Replication Stress: A Lifetime of Epigenetic Change
Abstract
:1. Introduction
2. The Cellular Response to Replication Stress
3. Sources of Replication Stress
4. Replication Stress and DNA Damage Signaling
5. Replication Stress Signaling Drives Senescence
6. A Role for Chromatin in the Replication Stress Response
7. Impact of Pre-Existing Chromatin on DNA Replication
7.1. Repressive Chromatin
7.2. H2B Ubiquitin
8. Replication Stress-Associated Chromatin Reorganization
| Chromatin Component | Modifiers/Interactors | Major Functions during RS | References |
|---|---|---|---|
| Histone modifications | |||
| γ-H2AX (phospho-S139) γ-H2A (phospho-S129) | ATM/ATR Mec1/Tel1 (yeast) | Replication fork progression, repair of collapsed forks. | [49,50,51,52] |
| H3 phospho-T45 | Cdc-DbF4 (yeast) | Increases resistance to RS. | [53] |
| H3K56ac | HAT: RTT109 (yeast) HDAC: Hst3/4 (yeast) | Pre-disposition mark on nascent chromatin, promotes replication fork stability. | [54,55] |
| H4K5ac, H4K12ac | HAT: HAT1 HDAC: HDAC1-3 | Pre-disposition mark on nascent chromatin, promotes replication fork stability. | [56,57,58] |
| H3K4me3 | HMT: Set1p (yeast) HMT: METNASE | Facilitates DSB repair at collapsed forks. Facilitates replication fork restart. | [59,60] |
| H3K9me | HMT:Clr4 (yeast) HMT: SUV39H1 KDM: KDM4A | Promotes HU induced S/M checkpoint. Controls replication timing and replication fork stability. | [40,41,61] |
| H3K36me3 | HMT: Set2p (yeast) | Promotes HU induced S/M checkpoint. Facilitates replication fork restart. | [61] |
| H3K27me1 | HMT: TXR1 (tetrahymena) | Protects from RS. | [42] |
| H3K79me3 | Dot1 (yeast) | Promotes sister chromatid recombination after RS. | [62] |
| H4K20me | HMT: WHSC1 | Identified in genetic screen for genes involved in RS. | [63] |
| H2Aub1 | DUB: USP3 | Facilitates DSB repair at collapsed forks. | [64] |
| H2Bub1 | Bre1 (yeast) | Nucleosome re-assembly, replisome stability | [44] |
| Poly(ADP-)ribose | PARP/PARG | Modulates RPA accumulation at collapsed forks. | [65] |
| Chaperones and remodelers | |||
| ASF1 | H3/H4 | H3/H4 chaperone, promotes nucleosome reassembly at sites of RS. | [48] |
| ATRX | Histone 3.3 | H3.3 chaperone, promotes nucleosome reassembly, fork progression. | [66,67] |
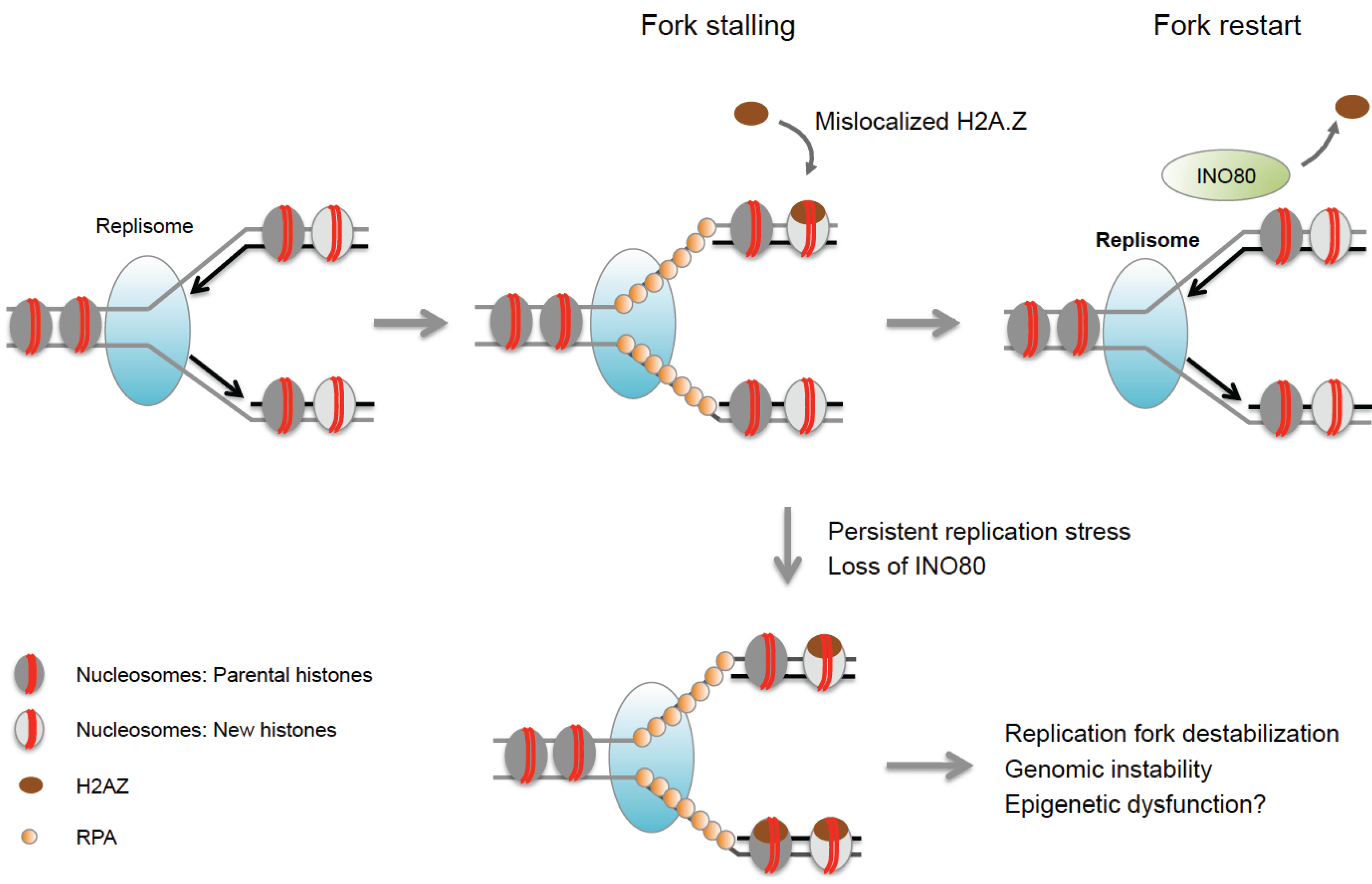
| INO80 | H2A.Z (yeast) | Removes H2A.Z from nucleosomes, promotes recovery of stalled forks. | [68,69,70] |
| RSC2/BAF180 | PCNA | Promotes recovery of stalled replication forks. | [71] |
| MMS22L | TONSL ASF1, FACT | Promotes recovery from RS, facilitates RAD51 loading and HR. Nucleosome reassembly | [72,73,74,75,76] |
| NASP | H3/H4 | H3/H4 chaperone, regulates histone metabolism during RS. | [77] |
| SAFB1 | Chromatin | Scaffold protein, regulates γ-H2AX-spreading during RS. | [78] |
8.1. γ-H2AX
8.2. Chromatin Remodeling
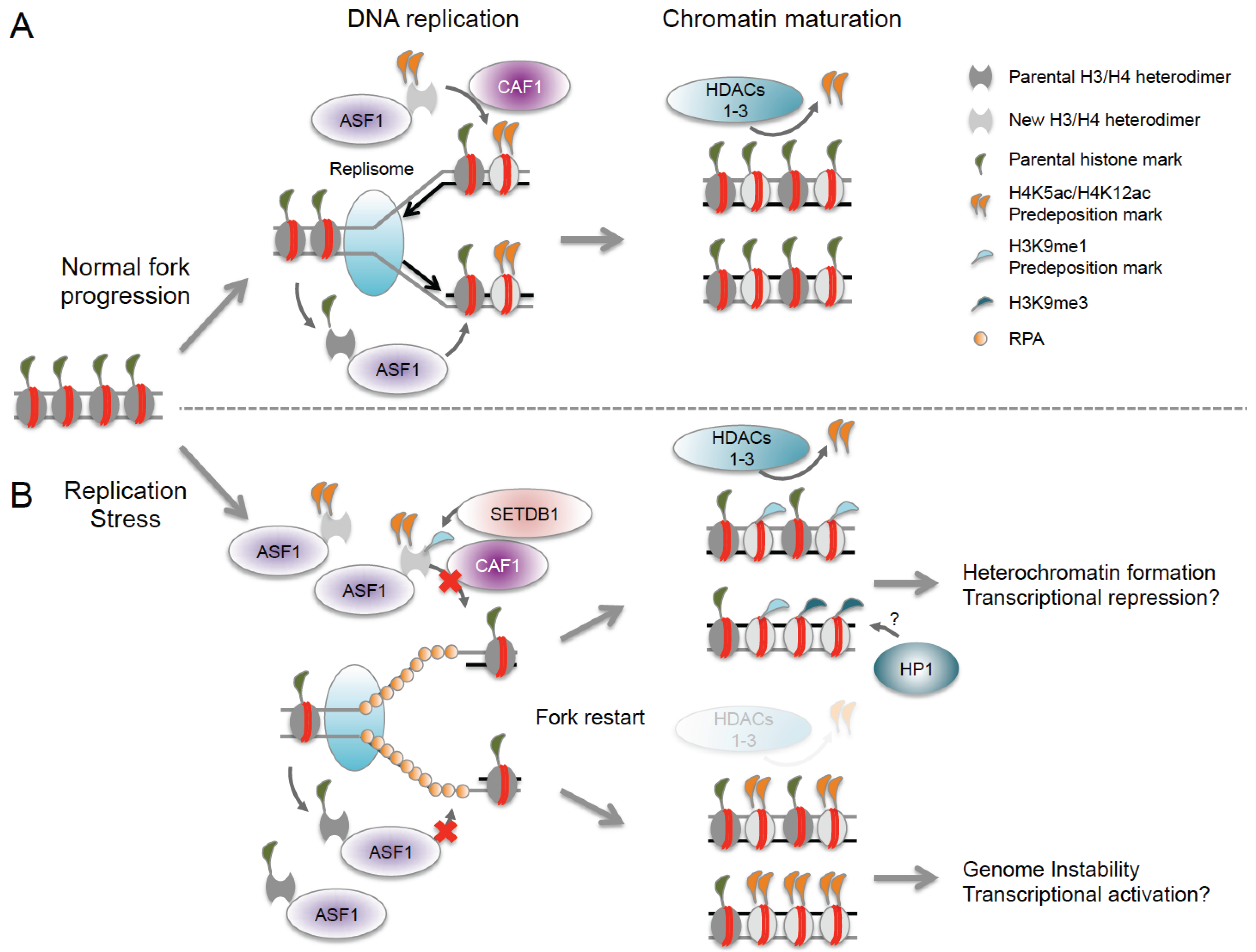
8.3. Histone Acetylation and Nucleosome Reassembly
9. Physiological Consequences of Replication Stress-Induced Chromatin Reorganization
10. Replication Stress and Aberrant Gene Expression
11. Replication Stress Induced Chromatin Reorganization in OIS
12. Conclusions and Perspective
Acknowledgments
Authors Contributions
Conflicts of Interest
References
- Sulli, G.; di Micco, R.; d’Adda di Fagagna, F. Crosstalk between chromatin state and DNA damage response in cellular senescence and cancer. Nat. Rev. Cancer 2012, 12, 709–720. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.P.; Donahue, G.; Otte, G.L.; Capell, B.C.; Nelson, D.M.; Cao, K.; Aggarwala, V.; Cruickshanks, H.A.; Rai, T.S.; McBryan, T.; et al. Lamin b1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev. 2013, 27, 1787–1799. [Google Scholar] [CrossRef] [PubMed]
- Sperka, T.; Wang, J.; Rudolph, K.L. DNA damage checkpoints in stem cells, ageing and cancer. Nat. Rev. Mol. Cell Biol. 2012, 13, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Zeman, M.K.; Cimprich, K.A. Causes and consequences of replication stress. Nat. Cell Biol. 2014, 16, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, A.; Garcia-Muse, T. Causes of genome instability. Annu. Rev. Genet. 2013, 47, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Barlow, J.H.; Faryabi, R.B.; Callen, E.; Wong, N.; Malhowski, A.; Chen, H.T.; Gutierrez-Cruz, G.; Sun, H.W.; McKinnon, P.; Wright, G.; et al. Identification of early replicating fragile sites that contribute to genome instability. Cell 2013, 152, 620–632. [Google Scholar] [CrossRef] [PubMed]
- Franchitto, A. Genome instability at common fragile sites: Searching for the cause of their instability. Biomed. Res. Int. 2013. [Google Scholar] [CrossRef] [PubMed]
- Letessier, A.; Millot, G.A.; Koundrioukoff, S.; Lachages, A.M.; Vogt, N.; Hansen, R.S.; Malfoy, B.; Brison, O.; Debatisse, M. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature 2011, 470, 120–123. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, A.; Garcia-Muse, T. R loops: From transcription byproducts to threats to genome stability. Mol. Cell 2012, 46, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Mazouzi, A.; Velimezi, G.; Loizou, J.I. DNA replication stress: Causes, resolution and disease. Exp. Cell Res. 2014, 329, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Cimprich, K.A.; Cortez, D. Atr: An essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008, 9, 616–627. [Google Scholar] [CrossRef] [PubMed]
- Walworth, N.C.; Bernards, R. Rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science 1996, 271, 353–356. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Guntuku, S.; Cui, X.S.; Matsuoka, S.; Cortez, D.; Tamai, K.; Luo, G.; Carattini-Rivera, S.; DeMayo, F.; Bradley, A.; et al. Chk1 is an essential kinase that is regulated by atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000, 14, 1448–1459. [Google Scholar] [PubMed]
- Shechter, D.; Costanzo, V.; Gautier, J. ATR and ATM regulate the timing of DNA replication origin firing. Nat. Cell Biol. 2004, 6, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Maya-Mendoza, A.; Petermann, E.; Gillespie, D.A.; Caldecott, K.W.; Jackson, D.A. Chk1 regulates the density of active replication origins during the vertebrate S phase. EMBO J. 2007, 26, 2719–2731. [Google Scholar] [CrossRef] [PubMed]
- Shirahige, K.; Hori, Y.; Shiraishi, K.; Yamashita, M.; Takahashi, K.; Obuse, C.; Tsurimoto, T.; Yoshikawa, H. Regulation of DNA-replication origins during cell-cycle progression. Nature 1998, 395, 618–621. [Google Scholar] [PubMed]
- Casper, A.M.; Nghiem, P.; Arlt, M.F.; Glover, T.W. Atr regulates fragile site stability. Cell 2002, 111, 779–789. [Google Scholar] [CrossRef]
- Ragland, R.L.; Arlt, M.F.; Hughes, E.D.; Saunders, T.L.; Glover, T.W. Mice hypomorphic for ATR have increased DNA damage and abnormal checkpoint response. Mamm. Genome 2009, 20, 375–385. [Google Scholar] [CrossRef] [PubMed]
- Ciccia, A.; Elledge, S.J. The DNA damage response: Making it safe to play with knives. Mol. Cell 2010, 40, 179–204. [Google Scholar] [CrossRef] [PubMed]
- Duursma, A.M.; Driscoll, R.; Elias, J.E.; Cimprich, K.A. A role for the MRN complex in ATR activation via TOPBP1 recruitment. Mol. Cell 2013, 50, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Dunphy, W.G. The Mre11-Rad50-Nbs1 (MRN) complex has a specific role in the activation of Chk1 in response to stalled replication forks. Mol. Biol. Cell 2013, 24, 1343–1353. [Google Scholar] [CrossRef] [PubMed]
- Trenz, K.; Smith, E.; Smith, S.; Costanzo, V. ATM and ATR promote Mre11 dependent restart of collapsed replication forks and prevent accumulation of DNA breaks. EMBO J. 2006, 25, 1764–1774. [Google Scholar] [CrossRef] [PubMed]
- Ammazzalorso, F.; Pirzio, L.M.; Bignami, M.; Franchitto, A.; Pichierri, P. ATR and ATM differently regulate WRN to prevent DSBs at stalled replication forks and promote replication fork recovery. EMBO J. 2010, 29, 3156–3169. [Google Scholar] [CrossRef] [PubMed]
- Ozeri-Galai, E.; Schwartz, M.; Rahat, A.; Kerem, B. Interplay between atm and atr in the regulation of common fragile site stability. Oncogene 2008, 27, 2109–2117. [Google Scholar] [CrossRef] [PubMed]
- Campisi, J.; d’Adda di Fagagna, F. Cellular senescence: When bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 2007, 8, 729–740. [Google Scholar] [CrossRef] [PubMed]
- D’Adda di Fagagna, F. Living on a break: Cellular senescence as a DNA-damage response. Nat. Rev. Cancer 2008, 8, 512–522. [Google Scholar] [CrossRef] [PubMed]
- Acosta, J.C.; O’Loghlen, A.; Banito, A.; Guijarro, M.V.; Augert, A.; Raguz, S.; Fumagalli, M.; da Costa, M.; Brown, C.; Popov, N.; et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell 2008, 133, 1006–1018. [Google Scholar] [CrossRef] [PubMed]
- Coppe, J.P.; Patil, C.K.; Rodier, F.; Sun, Y.; Munoz, D.P.; Goldstein, J.; Nelson, P.S.; Desprez, P.Y.; Campisi, J. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008, 6, 2853–2868. [Google Scholar] [CrossRef] [PubMed]
- Kuilman, T.; Michaloglou, C.; Vredeveld, L.C.; Douma, S.; van Doorn, R.; Desmet, C.J.; Aarden, L.A.; Mooi, W.J.; Peeper, D.S. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell 2008, 133, 1019–1031. [Google Scholar] [CrossRef] [PubMed]
- Coppe, J.P.; Desprez, P.Y.; Krtolica, A.; Campisi, J. The senescence-associated secretory phenotype: The dark side of tumor suppression. Annu. Rev. Pathol. 2010, 5, 99–118. [Google Scholar] [CrossRef] [PubMed]
- Rodier, F.; Coppe, J.P.; Patil, C.K.; Hoeijmakers, W.A.; Munoz, D.P.; Raza, S.R.; Freund, A.; Campeau, E.; Davalos, A.R.; Campisi, J. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat. Cell Biol. 2009, 11, 973–979. [Google Scholar] [CrossRef] [PubMed]
- Michaloglou, C.; Vredeveld, L.C.; Soengas, M.S.; Denoyelle, C.; Kuilman, T.; van der Horst, C.M.; Majoor, D.M.; Shay, J.W.; Mooi, W.J.; Peeper, D.S. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 2005, 436, 720–724. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Sola, D.; Ying, C.Y.; Grandori, C.; Ruggiero, L.; Chen, B.; Li, M.; Galloway, D.A.; Gu, W.; Gautier, J.; Dalla-Favera, R. Non-transcriptional control of DNA replication by c-Myc. Nature 2007, 448, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Di Micco, R.; Fumagalli, M.; Cicalese, A.; Piccinin, S.; Gasparini, P.; Luise, C.; Schurra, C.; Garre, M.; Nuciforo, P.G.; Bensimon, A.; et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 2006, 444, 638–642. [Google Scholar] [CrossRef] [PubMed]
- Bartkova, J.; Rezaei, N.; Liontos, M.; Karakaidos, P.; Kletsas, D.; Issaeva, N.; Vassiliou, L.V.; Kolettas, E.; Niforou, K.; Zoumpourlis, V.C.; et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 2006, 444, 633–637. [Google Scholar] [CrossRef] [PubMed]
- Di Micco, R.; Sulli, G.; Dobreva, M.; Liontos, M.; Botrugno, O.A.; Gargiulo, G.; dal Zuffo, R.; Matti, V.; d’Ario, G.; Montani, E.; et al. Interplay between oncogene-induced DNA damage response and heterochromatin in senescence and cancer. Nat. Cell Biol. 2011, 13, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Aird, K.M.; Zhang, G.; Li, H.; Tu, Z.; Bitler, B.G.; Garipov, A.; Wu, H.; Wei, Z.; Wagner, S.N.; Herlyn, M.; et al. Suppression of nucleotide metabolism underlies the establishment and maintenance of oncogene-induced senescence. Cell Rep. 2013, 3, 1252–1265. [Google Scholar] [CrossRef] [PubMed]
- Aird, K.M.; Worth, A.J.; Snyder, N.W.; Lee, J.V.; Sivanand, S.; Liu, Q.; Blair, I.A.; Wellen, K.E.; Zhang, R. ATM couples replication stress and metabolic reprogramming during cellular senescence. Cell. Rep 2015, 11, 893–901. [Google Scholar] [CrossRef] [PubMed]
- Mechali, M.; Yoshida, K.; Coulombe, P.; Pasero, P. Genetic and epigenetic determinants of DNA replication origins, position and activation. Curr. Opin. Genet. Dev. 2013, 23, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Black, J.C.; Allen, A.; Van Rechem, C.; Forbes, E.; Longworth, M.; Tschop, K.; Rinehart, C.; Quiton, J.; Walsh, R.; Smallwood, A.; et al. Conserved antagonism between JMJD2A/KDM4A and HP1gamma during cell cycle progression. Mol. Cell 2010, 40, 736–748. [Google Scholar] [CrossRef] [PubMed]
- Black, J.C.; Manning, A.L.; Van Rechem, C.; Kim, J.; Ladd, B.; Cho, J.; Pineda, C.M.; Murphy, N.; Daniels, D.L.; Montagna, C.; et al. KDM4A lysine demethylase induces site-specific copy gain and rereplication of regions amplified in tumors. Cell 2013, 154, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Xiong, J.; Zhang, C.; Berquist, B.R.; Yang, R.; Zhao, M.; Molascon, A.J.; Kwiatkowski, S.Y.; Yuan, D.; Qin, Z.; et al. Impaired replication elongation in Tetrahymena mutants deficient in histone H3 Lys 27 monomethylation. Genes Dev. 2013, 27, 1662–1679. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; de Vries, H.I.; Siudeja, K.; Rana, A.; Lemstra, W.; Brunsting, J.F.; Kok, R.M.; Smulders, Y.M.; Schaefer, M.; Dijk, F.; et al. Stwl modifies chromatin compaction and is required to maintain DNA integrity in the presence of perturbed DNA replication. Mol. Biol. Cell 2009, 20, 983–994. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, K.M.; Osley, M.A. A role for H2B ubiquitylation in DNA replication. Mol. Cell 2012, 48, 734–746. [Google Scholar] [CrossRef] [PubMed]
- Alvino, G.M.; Collingwood, D.; Murphy, J.M.; Delrow, J.; Brewer, B.J.; Raghuraman, M.K. Replication in hydroxyurea: It’s a matter of time. Mol. Cell Biol. 2007, 27, 6396–6406. [Google Scholar] [CrossRef] [PubMed]
- Pavri, R.; Zhu, B.; Li, G.; Trojer, P.; Mandal, S.; Shilatifard, A.; Reinberg, D. Histone H2B monoubiquitination functions cooperatively with fact to regulate elongation by RNA polymerase II. Cell 2006, 125, 703–717. [Google Scholar] [CrossRef] [PubMed]
- Chernikova, S.B.; Razorenova, O.V.; Higgins, J.P.; Sishc, B.J.; Nicolau, M.; Dorth, J.A.; Chernikova, D.A.; Kwok, S.; Brooks, J.D.; Bailey, S.M.; et al. Deficiency in mammalian histone H2B ubiquitin ligase Bre1 (Rnf20/Rnf40) leads to replication stress and chromosomal instability. Cancer Res. 2012, 72, 2111–2119. [Google Scholar] [CrossRef] [PubMed]
- Alabert, C.; Groth, A. Chromatin replication and epigenome maintenance. Nat. Rev. Mol. Cell Biol. 2012, 13, 153–167. [Google Scholar] [CrossRef] [PubMed]
- Sirbu, B.M.; Couch, F.B.; Feigerle, J.T.; Bhaskara, S.; Hiebert, S.W.; Cortez, D. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 2011, 25, 1320–1327. [Google Scholar] [CrossRef] [PubMed]
- Chanoux, R.A.; Yin, B.; Urtishak, K.A.; Asare, A.; Bassing, C.H.; Brown, E.J. ATR and H2AX cooperate in maintaining genome stability under replication stress. J. Biol. Chem. 2009, 284, 5994–6003. [Google Scholar] [CrossRef] [PubMed]
- Szilard, R.K.; Jacques, P.E.; Laramee, L.; Cheng, B.; Galicia, S.; Bataille, A.R.; Yeung, M.; Mendez, M.; Bergeron, M.; Robert, F.; et al. Systematic identification of fragile sites via genome-wide location analysis of gamma-H2AX. Nat. Struct. Mol. Biol. 2010, 17, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Cha, R.S.; Kleckner, N. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 2002, 297, 602–606. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.P.; Phillips, J.; Anderson, S.; Qiu, Q.; Shabanowitz, J.; Smith, M.M.; Yates, J.R., 3rd; Hunt, D.F.; Grant, P.A. Histone H3 Thr 45 phosphorylation is a replication-associated post-translational modification in S. cerevisiae. Nat. Cell Biol. 2010, 12, 294–298. [Google Scholar] [CrossRef] [PubMed]
- Driscoll, R.; Hudson, A.; Jackson, S.P. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 2007, 315, 649–652. [Google Scholar] [CrossRef] [PubMed]
- Celic, I.; Masumoto, H.; Griffith, W.P.; Meluh, P.; Cotter, R.J.; Boeke, J.D.; Verreault, A. The sirtuins Hst3 and Hst4p preserve genome integrity by controlling histone H3 lysine 56 deacetylation. Curr. Biol. 2006, 16, 1280–1289. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, P.; Ge, Z.; Sirbu, B.; Doughty, C.; Agudelo Garcia, P.A.; Schlederer, M.; Annunziato, A.T.; Cortez, D.; Kenner, L.; Parthun, M.R. Histone acetyl transferase 1 is essential for mammalian development, genome stability, and the processing of newly synthesized histones H3 and H4. PLoS Genet. 2013, 9, e1003518. [Google Scholar] [CrossRef] [PubMed]
- Bhaskara, S.; Jacques, V.; Rusche, J.R.; Olson, E.N.; Cairns, B.R.; Chandrasekharan, M.B. Histone deacetylases 1 and 2 maintain S-phase chromatin and DNA replication fork progression. Epigenet. Chromatin. 2013. [Google Scholar] [CrossRef] [PubMed]
- Bhaskara, S.; Knutson, S.K.; Jiang, G.; Chandrasekharan, M.B.; Wilson, A.J.; Zheng, S.; Yenamandra, A.; Locke, K.; Yuan, J.L.; Bonine-Summers, A.R.; et al. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell 2010, 18, 436–447. [Google Scholar] [CrossRef] [PubMed]
- Faucher, D.; Wellinger, R.J. Methylated H3K4, a transcription-associated histone modification, is involved in the DNA damage response pathway. PLoS Genet. 2010. [Google Scholar] [CrossRef] [PubMed]
- De Haro, L.P.; Wray, J.; Williamson, E.A.; Durant, S.T.; Corwin, L.; Gentry, A.C.; Osheroff, N.; Lee, S.H.; Hromas, R.; Nickoloff, J.A. Metnase promotes restart and repair of stalled and collapsed replication forks. Nucleic Acids Res. 2010, 38, 5681–5691. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Rhee, D.K.; Jang, Y.K. Methylations of histone H3 lysine 9 and lysine 36 are functionally linked to DNA replication checkpoint control in fission yeast. Biochem. Biophys. Res. Commun. 2008, 368, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Levesque, N.; Leung, G.P.; Fok, A.K.; Schmidt, T.I.; Kobor, M.S. Loss of h3 k79 trimethylation leads to suppression of Rtt107-dependent DNA damage sensitivity through the translesion synthesis pathway. J. Biol. Chem. 2010, 285, 35113–35122. [Google Scholar] [CrossRef] [PubMed]
- Hajdu, I.; Ciccia, A.; Lewis, S.M.; Elledge, S.J. Wolf-hirschhorn syndrome candidate 1 is involved in the cellular response to DNA damage. Proc. Natl. Acad. Sci. USA 2011, 108, 13130–13134. [Google Scholar] [CrossRef] [PubMed]
- Nicassio, F.; Corrado, N.; Vissers, J.H.; Areces, L.B.; Bergink, S.; Marteijn, J.A.; Geverts, B.; Houtsmuller, A.B.; Vermeulen, W.; di Fiore, P.P.; et al. Human usp3 is a chromatin modifier required for s phase progression and genome stability. Curr. Biol. 2007, 17, 1972–1977. [Google Scholar] [CrossRef] [PubMed]
- Illuzzi, G.; Fouquerel, E.; Ame, J.C.; Noll, A.; Rehmet, K.; Nasheuer, H.P.; Dantzer, F.; Schreiber, V. Parg is dispensable for recovery from transient replicative stress but required to prevent detrimental accumulation of poly(adp-ribose) upon prolonged replicative stress. Nucleic Acids Res. 2014, 42, 7776–7792. [Google Scholar] [CrossRef] [PubMed]
- Leung, J.W.; Ghosal, G.; Wang, W.; Shen, X.; Wang, J.; Li, L.; Chen, J. Alpha thalassemia/mental retardation syndrome x-linked gene product atrx is required for proper replication restart and cellular resistance to replication stress. J. Biol. Chem. 2013, 288, 6342–6350. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, A.D.; Banaszynski, L.A.; Noh, K.M.; Lewis, P.W.; Elsaesser, S.J.; Stadler, S.; Dewell, S.; Law, M.; Guo, X.; Li, X.; et al. Distinct factors control histone variant h3.3 localization at specific genomic regions. Cell 2010, 140, 678–691. [Google Scholar] [CrossRef] [PubMed]
- Papamichos-Chronakis, M.; Peterson, C.L. The ino80 chromatin-remodeling enzyme regulates replisome function and stability. Nat. Struct. Mol. Biol. 2008, 15, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Papamichos-Chronakis, M.; Watanabe, S.; Rando, O.J.; Peterson, C.L. Global regulation of h2a.Z localization by the ino80 chromatin-remodeling enzyme is essential for genome integrity. Cell 2011, 144, 200–213. [Google Scholar] [CrossRef] [PubMed]
- Shimada, K.; Oma, Y.; Schleker, T.; Kugou, K.; Ohta, K.; Harata, M.; Gasser, S.M. Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Curr. Biol. 2008, 18, 566–575. [Google Scholar] [CrossRef] [PubMed]
- Niimi, A.; Chambers, A.L.; Downs, J.A.; Lehmann, A.R. A role for chromatin remodellers in replication of damaged DNA. Nucleic Acids Res. 2012, 40, 7393–7403. [Google Scholar] [CrossRef] [PubMed]
- Duro, E.; Lundin, C.; Ask, K.; Sanchez-Pulido, L.; MacArtney, T.J.; Toth, R.; Ponting, C.P.; Groth, A.; Helleday, T.; Rouse, J. Identification of the MMS22L-TONSL complex that promotes homologous recombination. Mol. Cell 2010, 40, 632–644. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, L.; Panier, S.; Wildenhain, J.; Tkach, J.M.; al-Hakim, A.; Landry, M.C.; Escribano-Diaz, C.; Szilard, R.K.; Young, J.T.; Munro, M.; et al. The MMS22L-TONSL complex mediates recovery from replication stress and homologous recombination. Mol. Cell 2010, 40, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Piwko, W.; Olma, M.H.; Held, M.; Bianco, J.N.; Pedrioli, P.G.; Hofmann, K.; Pasero, P.; Gerlich, D.W.; Peter, M. RNAi-based screening identifies the MMS22L-Nfkbil2 complex as a novel regulator of DNA replication in human cells. EMBO J. 2010, 29, 4210–4222. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, B.C.; Adamson, B.; Lydeard, J.R.; Sowa, M.E.; Ciccia, A.; Bredemeyer, A.L.; Schlabach, M.; Gygi, S.P.; Elledge, S.J.; Harper, J.W. A genome-wide camptothecin sensitivity screen identifies a mammalian MMS22l-Nfkbil2 complex required for genomic stability. Mol. Cell 2010, 40, 645–657. [Google Scholar] [CrossRef] [PubMed]
- Duro, E.; Vaisica, J.A.; Brown, G.W.; Rouse, J. Budding yeast MMS22 and MMS1 regulate homologous recombination induced by replisome blockage. DNA Repair. 2008, 7, 811–818. [Google Scholar] [CrossRef] [PubMed]
- Cook, A.J.; Gurard-Levin, Z.A.; Vassias, I.; Almouzni, G. A specific function for the histone chaperone nasp to fine-tune a reservoir of soluble H3-H4 in the histone supply chain. Mol. Cell 2011, 44, 918–927. [Google Scholar] [CrossRef] [PubMed]
- Altmeyer, M.; Toledo, L.; Gudjonsson, T.; Grofte, M.; Rask, M.B.; Lukas, C.; Akimov, V.; Blagoev, B.; Bartek, J.; Lukas, J. The chromatin scaffold protein SAFB1 renders chromatin permissive for DNA damage signaling. Mol. Cell 2013, 52, 206–220. [Google Scholar] [CrossRef] [PubMed]
- Ward, I.M.; Chen, J. Histone h2ax is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 2001, 276, 47759–47762. [Google Scholar] [PubMed]
- Petermann, E.; Orta, M.L.; Issaeva, N.; Schultz, N.; Helleday, T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell 2010, 37, 492–502. [Google Scholar] [CrossRef] [PubMed]
- Iacovoni, J.S.; Caron, P.; Lassadi, I.; Nicolas, E.; Massip, L.; Trouche, D.; Legube, G. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 2010, 29, 1446–1457. [Google Scholar] [CrossRef] [PubMed]
- Min, J.N.; Tian, Y.; Xiao, Y.; Wu, L.; Li, L.; Chang, S. The mino80 chromatin remodeling complex is required for efficient telomere replication and maintenance of genome stability. Cell Res. 2013, 23, 1396–1413. [Google Scholar] [CrossRef] [PubMed]
- Vassileva, I.; Yanakieva, I.; Peycheva, M.; Gospodinov, A.; Anachkova, B. The mammalian ino80 chromatin remodeling complex is required for replication stress recovery. Nucleic Acids Res. 2014, 42, 9074–9086. [Google Scholar] [CrossRef] [PubMed]
- Alabert, C.; Bukowski-Wills, J.C.; Lee, S.B.; Kustatscher, G.; Nakamura, K.; de Lima Alves, F.; Menard, P.; Mejlvang, J.; Rappsilber, J.; Groth, A. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell Biol. 2014, 16, 281–293. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Contreras, A.J.; Ruppen, I.; Nieto-Soler, M.; Murga, M.; Rodriguez-Acebes, S.; Remeseiro, S.; Rodrigo-Perez, S.; Rojas, A.M.; Mendez, J.; Munoz, J.; et al. A proteomic characterization of factors enriched at nascent DNA molecules. Cell Rep. 2013, 3, 1105–1116. [Google Scholar] [CrossRef] [PubMed]
- Sansoni, V.; Casas-Delucchi, C.S.; Rajan, M.; Schmidt, A.; Bonisch, C.; Thomae, A.W.; Staege, M.S.; Hake, S.B.; Cardoso, M.C.; Imhof, A. The histone variant H2A.Bbd is enriched at sites of DNA synthesis. Nucleic Acids Res. 2014, 42, 6405–6420. [Google Scholar] [CrossRef] [PubMed]
- Khurana, S.; Kruhlak, M.J.; Kim, J.; Tran, A.D.; Liu, J.; Nyswaner, K.; Shi, L.; Jailwala, P.; Sung, M.H.; Hakim, O.; et al. A macrohistone variant links dynamic chromatin compaction to brca1-dependent genome maintenance. Cell Rep. 2014, 8, 1049–1062. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.H.; McGhie, J.D.; Sim, M.; Anderson, M.A.; Ahn, S.; Hannan, R.D.; George, A.J.; Morgan, K.A.; Mann, J.R.; Choo, K.H. Atrx interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010, 20, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Perry, C.A.; Annunziato, A.T. Influence of histone acetylation on the solubility, H1 content and DNAse I sensitivity of newly assembled chromatin. Nucleic Acids Res. 1989, 17, 4275–4291. [Google Scholar] [CrossRef] [PubMed]
- Clemente-Ruiz, M.; Gonzalez-Prieto, R.; Prado, F. Histone H3k56 acetylation, Caf1, and Rtt106 coordinate nucleosome assembly and stability of advancing replication forks. PLoS Genet. 2011, 7, e1002376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Myung, K.; Pennaneach, V.; Kats, E.S.; Kolodner, R.D. Saccharomyces cerevisiae chromatin-assembly factors that act during DNA replication function in the maintenance of genome stability. Proc. Natl. Acad. Sci. USA 2003, 100, 6640–6645. [Google Scholar] [CrossRef] [PubMed]
- Das, C.; Lucia, M.S.; Hansen, K.C.; Tyler, J.K. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature 2009, 459, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Conti, C.; Leo, E.; Eichler, G.S.; Sordet, O.; Martin, M.M.; Fan, A.; Aladjem, M.I.; Pommier, Y. Inhibition of histone deacetylase in cancer cells slows down replication forks, activates dormant origins, and induces DNA damage. Cancer Res. 2010, 70, 4470–4480. [Google Scholar] [CrossRef] [PubMed]
- Taddei, A.; Maison, C.; Roche, D.; Almouzni, G. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat. Cell Biol. 2001, 3, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Ask, K.; Jasencakova, Z.; Menard, P.; Feng, Y.; Almouzni, G.; Groth, A. Codanin-1, mutated in the anaemic disease CDAI, regulates Asf1 function in S-phase histone supply. EMBO J. 2012, 31, 2013–2023. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S.; Tadele, Z.; Hofmann, I.; Probst, A.V.; Angelis, K.J.; Kaya, H.; Araki, T.; Mengiste, T.; Mittelsten Scheid, O.; Shibahara, K.; et al. Bru1, a novel link between responses to DNA damage and epigenetic gene silencing in Arabidopsis. Genes Dev. 2004, 18, 782–793. [Google Scholar] [CrossRef] [PubMed]
- Jasencakova, Z.; Scharf, A.N.; Ask, K.; Corpet, A.; Imhof, A.; Almouzni, G.; Groth, A. Replication stress interferes with histone recycling and predeposition marking of new histones. Mol. Cell 2010, 37, 736–743. [Google Scholar] [CrossRef] [PubMed]
- Loyola, A.; Bonaldi, T.; Roche, D.; Imhof, A.; Almouzni, G. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol. Cell 2006, 24, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Sarkies, P.; Reams, C.; Simpson, L.J.; Sale, J.E. Epigenetic instability due to defective replication of structured DNA. Mol. Cell 2010, 40, 703–713. [Google Scholar] [CrossRef] [PubMed]
- Zane, L.; Sharma, V.; Misteli, T. Common features of chromatin in aging and cancer: Cause or coincidence? Trends Cell Biol. 2014, 24, 686–694. [Google Scholar] [CrossRef] [PubMed]
- Oberdoerffer, P.; Sinclair, D.A. The role of nuclear architecture in genomic instability and ageing. Nat. Rev. Mol. Cell Biol. 2007, 8, 692–702. [Google Scholar] [CrossRef] [PubMed]
- Im, J.S.; Keaton, M.; Lee, K.Y.; Kumar, P.; Park, J.; Dutta, A. ATR checkpoint kinase and CRL1βTRCP collaborate to degrade ASF1a and thus repress genes overlapping with clusters of stalled replication forks. Genes Dev. 2014, 28, 875–887. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Klar, A.J. Mutations in deoxyribonucleotide biosynthesis pathway cause spreading of silencing across heterochromatic barriers at the mating-type region of the fission yeast. Yeast 2008, 25, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Dubarry, M.; Loiodice, I.; Chen, C.L.; Thermes, C.; Taddei, A. Tight protein-DNA interactions favor gene silencing. Genes Dev. 2011, 25, 1365–1370. [Google Scholar] [CrossRef] [PubMed]
- Shankaranarayana, G.D.; Motamedi, M.R.; Moazed, D.; Grewal, S.I. Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Curr. Biol. 2003, 13, 1240–1246. [Google Scholar] [CrossRef] [PubMed]
- Oberdoerffer, P.; Michan, S.; McVay, M.; Mostoslavsky, R.; Vann, J.; Park, S.K.; Hartlerode, A.; Stegmuller, J.; Hafner, A.; Loerch, P.; et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell 2008, 135, 907–918. [Google Scholar] [CrossRef] [PubMed]
- Law, M.J.; Lower, K.M.; Voon, H.P.; Hughes, J.R.; Garrick, D.; Viprakasit, V.; Mitson, M.; de Gobbi, M.; Marra, M.; Morris, A.; et al. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell 2010, 143, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Ratnakumar, K.; Bernstein, E. ATRX: The case of a peculiar chromatin remodeler. Epigenetics 2013, 8, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Lopes, J.; Piazza, A.; Bermejo, R.; Kriegsman, B.; Colosio, A.; Teulade-Fichou, M.P.; Foiani, M.; Nicolas, A. G-quadruplex-induced instability during leading-strand replication. EMBO J. 2011, 30, 4033–4046. [Google Scholar] [CrossRef] [PubMed]
- Lansdorp, P.M. Immortal strands? Give me a break. Cell 2007, 129, 1244–1247. [Google Scholar] [CrossRef] [PubMed]
- Saveliev, A.; Everett, C.; Sharpe, T.; Webster, Z.; Festenstein, R. DNA triplet repeats mediate heterochromatin-protein-1-sensitive variegated gene silencing. Nature 2003, 422, 909–913. [Google Scholar] [CrossRef] [PubMed]
- Narita, M.; Nunez, S.; Heard, E.; Lin, A.W.; Hearn, S.A.; Spector, D.L.; Hannon, G.J.; Lowe, S.W. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 2003, 113, 703–716. [Google Scholar] [CrossRef]
- Zhang, R.; Chen, W.; Adams, P.D. Molecular dissection of formation of senescence-associated heterochromatin foci. Mol. Cell Biol. 2007, 27, 2343–2358. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Poustovoitov, M.V.; Ye, X.; Santos, H.A.; Chen, W.; Daganzo, S.M.; Erzberger, J.P.; Serebriiskii, I.G.; Canutescu, A.A.; Dunbrack, R.L.; et al. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell 2005, 8, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Narita, M.; Krizhanovsky, V.; Nunez, S.; Chicas, A.; Hearn, S.A.; Myers, M.P.; Lowe, S.W. A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell 2006, 126, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Smith, O.K.; Aladjem, M.I. Chromatin structure and replication origins: Determinants of chromosome replication and nuclear organization. J. Mol. Biol. 2014, 426, 3330–3341. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khurana, S.; Oberdoerffer, P. Replication Stress: A Lifetime of Epigenetic Change. Genes 2015, 6, 858-877. https://doi.org/10.3390/genes6030858
Khurana S, Oberdoerffer P. Replication Stress: A Lifetime of Epigenetic Change. Genes. 2015; 6(3):858-877. https://doi.org/10.3390/genes6030858
Chicago/Turabian StyleKhurana, Simran, and Philipp Oberdoerffer. 2015. "Replication Stress: A Lifetime of Epigenetic Change" Genes 6, no. 3: 858-877. https://doi.org/10.3390/genes6030858





