within the rcsC gene. J. Bacteriol. 1992, 174, 7854–7858. [Google Scholar] [PubMed]
- Hill, T.M. Features of the chromosomal terminus region. In Escherichia coli and Salmonella Cellular and Molecular Biology; ASM Press: Washington, DC, USA, 1996; pp. 1602–1614. [Google Scholar]
- Duggin, I.G.; Bell, S.D. Termination structures in the Escherichia coli chromosome replication fork trap. J. Mol. Biol. 2009, 387, 532–539. [Google Scholar] [CrossRef] [PubMed]
- Berghuis, B.A.; Dulin, D.; Xu, Z.-Q.; van Laar, T.; Cross, B.; Janissen, R.; Jergic, S.; Dixon, N.E.; Depken, M.; Dekker, N.H. Strand separation establishes a sustained lock at the Tus-Ter replication fork barrier. Nat. Chem. Biol. 2015, 11, 579–585. [Google Scholar] [CrossRef] [PubMed]
- Elshenawy, M.M.; Jergic, S.; Xu, Z.-Q.; Sobhy, M.A.; Takahashi, M.; Oakley, A.J.; Dixon, N.E.; Hamdan, S.M. Replisome speed determines the efficiency of the Tus-Ter replication termination barrier. Nature 2015, 525, 394–398. [Google Scholar] [CrossRef] [PubMed]
- Mulcair, M.D.; Schaeffer, P.M.; Oakley, A.J.; Cross, H.F.; Neylon, C.; Hill, T.M.; Dixon, N.E. A molecular mousetrap determines polarity of termination of DNA replication in E. coli. Cell 2006, 125, 1309–1319. [Google Scholar] [CrossRef] [PubMed]
- Bidnenko, V.; Ehrlich, S.D.; Michel, B. Replication fork collapse at replication terminator sequences. EMBO J. 2002, 21, 3898–3907. [Google Scholar] [CrossRef] [PubMed]
- Bidnenko, V.; Lestini, R.; Michel, B. The Escherichia coli UvrD helicase is essential for Tus removal during recombination-dependent replication restart from Ter sites. Mol. Microbiol. 2006, 62, 382–396. [Google Scholar] [CrossRef] [PubMed]
- Michel, B.; Grompone, G.; Florès, M.-J.; Bidnenko, V. Multiple pathways process stalled replication forks. Proc. Natl. Acad. Sci. USA 2004, 101, 12783–12788. [Google Scholar] [CrossRef] [PubMed]
- Moolman, M.C.; Tiruvadi Krishnan, S.; Kerssemakers, J.W.J.; de Leeuw, R.; Lorent, V.; Sherratt, D.J.; Dekker, N.H. The progression of replication forks at natural replication barriers in live bacteria. Nucleic Acids Res. 2016. [Google Scholar] [CrossRef] [PubMed]
- Maisnier-Patin, S.; Nordström, K.; Dasgupta, S. RecA-mediated rescue of Escherichia coli strains with replication forks arrested at the terminus. J. Bacteriol. 2001, 183, 6065–6073. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.; Hill, T.M. Insertion of inverted Ter sites into the terminus region of the Escherichia coli chromosome delays completion of DNA replication and disrupts the cell cycle. Mol. Microbiol. 1995, 18, 45–61. [Google Scholar] [CrossRef] [PubMed]
- Moreau, M.J.J.; Schaeffer, P.M. Differential Tus-Ter binding and lock formation: Implications for DNA replication termination in Escherichia coli. Mol. Biosyst. 2012, 8, 2783–2791. [Google Scholar] [CrossRef] [PubMed]
- Müller, C.A.; Hawkins, M.; Retkute, R.; Malla, S.; Wilson, R.; Blythe, M.J.; Nakato, R.; Komata, M.; Shirahige, K.; de Moura, A.P.S.; et al. The dynamics of genome replication using deep sequencing. Nucleic Acids Res. 2014, 42, e3. [Google Scholar] [CrossRef] [PubMed]
- Skovgaard, O.; Bak, M.; Løbner-Olesen, A.; Tommerup, N. Genome-wide detection of chromosomal rearrangements, indels, and mutations in circular chromosomes by short read sequencing. Genome Res. 2011, 21, 1388–1393. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lesterlin, C.; Reyes-Lamothe, R.; Ball, G.; Sherratt, D.J. Replication and segregation of an Escherichia coli chromosome with two replication origins. Proc. Natl. Acad. Sci. USA 2011, 108, E243–E250. [Google Scholar] [CrossRef] [PubMed]
- Boubakri, H.; de Septenville, A.L.; Viguera, E.; Michel, B. The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo. EMBO J. 2010, 29, 145–157. [Google Scholar] [CrossRef] [PubMed]
- De Septenville, A.L.; Duigou, S.; Boubakri, H.; Michel, B. Replication fork reversal after replication-transcription collision. PLoS Genet. 2012, 8, e1002622. [Google Scholar] [CrossRef] [PubMed]
- Srivatsan, A.; Tehranchi, A.; MacAlpine, D.M.; Wang, J.D. Co-orientation of replication and transcription preserves genome integrity. PLoS Genet. 2010, 6, e1000810. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.D.; Berkmen, M.B.; Grossman, A.D. Genome-wide coorientation of replication and transcription reduces adverse effects on replication in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2007, 104, 5608–5613. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Shatalin, K.; Epshtein, V.; Gottesman, M.E.; Nudler, E. Linking RNA polymerase backtracking to genome instability in E. coli. Cell 2011, 146, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Trautinger, B.W.; Jaktaji, R.P.; Rusakova, E.; Lloyd, R.G. RNA polymerase modulators and DNA repair activities resolve conflicts between DNA replication and transcription. Mol. Cell 2005, 19, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Maduike, N.Z.; Tehranchi, A.K.; Wang, J.D.; Kreuzer, K.N. Replication of the Escherichia coli chromosome in RNase HI-deficient cells: Multiple initiation regions and fork dynamics. Mol. Microbiol. 2014, 91, 39–56. [Google Scholar] [CrossRef] [PubMed]
- Arakawa, K.; Saito, R.; Tomita, M. Noise-reduction filtering for accurate detection of replication termini in bacterial genomes. FEBS Lett. 2007, 581, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Grigoriev, A. Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 1998, 26, 2286–2290. [Google Scholar] [CrossRef] [PubMed]
- Hendrickson, H.; Lawrence, J.G. Mutational bias suggests that replication termination occurs near the dif site, not at Ter sites. Mol. Microbiol. 2007, 64, 42–56. [Google Scholar] [CrossRef] [PubMed]
- Lobry, J.R. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol. 1996, 13, 660–665. [Google Scholar] [CrossRef] [PubMed]
- Fijalkowska, I.J.; Jonczyk, P.; Tkaczyk, M.M.; Bialoskorska, M.; Schaaper, R.M. Unequal fidelity of leading strand and lagging strand DNA replication on the Escherichia coli chromosome. Proc. Natl. Acad. Sci. USA 1998, 95, 10020–10025. [Google Scholar] [CrossRef] [PubMed]
- Lobry, J.R.; Sueoka, N. Asymmetric directional mutation pressures in bacteria. Genome Biol. 2002. [Google Scholar] [CrossRef]
- Kono, N.; Arakawa, K.; Tomita, M. Validation of bacterial replication termination models using simulation of genomic mutations. PLoS ONE 2012, 7, e34526. [Google Scholar] [CrossRef] [PubMed]
- Brewer, B.J. When polymerases collide: Replication and the transcriptional organization of the E. coli chromosome. Cell 1988, 53, 679–686. [Google Scholar] [CrossRef]
- French, S. Consequences of replication fork movement through transcription units in vivo. Science 1992, 258, 1362–1365. [Google Scholar] [CrossRef] [PubMed]
- Rocha, E.P.C.; Danchin, A. Gene essentiality determines chromosome organisation in bacteria. Nucleic Acids Res. 2003, 31, 6570–6577. [Google Scholar] [CrossRef] [PubMed]
- Vogel, U.; Jensen, K.F. The RNA chain elongation rate in Escherichia coli depends on the growth rate. J. Bacteriol. 1994, 176, 2807–2813. [Google Scholar] [PubMed]
- Dennis, P.P.; Ehrenberg, M.; Fange, D.; Bremer, H. Varying rate of RNA chain elongation during rrn transcription in Escherichia coli. J. Bacteriol. 2009, 191, 3740–3746. [Google Scholar] [CrossRef] [PubMed]
- Evertts, A.G.; Coller, H.A. Back to the origin: Reconsidering replication, transcription, epigenetics, and cell cycle control. Genes Cancer 2012, 3, 678–696. [Google Scholar] [CrossRef] [PubMed]
- McLean, M.J.; Wolfe, K.H.; Devine, K.M. Base composition skews, replication orientation, and gene orientation in 12 prokaryote genomes. J. Mol. Evol. 1998, 47, 691–696. [Google Scholar] [CrossRef] [PubMed]
- Hyrien, O. Peaks cloaked in the mist: The landscape of mammalian replication origins. J. Cell Biol. 2015, 208, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Hyrien, O. Mechanisms and consequences of replication fork arrest. Biochimie 2000, 82, 5–17. [Google Scholar] [CrossRef]
- Mirkin, E.V.; Mirkin, S.M. Replication fork stalling at natural impediments. Microbiol. Mol. Biol. Rev. MMBR 2007, 71, 13–35. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Jinks-Robertson, S. Transcription as a source of genome instability. Nat. Rev. Genet. 2012, 13, 204–214. [Google Scholar] [CrossRef] [PubMed]
- McGlynn, P.; Savery, N.J.; Dillingham, M.S. The conflict between DNA replication and transcription. Mol. Microbiol. 2012, 85, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Rudolph, C.J.; Dhillon, P.; Moore, T.; Lloyd, R.G. Avoiding and resolving conflicts between DNA replication and transcription. DNA Repair 2007, 6, 981–993. [Google Scholar] [CrossRef] [PubMed]
- Merrikh, H.; Zhang, Y.; Grossman, A.D.; Wang, J.D. Replication-transcription conflicts in bacteria. Nat. Rev. Microbiol. 2012, 10, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Million-Weaver, S.; Samadpour, A.N.; Merrikh, H. Replication Restart after replication-transcription conflicts requires RecA in Bacillus subtilis. J. Bacteriol. 2015, 197, 2374–2382. [Google Scholar] [CrossRef] [PubMed]
- Kogoma, T. Stable DNA replication: Interplay between DNA replication, homologous recombination, and transcription. Microbiol. Mol. Biol. Rev. MMBR 1997, 61, 212–238. [Google Scholar] [PubMed]
- De Massy, B.; Fayet, O.; Kogoma, T. Multiple origin usage for DNA replication in sdrA(rnh) mutants of Escherichia coli K-12. Initiation in the absence of oriC. J. Mol. Biol. 1984, 178, 227–236. [Google Scholar] [CrossRef]
- Merrikh, H.; Machón, C.; Grainger, W.H.; Grossman, A.D.; Soultanas, P. Co-directional replication-transcription conflicts lead to replication restart. Nature 2011, 470, 554–557. [Google Scholar] [CrossRef] [PubMed]
- Horiuchi, T.; Fujimura, Y.; Nishitani, H.; Kobayashi, T.; Hidaka, M. The DNA replication fork blocked at the Ter site may be an entrance for the RecBCD enzyme into duplex DNA. J. Bacteriol. 1994, 176, 4656–4663. [Google Scholar] [PubMed]
- Rothstein, R.; Michel, B.; Gangloff, S. Replication fork pausing and recombination or “gimme a break”. Genes Dev. 2000, 14, 1–10. [Google Scholar] [PubMed]
- Hiasa, H.; Marians, K.J. Tus prevents overreplication of oriC plasmid DNA. J. Biol. Chem. 1994, 269, 26959–26968. [Google Scholar] [PubMed]
- Krabbe, M.; Zabielski, J.; Bernander, R.; Nordström, K. Inactivation of the replication-termination system affects the replication mode and causes unstable maintenance of plasmid R1. Mol. Microbiol. 1997, 24, 723–735. [Google Scholar] [CrossRef] [PubMed]
- Nordström, K. Plasmid R1—Replication and its control. Plasmid 2006, 55, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Markovitz, A. A new in vivo termination function for DNA polymerase I of Escherichia coli K12. Mol. Microbiol. 2005, 55, 1867–1882. [Google Scholar] [CrossRef] [PubMed]
- Lemon, K.P.; Kurtser, I.; Grossman, A.D. Effects of replication termination mutants on chromosome partitioning in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2001, 98, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Hong, X.; Cadwell, G.W.; Kogoma, T. Escherichia coli RecG and RecA proteins in R-loop formation. EMBO J. 1995, 14, 2385–2392. [Google Scholar] [PubMed]
- Wallet, C.; le Ret, M.; Bergdoll, M.; Bichara, M.; Dietrich, A.; Gualberto, J.M. The RECG1 DNA translocase is a key factor in recombination surveillance, repair, and segregation of the mitochondrial DNA in Arabidopsis. Plant Cell 2015, 27, 2907–2925. [Google Scholar] [CrossRef] [PubMed]
- Wendel, B.M.; Courcelle, C.T.; Courcelle, J. Completion of DNA replication in Escherichia coli. Proc. Natl. Acad. Sci. USA 2014, 111, 16454–16459. [Google Scholar] [CrossRef] [PubMed]
- Gregg, A.V.; McGlynn, P.; Jaktaji, R.P.; Lloyd, R.G. Direct rescue of stalled DNA replication forks via the combined action of PriA and RecG helicase activities. Mol. Cell 2002, 9, 241–251. [Google Scholar] [CrossRef]
- LeBowitz, J.H.; McMacken, R. The Escherichia coli dnaB replication protein is a DNA helicase. J. Biol. Chem. 1986, 261, 4738–4748. [Google Scholar] [PubMed]
- Kaplan, D.L.; O’Donnell, M. DnaB drives DNA branch migration and dislodges proteins while encircling two DNA strands. Mol. Cell 2002, 10, 647–657. [Google Scholar] [CrossRef]
- Bianco, P.R. I came to a fork in the DNA and there was RecG. Prog. Biophys. Mol. Biol. 2015, 117, 166–173. [Google Scholar] [CrossRef] [PubMed]
- McGlynn, P.; Lloyd, R.G.; Marians, K.J. Formation of Holliday junctions by regression of nascent DNA in intermediates containing stalled replication forks: RecG stimulates regression even when the DNA is negatively supercoiled. Proc. Natl. Acad. Sci. USA 2001, 98, 8235–8240. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Masai, H. Stabilization of a stalled replication fork by concerted actions of two helicases. J. Biol. Chem. 2006, 281, 3484–3493. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, R.G.; Rudolph, C.J. 25 Years on and no end in sight: A perspective on the role of RecG protein. Curr. Genet. 2016. [Google Scholar] [CrossRef] [PubMed]
- Kowalczykowski, S.C. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 2000, 25, 156–165. [Google Scholar] [CrossRef]
- Fukuoh, A.; Iwasaki, H.; Ishioka, K.; Shinagawa, H. ATP-dependent resolution of R-loops at the ColE1 replication origin by Escherichia coli RecG protein, a Holliday junction-specific helicase. EMBO J. 1997, 16, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Horiuchi, T.; Maki, H.; Sekiguchi, M. RNase H-defective mutants of Escherichia coli: A possible discriminatory role of RNase H in initiation of DNA replication. Mol. Gen. Genet. MGG 1984, 195, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Tadokoro, T.; Kanaya, S. Ribonuclease H: Molecular diversities, substrate binding domains, and catalytic mechanism of the prokaryotic enzymes. FEBS J. 2009, 276, 1482–1493. [Google Scholar] [CrossRef] [PubMed]
- Vincent, S.D.; Mahdi, A.A.; Lloyd, R.G. The RecG branch migration protein of Escherichia coli dissociates R-loops. J. Mol. Biol. 1996, 264, 713–721. [Google Scholar] [CrossRef] [PubMed]
- Gowrishankar, J. End of the beginning: elongation and termination features of alternative modes of chromosomal replication initiation in bacteria. PLoS Genet. 2015, 11, e1004909. [Google Scholar] [CrossRef] [PubMed]
- Mahdi, A.A.; Briggs, G.S.; Lloyd, R.G. Modulation of DNA damage tolerance in Escherichia coli recG and ruv strains by mutations affecting PriB, the ribosome and RNA polymerase. Mol. Microbiol. 2012, 86, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Duggin, I.G.; Dubarry, N.; Bell, S.D. Replication termination and chromosome dimer resolution in the archaeon Sulfolobus solfataricus. EMBO J. 2011, 30, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Dewar, J.M.; Budzowska, M.; Walter, J.C. The mechanism of DNA replication termination in vertebrates. Nature 2015, 525, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Maric, M.; Maculins, T.; de Piccoli, G.; Labib, K. Cdc48 and a ubiquitin ligase drive disassembly of the CMG helicase at the end of DNA replication. Science 2014. [Google Scholar] [CrossRef] [PubMed]
- Moreno, S.P.; Bailey, R.; Campion, N.; Herron, S.; Gambus, A. Polyubiquitylation drives replisome disassembly at the termination of DNA replication. Science 2014, 346, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Fachinetti, D.; Bermejo, R.; Cocito, A.; Minardi, S.; Katou, Y.; Kanoh, Y.; Shirahige, K.; Azvolinsky, A.; Zakian, V.A.; Foiani, M. Replication termination at eukaryotic chromosomes is mediated by Top2 and occurs at genomic loci containing pausing elements. Mol. Cell 2010, 39, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Steinacher, R.; Osman, F.; Dalgaard, J.Z.; Lorenz, A.; Whitby, M.C. The DNA helicase Pfh1 promotes fork merging at replication termination sites to ensure genome stability. Genes Dev. 2012, 26, 594–602. [Google Scholar] [CrossRef] [PubMed]
- Moreno, S.P.; Gambus, A. Regulation of unperturbed DNA replication by ubiquitylation. Genes 2015, 6, 451–468. [Google Scholar] [CrossRef] [PubMed]
- Whitby, M.C. The FANCM family of DNA helicases/translocases. DNA Repair 2010, 9, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Ralf, C.; Hickson, I.D.; Wu, L. The Bloom’s syndrome helicase can promote the regression of a model replication fork. J. Biol. Chem. 2006, 281, 22839–22846. [Google Scholar] [CrossRef] [PubMed]
- Bétous, R.; Mason, A.C.; Rambo, R.P.; Bansbach, C.E.; Badu-Nkansah, A.; Sirbu, B.M.; Eichman, B.F.; Cortez, D. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev. 2012, 26, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Killen, M.W.; Stults, D.M.; Wilson, W.A.; Pierce, A.J. Escherichia coli RecG functionally suppresses human Bloom syndrome phenotypes. BMC Mol. Biol. 2012. [Google Scholar] [CrossRef] [PubMed]
- Diffley, J.F.X. Quality control in the initiation of eukaryotic DNA replication. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011, 366, 3545–3553. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.L.; Huang, J.; Edwards, Y.J.K.; Ulloa, R.H.; Dillon, L.M.; Prolla, T.A.; Vance, J.M.; Moraes, C.T.; Züchner, S. The mtDNA mutation spectrum of the progeroid Polg mutator mouse includes abundant control region multimers. Cell Metab. 2010, 12, 675–682. [Google Scholar] [CrossRef] [PubMed]
- Kornblum, C.; Nicholls, T.J.; Haack, T.B.; Schöler, S.; Peeva, V.; Danhauser, K.; Hallmann, K.; Zsurka, G.; Rorbach, J.; Iuso, A.; et al. Loss-of-function mutations in MGME1 impair mtDNA replication and cause multisystemic mitochondrial disease. Nat. Genet. 2013, 45, 214–219. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dimude, J.U.; Midgley-Smith, S.L.; Stein, M.; Rudolph, C.J. Replication Termination: Containing Fork Fusion-Mediated Pathologies in Escherichia coli. Genes 2016, 7, 40. https://doi.org/10.3390/genes7080040
Dimude JU, Midgley-Smith SL, Stein M, Rudolph CJ. Replication Termination: Containing Fork Fusion-Mediated Pathologies in Escherichia coli. Genes. 2016; 7(8):40. https://doi.org/10.3390/genes7080040
Chicago/Turabian StyleDimude, Juachi U., Sarah L. Midgley-Smith, Monja Stein, and Christian J. Rudolph. 2016. "Replication Termination: Containing Fork Fusion-Mediated Pathologies in Escherichia coli" Genes 7, no. 8: 40. https://doi.org/10.3390/genes7080040
APA StyleDimude, J. U., Midgley-Smith, S. L., Stein, M., & Rudolph, C. J. (2016). Replication Termination: Containing Fork Fusion-Mediated Pathologies in Escherichia coli. Genes, 7(8), 40. https://doi.org/10.3390/genes7080040






