The Impact of Weir Construction in Korea’s Nakdong River on the Population Genetic Variability of the Endangered Fish Species, Rapid Small Gudgeon (Microphysogobio rapidus)
Abstract
:1. Introduction
2. Materials and Methods
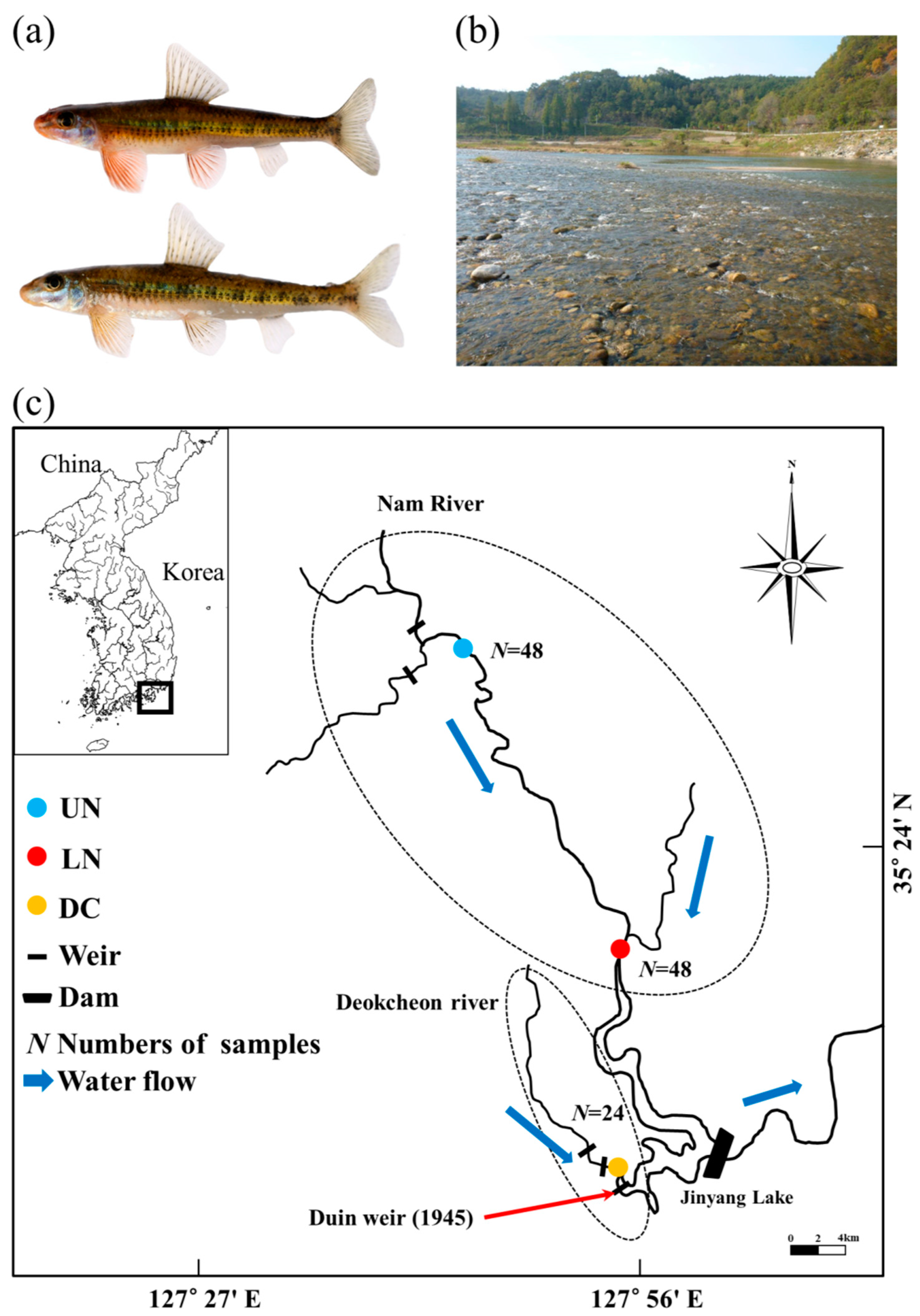
2.1. Sample Collection
2.2. Genomic DNA Extraction
2.3. Microsatellite Marker Development
2.3.1. DNA Cutting and Ligation
2.3.2. Enrichment of Microsatellite DNA Libraries
2.3.3. Cloning and Selection
2.4. PCR and Genotyping
2.5. Genetic Diversity and Structure Analysis
3. Results
3.1. Development of Microsatellite Markers
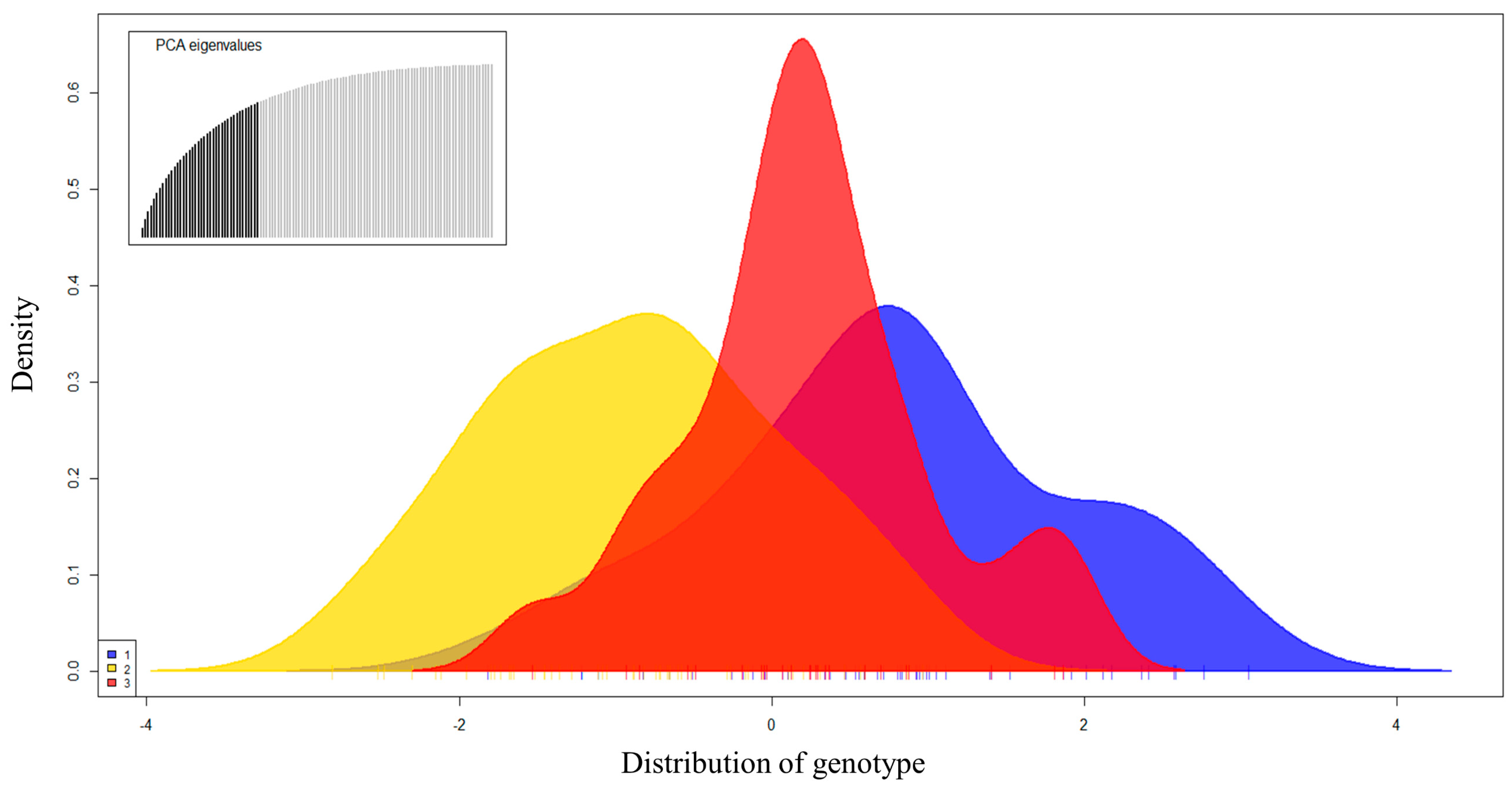
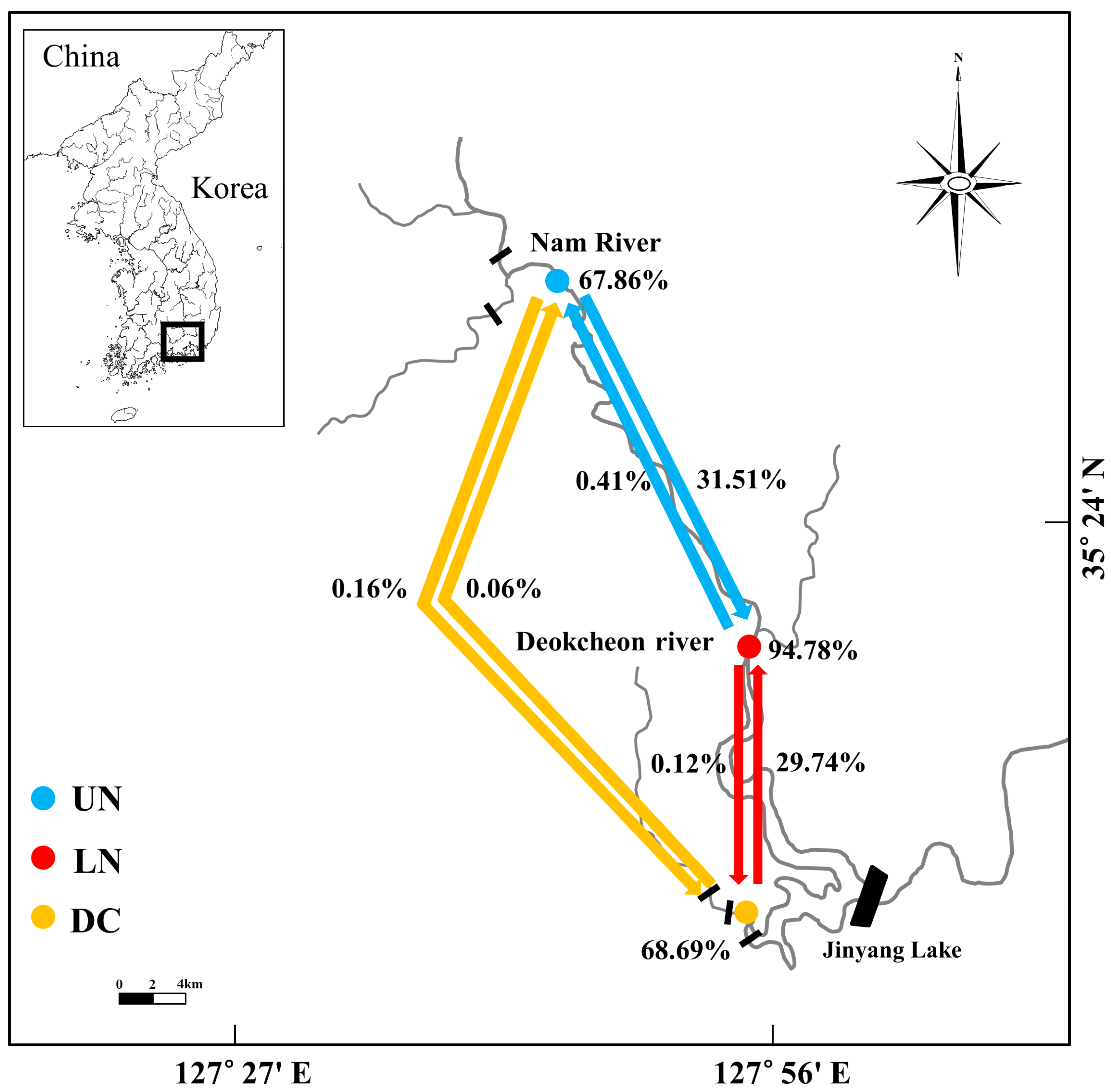
3.2. Genetic Diversity and Population Structure
4. Discussion
5. Conservation Implications
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Hedrick, P.W.; Miller, P.S. Conservation genetics: Techniques and fundamentals. Ecol. Appl. 1992, 2, 30–46. [Google Scholar] [CrossRef] [PubMed]
- Northcote, D.H. Aspects of vascular tissue differentiation in plants: Parameters that may be used to monitor the process. Int. J. Plant Sci. 1995, 156, 245–256. [Google Scholar] [CrossRef]
- Meldgaard, T.; Nielsen, E.E.; Loeschcke, V. Fragmentation by weirs in a riverine system: A study of genetic variation in time and space among populations of European grayling (Thymallus thymallus) in a Danish river system. Conserv. Genet. 2003, 4, 735–747. [Google Scholar] [CrossRef]
- Yamamoto, S.; Morita, K.; Koizumi, I.; Maekawa, K. Genetic differentiation of white-spotted charr (Salvelinus leucomaenis) populations after habitat fragmentation: Spatial-temporal changes in gene frequencies. Conserv. Genet. 2004, 5, 529–538. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Yamano, A.; Oura, K. Recent microscale disturbance of gene flow in threatened fluvial lamprey, Lethenteron sp. N, living in a paddy water system. Conserv. Genet. 2011, 12, 1373–1377. [Google Scholar] [CrossRef]
- Frankham, R.; Ballou, J.D.; Briscoe, D.A.A. Primer of Conservation Genetics; Cambridge University Press: Cambridge, UK, 2004. [Google Scholar]
- Chae, B.S.; Yang, H.J. Microphysogobio rapidus, a new species of gudgeon (Cyprinidae, Pisces) from Korea, with revised key to species of the genus Microphysogobio from Korea. Korean J. Biol. Sci. 1999, 3, 17–21. [Google Scholar] [CrossRef]
- Kim, I.S.; Yang, H. A revision of the genus Microphysogobio in Korea with description of a new species. Korean J. Ichthyol. 1999, 11, 1–11. [Google Scholar]
- NIBR (National Institute of Biological Resources). Red Data Book of Endangered Fishes in Korea; National Institute Of Biological Resources: Incheon, Republic of Korea, 2011; pp. 1–202. [Google Scholar]
- ME (Ministry of Environment). Conservation and Management Laws of Wildlife (Law No. 10977). 2012. Available online: https://elaw.klri.re.kr/eng_mobile/ganadaDetail.do?hseq=37570&type=abc&key=WILDLIFE%20PROTECTION%20AND%20MANAGEMENT%20ACT¶m=W (accessed on 1 January 2018).
- Chae, B.S.; Kang, Y.H.; Lee, Y.H. Ecological Report of Nakdong River: The Ichthyofauna and Structure of Fish Community around the Nakdong River System Korea; Youngnam Man and Nature Society, 1996; pp. 287–402. [Google Scholar]
- Hong, Y.K.; Sung, H.C.; Ko, M.H.; Kim, K.S.; Bang, I.C. Distribution status and habitat characteristics of the endangered freshwater fish, Microphysogobio rapidus (Cyprinidae). Anim. Cells Syst. 2017, 21, 286–293. [Google Scholar] [CrossRef]
- Ministry of Environment (ME). 1997–2008. National Environment Investigation of Freshwater Fish; Ministry of Environment (ME): Sejong, Republic of Korea, 1997. [Google Scholar]
- Kim, K.Y.; Bang, I.C. Molecular phylogenetic position of Abbottina springeri (Cypriniformes; Cyprinidae) based on nucleotide sequences of RAG1 gene. Korean J. Ichthyol. 2010, 22, 273–278. [Google Scholar]
- Hong, Y.K.; Ko, M.H.; Park, S.Y.; Bang, I.C. Egg Development and Early Life History of the Endangered Species, Microphysogobio rapidus (Cyprinidae). Korean J. Ichthyol. 2015, 27, 86–94. [Google Scholar]
- Kim, K.R.; Bang, I.C. Complete mitochondrial genome of the endangered freshwater fish Microphysogobio rapidus (Cypriniformes, Cyprinidae) from Korea. Mitochondrial DNA Part B Resour. 2020, 5, 447–448. [Google Scholar] [CrossRef] [PubMed]
- Karp, A.; Seberg, O.L.E.; Buiatti, M. Molecular techniques in the assessment of botanical diversity. Ann. Bot. 1996, 78, 143–149. [Google Scholar] [CrossRef]
- Kwak, Y.H.; Kim, K.R.; Kim, M.S.; Bang, I.C. Genetic diversity and population structure of the endangered fish Pseudobagrus brevicorpus (Bagridae) using a newly developed 12-microsatellite marker. Genes Genom. 2020, 42, 1291–1298. [Google Scholar] [CrossRef]
- Kaczmarczyk, D. Techniques based on the polymorphism of microsatellite DNA as tools for conservation of endangered populations. Appl. Ecol. Environ. Res. 2019, 17, 1599–1615. [Google Scholar] [CrossRef]
- Asahida, T.; Kobayashi, T.; Saitoh, K.; Nakayama, I. Tissue preservation and total DNA extraction form fish stored at ambient temperature using buffers containing high concentration of urea. Fish. Sci. 1996, 62, 727–730. [Google Scholar] [CrossRef]
- Hamilton, M.B.; Pincus, E.L.; Di Fiore, A.; Fleischer, R.C. Universal linker and ligation procedures for construction of genomic DNA libraries enriched for microsatellites. Biotechniques 1999, 27, 500–507. [Google Scholar] [CrossRef]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 1999, 132, 365–386. [Google Scholar] [CrossRef]
- Kim, K.S. Studies on Conservation Genetics of Endangered Gobioninae Fishes in Korea; Soonchunhyang University Press: Chungnam, Republic of Korea, 2012. [Google Scholar]
- Van Oosterhout, C.; Hutchinson, W.; Willds, D.; Shipley, P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Marshall, T.C.; Slate, J.; Kruuk, L.E.B.; Pemberton, J.M. Statistical confidence for likelihood-based paternity inference in natural populations. Mol. Ecol. 1998, 7, 639–655. [Google Scholar] [CrossRef]
- Goudet, J. FSTAT version 1.2: A computer program to calculate F-statistics. J. Hered. 1995, 86, 485–486. [Google Scholar] [CrossRef]
- Raymond, M.; Rousset, F. GENEPOP (ver. 1.2): Population genetics software for exact tests and ecumenicism. J. Hered. 1995, 86, 248–249. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Waples, R.S.; Do, C. LDNE: A program estimating effective population size from data on linkage disequilibrium. Mol. Ecol. 2008, 8, 753–756. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 89, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.P. Tools for Population Genetic Analysis; Version 1.3; Northern Arizona University: Flagstaff, AZ, USA, 1997. [Google Scholar]
- Wilson, G.A.; Rannala, B. Bayesian inference of recent migration rates using multilocus genotypes. Genetics 2003, 163, 1177–1191. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Jombart, T. adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef]
- Escalona, T.; Engstrom, T.N.; Hernandez, O.E.; Bock, B.C.; Vogt, R.C.; Valenzuela, N. Population genetics of the endangered south american freshwater turtle, Podocnemis unifilis, inferred from microsatellite DNA data. Conserv. Genet. 2009, 10, 1683–1696. [Google Scholar] [CrossRef]
- Frankham, R.; Bradshaw, C.J.; Brook, B.W. Genetics in conservation management: Revised recommendations for the 50/500 rules, Red List criteria and population viability analyses. Biol. Conserv. 2014, 170, 56–63. [Google Scholar] [CrossRef]
- Palstra, F.P.; Ruzzante, D.E. Genetic estimates of contemporary effective population size: What can they tell us about the importance of genetic stochasticity for wild population persistence? Mol. Ecol. 2008, 17, 3428–3447. [Google Scholar] [CrossRef]
- Sharma, S.P.; Ghazi, M.G.; Katdare, S.; Dasgupta, N.; Mondol, S.; Gupta, S.K.; Hussain, S.A. Microsatellite analysis reveals low genetic diversity in managed populations of the critically endangered gharial (Gavialis gangeticus) in India. Sci. Rep. 2021, 11, 5627. [Google Scholar] [CrossRef] [PubMed]


| Locus | Motif Repeats | Primers (5′ → 3′) | Ta (°C) | Label | References |
|---|---|---|---|---|---|
| PNms172 | (GT)11 | F: TCAACCTTGTGAGTGTATGTGAGGA R: GGCGAGACACAGGCCTCTTA | 56 | HEX | [23] |
| MKms205 | (CA)12 | F: TCAGCAGGTCCACAGCTTGC R: CAGTGAGCTCCCATTTACTGTGAC | 58 | NED | |
| MKms633 | (CA)16 | F: TCGTAATTTACCCCAGCACAACA R: TTCATTTCGCCACCGAAAAA | 58 | 6-FAM | |
| MKms747 | (CA)19 | F: CGAAATAATCCGCTCCCTGG R: CCCACAGGACTTTCCCTCTTG | 58 | HEX | |
| GBms157 | (CA)6 (CA)16 | F: GGAGTTATGAACGATAGAGCAGAC R: GAGCCTCATCAGCGACAACG | 58 | 6-FAM | |
| GBms481 | (GT)13 | F: TGCTGAGCGAGAGAAGCAAT R: CCCAGACTAACACCTCATTTTTATG | 58 | 6-FAM | |
| MRms245-1 * | (GT)6 | F: GGTTACTATAAATCTCTGGTGTTACGC R: CCATCTGAGCCACGGTGAAG | 58 | 6-FAM | Present study |
| MRms245-2 * | (GT)14 | F: TCGATGCCTACGTGGAGGTC R: ATCATCAGCGTCCGCTCGTA | 58 | HEX | |
| MRms245-3 * | (GT)4 (GT)8 | F: AGATGCTCCGACAGATGCGT R: CGCTTCAGAATGAGCCCAGA | 58 | NED | |
| MRms637 * | (GT)21 | F: TGTGAGTTGAGTGCTAACGCTTG R: TCACAAGAGTGAAGGGGTGAATC | 58 | 6-FAM |
| Locus | Subpopulation | All | |||
|---|---|---|---|---|---|
| UN | LN | DC | |||
| PNms172 | n | 48 | 48 | 24 | 120 |
| A | 3 | 3 | 2 | 4 | |
| HO | 0.208 | 0.250 | 0.083 | 0.200 | |
| HE | 0.225 | 0.223 | 0.082 | 0.196 | |
| PHWE | 0.017 * | 1.000 | 1.000 | 0.229 | |
| FIS | 0.077 | −0.120 | −0.022 | −0.022 | |
| MKms205 | N | 48 | 46 | 24 | 118 |
| A | 7 | 6 | 7 | 8 | |
| HO | 0.813 | 0.565 | 0.792 | 0.712 | |
| HE | 0.702 | 0.686 | 0.771 | 0.708 | |
| PHWE | 0.531 | 0.172 | 0.385 | 0.349 | |
| FIS | −0.159 | 0.178 | −0.027 | −0.003 | |
| MKms633 | N | 48 | 48 | 24 | 120 |
| A | 21 | 22 | 17 | 25 | |
| HO | 0.896 | 0.958 | 0.875 | 0.917 | |
| HE | 0.938 | 0.921 | 0.927 | 0.932 | |
| PHWE | 0.386 | 0.417 | 0.199 | 0.332 | |
| FIS | 0.045 | −0.041 | 0.058 | 0.021 | |
| MKms747 | N | 48 | 48 | 24 | 120 |
| A | 16 | 15 | 10 | 16 | |
| HO | 0.938 | 0.833 | 0.792 | 0.867 | |
| HE | 0.922 | 0.894 | 0.812 | 0.900 | |
| PHWE | 0.099 | 0.009 ** | 0.701 | 0.023 * | |
| FIS | −0.017 | 0.068 | 0.026 | 0.026 | |
| MRms245-1 | N | 47 | 46 | 22 | 115 |
| A | 24 | 28 | 14 | 36 | |
| HO | 0.660 | 0.674 | 0.864 | 0.704 | |
| HE | 0.946 | 0.945 | 0.850 | 0.944 | |
| PHWE | 0.000 *** | 0.000 *** | 0.389 | 0.000 *** | |
| FIS | 0.305 | 0.289 | −0.017 | 0.192 | |
| MRms245-2 | N | 48 | 47 | 24 | 119 |
| A | 19 | 18 | 13 | 22 | |
| HO | 0.833 | 0.894 | 0.792 | 0.849 | |
| HE | 0.888 | 0.907 | 0.879 | 0.900 | |
| PHWE | 0.079 | 0.922 | 0.138 | 0.163 | |
| FIS | 0.062 | 0.015 | 0.102 | 0.060 | |
| MRms245-3 | n | 48 | 48 | 23 | 119 |
| A | 28 | 28 | 14 | 34 | |
| HO | 0.917 | 0.958 | 0.957 | 0.941 | |
| HE | 0.930 | 0.932 | 0.895 | 0.925 | |
| PHWE | 0.518 | 0.049 * | 0.072 | 0.050 | |
| MRms637 | n | 48 | 46 | 24 | 118 |
| A | 24 | 22 | 12 | 32 | |
| HO | 0.792 | 0.761 | 0.917 | 0.805 | |
| HE | 0.897 | 0.846 | 0.895 | 0.880 | |
| PHWE | 0.194 | 0.270 | 0.174 | 0.152 | |
| FIS | 0.119 | 0.101 | −0.024 | 0.065 | |
| GBms157 | n | 48 | 48 | 24 | 120 |
| A | 36 | 36 | 24 | 44 | |
| HO | 0.896 | 0.917 | 0.875 | 0.900 | |
| HE | 0.966 | 0.971 | 0.961 | 0.967 | |
| PHWE | 0.013 * | 0.064 | 0.024 * | 0.001 ** | |
| FIS | 0.074 | 0.057 | 0.091 | 0.074 | |
| GBms481 | n | 48 | 48 | 24 | 120 |
| A | 5 | 6 | 5 | 6 | |
| HO | 0.667 | 0.667 | 0.708 | 0.675 | |
| HE | 0.678 | 0.697 | 0.681 | 0.685 | |
| PHWE | 0.970 | 0.243 | 0.119 | 0.308 | |
| FIS | 0.017 | 0.044 | −0.041 | 0.007 | |
| Overall | A | 18.3 | 18.4 | 11.8 | 22.7 |
| AR (n = 22) | 13.8 | 13.8 | 11.5 | 13.7 | |
| HO | 0.762 | 0.748 | 0.766 | 0.757 | |
| HE | 0.809 | 0.802 | 0.775 | 0.804 | |
| PHWE | 0.000 *** | 0.000 *** | 0.041 * | 0.000 *** | |
| FIS | 0.056 ** | 0.053 ** | 0.000 | 0.044 ** | |
| Subpopulation | BOTTLENECK Tests | Ne (95% CI) | M-Ratio | |||
|---|---|---|---|---|---|---|
| PIAM | PTPM | PSMM | Mode-Shift | |||
| UN | 0.009 ** | 0.903 | 0.958 | L-shaped | 175 (93–804) | 0.355 |
| LN | 0.065 | 0.998 | 0.999 | L-shaped | 157 (79–1077) | 0.372 |
| DC | 0.016 * | 0.935 | 0.998 | L-shaped | 61 (29–571) | 0.341 |
| UN | LN | DC | |
|---|---|---|---|
| UN | - | 0.026 | 0.087 |
| LN | 0.001 | - | 0.082 |
| DC | 0.014 *** | 0.013 *** | - |
| Source of Variation | d.f. | Sum of Squares | Variance Components | Total Variance (%) | F-Statistics |
|---|---|---|---|---|---|
| Microsatellite Nam River region vs. Deokcheon River region (UN, LN vs. DC) | |||||
| Among groups | 1 | 7.790 | 0.04772 | 1.20 | FCT = 0.012 |
| Among populations within groups | 1 | 4.260 | 0.00353 | 0.09 | FSC = 0.001 |
| Within populations | 237 | 929.396 | 3.92150 | 98.71 | FST = 0.013 *** |
| Total | 239 | 941.446 | 3.97275 | 100.00 | |
| Nakdong River water system (UN, LN, DC) | |||||
| Among groups | 2 | 12.050 | 0.02739 | 0.69 | FST = 0.007 *** |
| Within populations | 237 | 929.396 | 3.92150 | 99.31 | |
| Total | 239 | 941.446 | 3.94889 | 100.00 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, Y.-K.; Kim, K.-R.; Kim, K.-S.; Bang, I.-C. The Impact of Weir Construction in Korea’s Nakdong River on the Population Genetic Variability of the Endangered Fish Species, Rapid Small Gudgeon (Microphysogobio rapidus). Genes 2023, 14, 1611. https://doi.org/10.3390/genes14081611
Hong Y-K, Kim K-R, Kim K-S, Bang I-C. The Impact of Weir Construction in Korea’s Nakdong River on the Population Genetic Variability of the Endangered Fish Species, Rapid Small Gudgeon (Microphysogobio rapidus). Genes. 2023; 14(8):1611. https://doi.org/10.3390/genes14081611
Chicago/Turabian StyleHong, Yang-Ki, Kang-Rae Kim, Keun-Sik Kim, and In-Chul Bang. 2023. "The Impact of Weir Construction in Korea’s Nakdong River on the Population Genetic Variability of the Endangered Fish Species, Rapid Small Gudgeon (Microphysogobio rapidus)" Genes 14, no. 8: 1611. https://doi.org/10.3390/genes14081611






