Abstract
The Yangtze River Delta white goat is a rare goat species capable of producing high-quality brush hair. Dual specificity protein phosphatase 1 (DUSP1) may play a role in the formation of high-quality brush hair, as evidenced by our previous research. We investigated the potential mechanisms that regulate the proliferation and apoptosis of goat hair follicle stem cells. We particularly focused on the relationship between DUSP1 and miR-101, which directly targets DUSP1, predicted and screened through bioinformatics websites. Then, fluorescence assays, flow cytometry, RT-qPCR, and Western blotting were used to investigate the effects of miR-101 on the proliferation and apoptosis of hair follicle stem cells. We found that miR-101 overexpression significantly decreased (p < 0.01) apoptosis and promoted the proliferation of hair follicle stem cells. Furthermore, the overexpression of miR-101 increased (p < 0.05) the mRNA and protein expression levels of the proliferation-related gene (PCNA) and anti-apoptotic gene (Bcl-2), and it decreased (p < 0.05) the mRNA and protein expression levels of the apoptotic gene (Bax). In conclusion, miR-101 can promote the proliferation of and inhibit the apoptosis of hair follicle stem cells by targeting DUSP1, which provides a theoretical basis for further elucidating the molecular mechanism that regulates the production of high-quality brush hair of Yangtze River Delta white goats.
1. Introduction
The Yangtze River Delta white goat, indigenous to China, produces high-quality brush hair, which is a unique raw material that is used to make top-grade writing brushes. Its unique superior-quality brush hair that is obtained mainly from the neck and ridge of the goat is valuable for many excellent characteristics, including its white color, straight peak, fine luster and elasticity [1]. Yangtze River Delta white goat brush hair of high quality, with prices ranging from 2500–3000 USD per kilogram, is popular in Japan, Korea, Singapore and other Southeast Asian nations influenced by Chinese culture, and it is typically a traditional Chinese export commodity [2]. The production of high-quality brush hair can be traced to the characteristic development of the development of hair follicles. The growth and development of hair follicles affect not only the growth, but also the quality of hair. Hair follicle development is a cyclic process, involving three stages: anagen, catagen and telogen, and it involves dynamic changes in gene regulation [3]. Dual-specificity phosphatase 1 (DUSP1), a two-way specific Sue/tyrosinase, is involved in inactivating different MAPK family isoforms [4], and it plays an indispensable role in cell cycle arrest and cell proliferation in tumor and normal cells [5,6]. DUSP1 has been recognized to play a role in tumorigenesis and tumor progression, as well as the proliferation, differentiation and apoptosis of normal cells [7]. Interestingly, in our previous study, we found that DUSP1 is significantly up-regulated in high-quality brush hair than in non-high-quality brush hair follicle cells [8,9]. Therefore, we hypothesized that DUSP1 may play an important role in the regulation of the growth and development of hair follicles, and consequently, of hair quality as well.
MicroRNAs are short (20–24 nt) single-stranded, non-coding RNAs that bind to the 3′-UTR of target mRNAs, translation repression or degradation of target mRNAs, resulting in the silencing of the target gene [10]. Studies have demonstrated that miRNAs participate in a variety of physiological processes, including proliferation, cell differentiation, apoptosis and even tissue and organogenesis development [11,12]. To date, several studies have shown that many miRNAs, such as miR-218-5p [13], miR-let7a [14] and miR-214 [15], are essential for hair follicle development and regeneration in skin tissue. MiR-101, encoded by two precursor transcripts (miR-101-1 and miR-101-2), is considered a tumor suppressor, as it targets mRNAs of critical oncogenes or anti-oncogenes, and its loss is associated with the occurrence and progression of various diseases [16]. It was manifested that miR-101 tends to inhibit cell proliferation by downregulating the expression of the enhancers of zeste homolog 2 (EZH2) in lung cancer [17], bladder transitional cell carcinoma [18] and embryonal rhabdomyosarcoma [19]. Furthermore, the study of Wang et al. revealed that the upregulation of miR-101-3p promotes apoptosis and inhibits the viability of oral cancer cells [20]. However, the function and mechanism of goat miR-101 in goat HFSCs in the Yangtze River Delta white goat have rarely been explored. Therefore, in this study, we explored the relationship between miR-101 and DUSP1, particularly focusing on their role in the production of superior-quality hair in Yangtze River Delta white goats.
2. Materials and Methods
2.1. Isolation and Culture of Cells
Hair follicle stem cells (HFSCs) from Yangtze River Delta white goats were extracted from the skin of the neck-spine of newborn lambs 120 days after birth at the Haimen State Goat Farm (Haimen City, Jiangsu Province, China), which was described and verified in our previous study [21]. Briefly, approximately 2 cm2 of skin was separated from newborn lambs and soaked in 75% alcohol for 30 s, followed by ringing in phosphate-buffered saline (PBS) three times. Next, the tissue blocks were cut into 2 × 2 mm pieces and were digested with 0.25% trypsin at 37 °C for 90 min. Next, the hair follicles were isolated and digested for 30 min. Eventually, HFSCs were obtained through crushing and sieving, and they were subsequently verified by confirming CD49f and CD34 double [22]. The sorted HFSCs and HEK293T cells were individually cultivated in 6-well plates (Corning, New York, NY, USA) with Dulbecco’s modified Eagle’s medium (DMEM/F12), supplemented with 10% fetal bovine serum (FBS) and 2% penicillin–streptomycin at 37 °C in a 5% CO2 atmosphere.
2.2. Prediction of miRNA Targeting DUSP1
TargetScan (http://www.targetscan.org/vert_72, accessed on 1 October 2021), RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/, accessed on 1 October 2021) and miRDB software were employed to predict miRNAs and their binding regions that could target DUSP1, and eventually, miR-101 was selected for further research.
2.3. Synthesis of Primers
According to the NCBI nucleic acid database (https://www.ncbi.nlm.nih.gov/nuccore/, accessed on 1 October 2021), the CDS and 3’-UTR of goat DUSP1 (registration number: XM_018065720.1) were obtained. All the primers were designed by Prime 5.0 software and the NCBI Primer-BLAST online website. GAPDH expression was used as the internal control. The primers for quantitative PCR were synthesized by Shanghai Bioengineering Co., Ltd. and are shown in Table 1. The Wt-DUSP1-miR101 and Mut-DUSP1-miR101 plasmids were constructed by the Gene Pharma of China.

Table 1.
Quantitative real-time PCR primer pairs.
2.4. Double Luciferase Reporter Assay
The wild-type 3′-untranslated region (UTR) of DUSP1 mRNA with a putative miR-101 binding site was engineered into pmirGLO, a dual-luciferase miRNA target expression vector (Promega, Madison, WI, USA) doubly digested with SacI and XhoI to generate Wt-DUSP1-miR101 plasmid. The Mut-DUSP1-miR101 plasmid was constructed by displacing the binding sites of miR-101 from AGTACTGTA to CCGAAATTT. The recombinant plasmids identified by sequencing were called Wt-DUSP1 and Mut-DUSP1. Subsequently, 293T cells were plated in 24-well plates when the cells reached 60%~70% confluence, and they were co-transfected with miR-101 mimics or miR-NC and Wt-DUSP1 or Mut-DUSP1 plasmid for 24 h using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. After transfection, the relative luciferase activity was determined using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions.
2.5. Construction and Transfection of Plasmid
The oligonucleotides of the negative control (NC), the miR-101 mimics, Inhibitor-NC and miR-101 inhibitors were synthesized by Gene pharma (Gene pharma Co., Ltd. Suzhou, China). For cell transfections, transient transfections were then performed using lipofectamine 3000 (Cat: L3000015, Invitrogen, CA, USA) according to the instructions of the manufacturer. In brief, after the stem cells were cultured for 24 h and reached 60%–70% confluence, miR-101 overexpression and knockdown in HFSCs were accomplished by transfecting with 100 nM miR-101 mimics and 200 nM miR-101 inhibitors, respectively. Total RNA or protein was harvested 48 h following transfection.
2.6. Quantitative Real-Time PCR
Total RNA was extracted from cultured HFSCs using the Trizol kit (Cat: 9109, Takara, Tokyo, Japan). The concentration and purity of extracted RNA were verified using Nanodrop spectrophotometry (Thermo Scientific, Shanghai, China), and the RNA was subsequently stored at −80 °C. Next, cDNA was synthesized using the EasyScript One-step gDNA Removal and cDNA Synthesis SuperMix kit (Cat: AE311, Transgen, Beijing, China) in accordance with the manufacturer’s instructions. The synthesized cDNA as a template was then used for the real-time PCR quantification of mRNAs on an ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) in light of the instructions of PerfectStartTM Green qPCR SuperMix Kit (Cat: AQ602, Transgen, Beijing, China) and the relative gene expression levels computed by the 2−ΔΔCt method. The related primers in this study were synthesized by Sangon Biotech (Shanghai, China), which are shown in Table 1.
2.7. EdU Incorporation Assay
For evaluating the proliferation potential of goat HFSCs, the Cell-Light TM EdU cell proliferation kit (Cat: C103101-1, RiboBio, Guangzhou, China) was used. Briefly, HFSCs were seeded in a 24-well culture plate at a density of 1 × 105 cells/well and were transfected with miR-101 oligo-nucleotides (Table 2) for 6 h. Then, the cells were transferred into a complete medium containing 10 μM 5-ethynyl-2′-deoxyuridine (EdU) reagent and were incubated for 24 h at 37 °C and 5% CO2. Next, the cells were fixed with 4% paraformaldehyde and were incubated with 1 × Apollo567 solution, and the cell nuclei were stained with 1 × Hoechst33342 solution for 30 min in the dark. Eventually, the fluorescence intensity of cells was observed immediately under a Leica fluorescence microscope (Leica, Wetzlar, Germany), and the percentage of EdU-positive nuclei (red) relative to the blue fluorescent nuclei was calculated using the Image J software.

Table 2.
RNA oligonucleotide sequence information.
2.8. Apoptosis Assay
After transfection, the apoptosis of different treatment groups was analyzed according to the instructions of the Annexin V-FITC apoptosis detection kit (Cat: C1062M, Beyotime, Shanghai, China). Flow cytometry (FACSAria SORP, BD Biosciences, CA, USA) was used to detect apoptosis rate. Flowjo software was used for the apoptosis rate.
2.9. Western Blotting
Total proteins were extracted 48 h after transient transfections according to the manufacturer’s instructions of efficient RIPA tissue/cell lysis buffer (Cat: R0010, Solarbio, Shanghai, China) containing 1% PMSF (Cat: P0100, Solarbio, Beijing, China) and were subjected to Western blotting. In brief, 10 μg proteins was separated by 10% or 12% SDS-polyacrylamide gel electrophoresis. Next, the membranes were rinsed three times with 1 × TBST (Tris-buffered saline buffer supplemented with Tween 20) for 10 min each and were blocked with 5% skim milk (Sangon Biotech, Shanghai, China) for 1 h at room temperature. Subsequently, the membranes were incubated at 4 °C overnight with appropriate dilutions of specific primary antibodies, namely, proliferating cell nuclear antigen (PCNA) (MW: 29kDa, Abcam, Cambridge, United Kingdom, 1:1000 dilution), cyclin dependent kinase 1 (CDK1), (MW: 34 kDa, Abcam, Cambridge, United Kingdom, 1:1000 dilution), cyclin D2 (CCND2) (MW: 33 kDa, Abcam, Cambridge, United Kingdom, 1:1000 dilution), BCL2 associated X, apoptosis regulator (Bax) (MW: 21 kDa, Abcam, Cambridge, United Kingdom, 1:5000 dilution), BCL2 apoptosis regulator (Bcl-2) (MW: 26 kDa, Proteintech, Rosemont, IL, United States, 1:1000 dilution) and β-actin (MW: 42 kDa, Bioworld, Nanjing, China, 1:5000 dilution). After washing, the membranes were incubated for 1 h at room temperature with the second antibodies, namely, goat-specific anti-rabbit IgG and goat-specific anti-mouse IgG ((Bioworld, Nanjing, China, 1:5000 dilution)). The immunoreactive bands were visualized using an ECL system (Biosharp, Hefei, China) and were analyzed using a FluorChem FC3 system (Protein-Simple, CA, USA).
2.10. Statistical Analysis
All data in this study were obtained from at least three independent experiments and are presented as the mean ± standard error (Mean ± SEM). The SPSS 24.0 (SPSS, Inc., Chicago, IL, USA) statistical software was used for one-way ANOVA analysis and independent-samples t-tests. The graphs were drawn using the GraphPad Prism 7.0 software. Differences are considered statistically significant when shown with an asterisk character. * means that p < 0.05 and that the difference is significant; ** means that p < 0.01 and that the difference is extremely significant.
3. Results
3.1. DUSP1 Is a Direct Target of miR-101
To confirm the biological regulation of miR-101 and DUSP1, it was discovered that miR-101 targets DUSP1 via a conserved region inside the 3′-UTR of DUSP1 (Figure 1A) using algorithm prediction. Subsequently, we verified the relationship between miR-101 and DUSP1 using a luciferase activity assay. As shown in Figure 1B and Figure S1, luciferase activity was significantly reduced in 293T cell lines transfected with miR-101 mimics compared to those transfected with miR-NC in Wt-DUSP1 plasmid (p < 0.05), whereas the difference was not significant in Mut-DUSP1 plasmid(Figure S2). These results confirm that miR-101 targets DUSP1 in hair follicle stem cells to regulate their function.
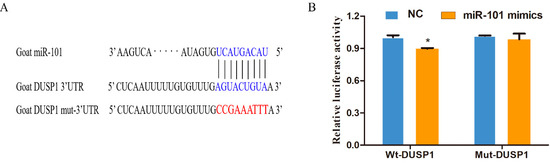
Figure 1.
MiR-101 can directly target the 3′UTR of the DUSP1 gene in hair follicle stem cells: (A) The predicted binding sites of miR-101 and DUSP1; the bases marked with blue represent the wild-type, and those in red represent the mutant-type; (B) Detection of dual-luciferase activity after co-transfection of NC or miR-101 with wild or mutant-type DUSP1. No asterisk, p > 0.05; *, p < 0.05.
3.2. Effect of miR-101 on the Proliferation of HFSCs
The proliferation potential of Goat HFSCs was determined using an EdU assay. Compared to the NC counterparts, the overexpression of miR-101 (miR-101 mimic-treated group) significantly enhanced the ratio of EdU-positive stem cells (p < 0.05), whereas the proportion was remarkably decreased after inhibiting the expression of miR-101 (miR-101 inhibitor-treated group) compared with inhibitor NC counterpart (p < 0.05), as shown in Figure 2B,C. The results above indicate that the overexpression of miR-101 can significantly promote the proliferation of hair follicle stem cells.
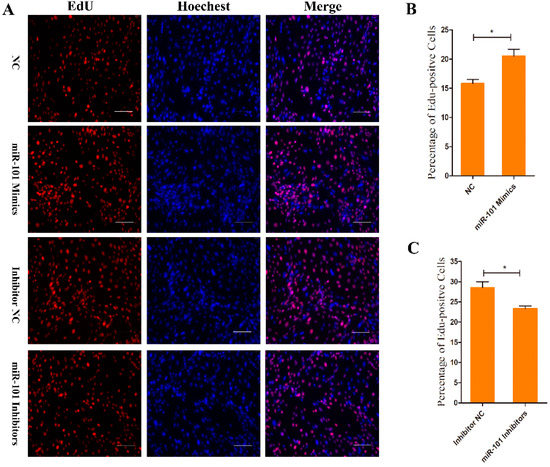
Figure 2.
EdU assay of HFSCs proliferation after transfection with NC, goat miR-101 mimics, inhibitor NC and goat miR-101 inhibitors: (A) Representative images after EdU assay of HFSCs at 24 h after transfection with NC, goat miR-101 mimics, inhibitor NC and goat miR-101 inhibitors. Bars = 100 μm; (B,C) Quantification of EdU-positive cells. The proportion of EdU-positive cells was calculated as (EdU-positive cells/Hoechst-stained cells) × 100%. No asterisk, p > 0.05; *, p < 0.05.
3.3. Effect of miR-101 on the Apoptosis of HFSCs
The Annexin V-FITC and PI double staining assay was performed to quantify HFSC apoptosis. The proportion of the specific cell population at various treatment groups is represented in Figure 3. As the result show, compared to the NC group, upregulating the expression of miR-101 can significantly increase the rate of living cell (98.5% vs. NC: 96.2%) and decrease the total apoptosis percentage (1.25% vs. NC: 3.57%) (p < 0.01) (Figure 3A,B). However, the total apoptosis rate of the inhibitor group (4.76%) was significantly higher than that of the inhibitor NC group (2.34%) (p < 0.05), and the living cell rate of the inhibitors group (96.1%) significantly declined (p < 0.05) compared with the inhibitor NC group (97.4%) (Figure 3C,D). The results above indicate that overexpression of miR-101 can significantly inhibit the apoptosis of hair follicle stem cells.
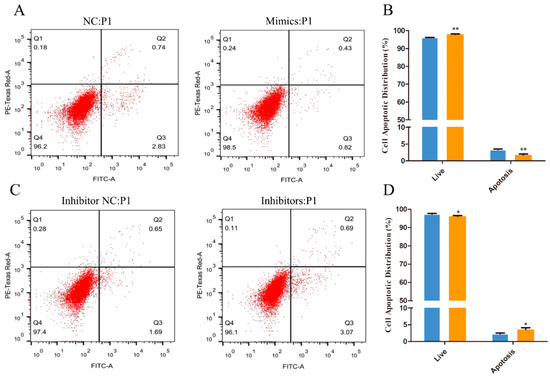
Figure 3.
The Annexin V-FITC/PI assay was performed to quantify HFSC apoptotic distribution: (A,B) The apoptosis levels of the cells transfected with NC and miR-101 mimics were tested by Annexin V-FITC/PI kit and flow cytometry; (C,D) The apoptosis levels of the cells transfected with inhibitor NC and miR-101 inhibitors were tested by Annexin V-FITC/PI kit and flow cytometry. Note: NC: negative control; Mimics: goat miR-101 mimic; INC: inhibitor NC; IN, inhibitor: goat miR-101 inhibitors. No asterisk, p > 0.05; *, p < 0.05; **, p < 0.01.
3.4. Effect of miR-101 on the Expression Levels of Proliferation and Apoptosis-Related Genes and Proteins
To investigate the effect of miR-101 on the expression levels of genes related to the proliferation and apoptosis of HFSCs, the expression levels of the proliferation-related genes (PCNA, CDK1 and CCND2) and apoptosis-related genes (Bax and Bcl2) were determined by RT-qPCR and Western blotting. As shown in Figure 4A, compared with the negative control group, the overexpression of miR-101 significantly decreased (p < 0.05) the mRNA expression of its target gene DUSP1, and the miR-101 mimic treatment significantly increased (p < 0.01) the mRNA expression levels of proliferation-related genes (PCNA and CDK1) and significantly decreased (p < 0.01) the mRNA expression of CCND2. Moreover, the overexpression of miR-101 led to a significant decline (p < 0.05) in the expression levels of pro-apoptotic genes (Bax) and an obvious augmentation (p < 0.05) of the expression levels of anti-apoptotic genes (Bcl-2). Combined with the Western blot test, the impact of miR-101 on proliferation and apoptosis-related protein expression was further determined. As shown in Figure 4B,C, compared with the control group, the relative protein expression of proliferation-related genes (PCNA) in the miR-101 mimic group was significantly enhanced (p < 0.05), and the relative protein expression of CDK1 tended to rise despite the lack of a difference. Moreover, concerning another proliferation-related gene (CCND2), its expression level was lower in the enhanced miR-101 treatment than it was in the negative control. Furthermore, the relative expression of pro-apoptotic genes (Bax) was dramatically reduced (p < 0.05), but the relative expression of anti-apoptotic genes (Bcl-2) was significantly raised (p < 0.05).
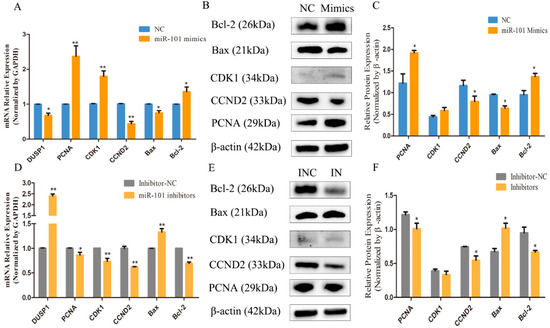
Figure 4.
Effect of miR-101 on the expression levels of proliferation and apoptosis-related genes and proteins: (A,D) The mRNA expression of DUSP1, proliferation-related genes (PCNA, CDK1 and CCND2) and apoptosis-related genes (Bax and Bcl2) at 48 h, after being transfected with NC, miR-101 mimics, inhibitor NC and miR-101 inhibitors, were determined by RT-qPCR. GAPDH was used as an internal control; (B,E) Protein expression of proliferation-related genes (PCNA, CDK1 and CCND2) and apoptosis-related genes (Bax and Bcl2) at 48 h, after being transfected with NC, mimics, INC and inhibitors, were determined by Western blotting; (C,F) The relative expression of related proteins from each group are shown as the mean ± SEM, which were obtained from at least three independent replicates (independent-samples t-tests). No asterisk, p > 0.05; *, p < 0.05; **, p < 0.01.
The results shown in Figure 4D reveal that the downregulation of the expression of miR-101 can significantly increase (p < 0.01) the mRNA expression of its target gene DUSP1 and can decrease (p < 0.05) the expression of the proliferation-related genes, PCNA, CDK1 and CCDN2, and the anti-apoptotic gene (Bcl-2), and it can greatly increase (p < 0.01) the expression of pro-apoptotic genes (Bax) and the Bax/Bcl-2 ratio compared with the negative control group. Furthermore, the results of the Western blot assay presented in Figure 4E,F manifest that the inhibition expression of miR-101 led to significantly decreasing the expression levels of proliferation-related genes (PCNA and CCDN2) and Bcl-2, and it significantly increased the expression levels of the Bax protein. In general, the results of RT-qPCR and Western blotting are consistent. Therefore, it can be speculated that the overexpression of miR-101 can enhance hair follicle stem cell growth and prevent hair follicle stem cell apoptosis.
4. Discussion
The results in our present study demonstrate that miR-101 promotes proliferation but inhibits the apoptosis of hair follicle stem cells. However, the mRNA and protein expression of CCND2, a proliferation-related gene in the cell cycle, tended to be decreased (p < 0.05). The cell cycle in most eukaryotic cells is a complex process, including a series of coordinated events, namely, preparation for cell growth (G1 phase), the replication of genetic material (S phase), the segregation of duplicated chromosomes (G2 phase) and cell division (M phase), which is regulated by cyclins, cyclin dependent kinases (CDKs) and CDK inhibitors [23]. The three D type cyclins (CCND1, CCND2 and CCND3) that modulate the cell cycle by engaging and activating CDKs, such as CDK4 and CDK6 and CDK-cyclin D complexes, are essential for G1 entrance [24]. Cyclin A combines with CDK1 in the late G2 and early M phase, and its protein levels stay constant throughout the cell cycle. Promoting entrance into mitosis is further governed by cyclin B in the complex with CDK1 [25,26]. These reports can verify our findings that enhanced miR-101 led to the reduction in the expression level of CCDN2 in the overexpression in the miR-101 treatment compared with the negative control group. It has been reported, furthermore, that miR-101 suppresses the expression of CCND2 in H. pylori related to gastric cancer [27], which is in accordance with our results.
DUSP1 as a phosphatase can dephosphorylate threonines and/or tyrosines of client proteins and inactivate mitogen-activated protein kinase kinase kinase 1 (MAP3K1), a member of the mitogen-activated protein kinase (MAPK) family and a signal transduction modulator to trigger MAPKs through dephosphorylation [28,29]. The activated MAP3K1 (also called p42/p44 MAPK) cascade promotes DUSP1 activity, which in turn attenuates MAP3K1-dependent events [30]. Thus, DUSP1 influences MAPK signaling and its targets through a negative feedback control mechanism [31,32]. Studies have shown that DUSP1 is specifically expressed, which may be associated with MAP3K1 phosphorylation [33], and reinforced DUSP1 expression results in the suppression of proliferation and altered cell apoptosis rates by suppressing the MAPK/ERK cascade, p-ERK, p-Elk-1 and Egr-1 [34].
Our previous results have manifested that some major genes are involved in the production of superior goat brush hair, including MAP3K1, CMTM3 (CKLF-like MARVEL transmembrane domain-containing family 3), DUSP1 (dual specificity protein phosphatase 1) and DUSP6 (dual specificity protein phosphatase 6) [35]. We also found that MAP3K1 expression was much higher in the skin tissue of superior-quality brush hair than their counterparts in normal-quality brush hair, and silencing MAP3K1 can greatly reduce proliferation and induce the death of superior goat hair follicle stem cells [36]. In this study, we confirmed that miR-101 can directly target DUSP1 by bioinformatics predictions and dual-luciferase reporter gene assays. Furthermore, the overexpression of miR-101 can facilitate proliferation and inhibit the apoptosis of hair follicle stem cells. Therefore, we postulate that miR-101 targets DUSP1 and reduces its expression, which, in turn, reduces the dephosphorylation of MAP3K1, and phosphorylated MAP3K1 serves as a positive regulator of proliferation and cell survival and inhibits the apoptosis of hair follicle stem cells in vitro. Furthermore, accumulating studies have illustrated that miR-101, as a negative regulator, can target DUSP1 and downregulate its expression [37,38,39,40].
Interestingly, previous high-throughput RNA-Seq results of our study have manifested that both MAP3K1 and DUSP1 are upregulated in skin samples of Yangtze River Delta white goats, which can generate high-quality hair compared with those that produce non-high-quality hair [8]. The potential molecular mechanisms behind this may likely include p-MAP3K1 upregulating the expression of DUSP1, which is vital for high-quality brush hair production. We will further explore the molecular mechanisms of DUSP1/MAP3K1 and how they regulate the production of superior-quality hair in Yangtze River Delta White in vitro and in vivo.
Taken together, this study identifies miR-101 as a positive regulator in the proliferation of hair follicle stem cells related to the production of high-quality brush hair, and downregulating miR-101 results in the inhibition of the proliferation and promotion of apoptosis of hair follicle stem cells. The above findings demonstrate that miR-101 may play an important role in regulating the formation of superior-quality brush hair traits in Yangtze River Delta white goats.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes13061035/s1, Figure S1: Sequence alignment analysis of the coding sequence of wild DUSP1 in the vector-matched coding sequence of DUSP1 in the DUSP1 database; Figure S2: Sequence alignment analysis of the coding sequence of Mut-DUSP1 in the vector-matched coding sequence of DUSP1 in the DUSP1 database.
Author Contributions
Y.L. and J.Q. conceived the idea and designed and performed the research. J.Q. designed and performed the research and drafted the figures and manuscript. X.W. collected and organized the investigation. Q.W. and J.W. collected the investigation and critically reviewed the manuscript. X.S. and D.J. critically reviewed the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (32172690 and 31572355), the Postgraduate Research and Practice Innovation Program of Jiangsu Province (SJCX19_0896) and the International Academic Exchange Special Foundation for Graduate students of Yangzhou University.
Institutional Review Board Statement
The animal study protocol was approved by the Institutional Ethics Committee of Yangzhou University (protocol code: SYXK [Su] IACUC2012-0029 and date of approval: 1 January 2012).
Acknowledgments
We would like to thank Bullet Edits Limited for their linguistic editing and proofreading of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- National livestock and Poultry Genetic Resources Committee. Chinese Livestock and Poultry Genetic Resources; China Agriculture Press: Beijing, China, 2011. [Google Scholar]
- Li, Y.; Huang, Y. The production of wool goat and writing brush in our country. China Herbiv. 2005, 25, 44–46. [Google Scholar]
- Gat, U.; DasGupta, L.; Fuchs, E. De Novo hair follicle morphogenesis and hair tumors in mice expressing a truncated β-catenin in skin. Cell 1998, 95, 605–614. [Google Scholar] [CrossRef]
- Theodosiou, A.; Ashworth, A. MAP kinase phosphatases. Genome Biol. 2002, 3, reviews3009.1. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Ling, Z.Q.; Ge, M.H. Expression of DUSP1 in salivary adenoid cystic carcinoma and its clinicopathological significance. Zhejiang Med. J. 2016, 38, 379–382. [Google Scholar]
- Celaya, A.M.; Sánchez-Pérez, I.; Bermúdez-Muñoz, J.M.; Rodríguez-de la Rosa, L.; Pintado-Berninches, L.; Perona, R.; Murillo-Cuesta, S.; Varela-Nieto, I. Deficit of mitogen-activated protein kinase phosphatase 1 (DUSP1) accelerates progressive hearing loss. eLife 2019, 8, e39159. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, Y.; Yu, H.; Shen, B.; Liang, Y.; Jin, R.; Liu, X.; Shi, L.; Cai, X. Role of DUSP1/MKP1 in tumorigenesis, tumor progression and therapy. Cancer Med. 2016, 5, 2061–2068. [Google Scholar] [CrossRef]
- Guo, H.; Cheng, G.; Li, Y. A Screen for Key Genes and Pathways Involved in High-Quality Brush Hair in the Yangtze River Delta White Goat. PLoS ONE 2017, 12, e0169820. [Google Scholar] [CrossRef]
- Li, Y.; Li, W.; Zhang, J.; Ji, D.; Zhang, G.; Yang, B. Identification of genes influencing formation of the Type III Brush Hair in Yangtze River Delta white goats by differential display of mRNA. Gene 2013, 526, 205–209. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef]
- Yi, R.; O’Caroll, D.; Pasolli, H.A.; Zhang, Z.; Dietrich, F.S.; Tarakhovsky, A.; Fuchs, E. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat. Genet. 2006, 38, 356–362. [Google Scholar] [CrossRef]
- Pal, A.S.; Kasinski, A.L. Animal Models to Study MicroRNA Function. Adv. Cancer Res. 2017, 135, 53–118. [Google Scholar] [PubMed]
- Zhao, B.; Chen, Y.; Yang, N.; Chen, Q.; Bao, Z.; Liu, M.; Hu, S.; Li, J.; Wu, X. miR-218-5p regulates skin and hair follicle development through Wnt/β-catenin signaling pathway by targeting SFRP2. J. Cell Physiol. 2019, 234, 20329–20341. [Google Scholar] [CrossRef]
- Ma, T.; Li, J.; Jiang, Q.; Wu, H.; Jiang, H.; Zhang, Q. Differential expression of miR-let7a in hair follicle cycle of Liaoning cashmere goats and identification of its targets. Funct. Integr. Genom. 2018, 18, 701–707. [Google Scholar] [CrossRef] [PubMed]
- Du, K.-T.; Deng, J.-Q.; He, X.-G.; Liu, Z.-p.; Peng, C.; Zhang, M.-S. MiR-214 Regulates the Human Hair Follicle Stem Cell Proliferation and Differentiation by Targeting EZH2 and Wnt/β-Catenin Signaling Way In Vitro. Tissue Eng. Regen. Med. 2018, 15, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, Y.; Wang, Y.; He, X.; Wang, J.; Cai, W.; Jia, Y.; Xiao, D.; Zhang, J.; Zhao, M.; et al. Overexpression of miR-101 suppresses collagen synthesis by targeting EZH2 in hypertrophic scar fibroblasts. Burn. Trauma 2021, 9, tkab038. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-g.; Guo, J.-F.; Liu, D.-L.; Liu, Q.; Wang, J.-J. MicroRNA-101 exerts tumor-suppressive functions in non-small cell lung cancer through directly targeting enhancer of zeste homolog 2. J. Thorac. Oncol. 2011, 6, 671–678. [Google Scholar] [CrossRef]
- Friedman, J.M.; Liang, G.; Liu, C.-C.; Wolff, E.M.; Tsai, Y.C.; Ye, W.; Zhou, X.; Jones, P.A. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009, 69, 2623–2629. [Google Scholar] [CrossRef]
- Vella, S.; Pomella, S.; Leoncini, P.P.; Colletti, M.; Conti, B.; Marquez, V.E.; Strillacci, A.; Roma, J.; Gallego, S.; Milano, G.M.; et al. MicroRNA-101 is repressed by EZH2 and its restoration inhibits tumorigenic features in embryonal rhabdomyosarcoma. Clin. Epigenetics 2015, 7, 82. [Google Scholar] [CrossRef]
- Wang, H.; Guo, Y.; Mi, N.; Zhou, L. miR-101-3p and miR-199b-5p promote cell apoptosis in oral cancer by targeting BICC1. Mol. Cell Probes 2020, 52, 101567. [Google Scholar] [CrossRef]
- Wang, Q.; Qu, J.; Li, Y.; Ji, D.; Zhang, H.; Yin, X.; Wang, J.; Niu, H. Hair follicle stem cells isolated from newborn Yangtze River Delta White Goats. Gene 2019, 698, 19–26. [Google Scholar] [CrossRef]
- Ouji, Y.; Yoshikawa, M.; Nishiofuku, M.; Ouji-Sageshima, N.; Kubo, A.; Ishizaka, S. Effects of Wnt-10b on proliferation and differentiation of adult murine skin-derived CD34 and CD49f double-positive cells. J. Biosci. Bioeng. 2010, 110, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Vermeulen, K.; Van Bockstaele, D.R.; Berneman, Z.N. The cell cycle: A review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif. 2003, 36, 131–149. [Google Scholar] [CrossRef] [PubMed]
- Sherr, C.J. G1 phase progression: Cycling on cue. Cell 1994, 79, 551–555. [Google Scholar] [CrossRef]
- King, R.W.; Jackson, P.K.; Kirschner, M.W. Mitosis in transition. Cell 1994, 79, 563–571. [Google Scholar] [CrossRef]
- Arellano, M.; Moreno, S. Regulation of CDK/cyclin complexes during the cell cycle. Int. J. Biochem. Cell Biol. 1997, 29, 559–573. [Google Scholar] [CrossRef]
- Zhou, X.; Xia, Y.; Li, L.; Zhang, G. MiR-101 inhibits cell growth and tumorigenesis of Helicobacter pylori related gastric cancer by repression of SOCS2. Cancer Biol. Ther. 2015, 16, 160–169. [Google Scholar] [CrossRef]
- Kraus, S.; Naor, Z.; Seger, R. Intracellular signaling pathways mediated by the gonadotropin-releasing hormone (GnRH) receptor. Arch. Med. Res. 2001, 32, 499–509. [Google Scholar] [CrossRef]
- Keyse, S.M. Protein phosphatases and the regulation of MAP kinase activity. Semin. Cell Dev. Biol. 1998, 9, 143–152. [Google Scholar] [CrossRef]
- Brondello, J.M.; Brunet, A.; Pouyssegur, J.; McKenzie, F.R. The dual specificity mitogen-activated protein kinase phosphatase-1 and -2 are induced by the p42/p44MAPK cascade. J. Biol. Chem. 1997, 272, 1368–1376. [Google Scholar] [CrossRef]
- Kondoh, K.; Nishida, E. Regulation of MAP kinases by MAP kinase phosphatases. Biochim. Biophys. Acta 2007, 1773, 1227–1237. [Google Scholar] [CrossRef]
- Farooq, A.; Zhou, M.M. Structure and regulation of MAPK phosphatases. Cell Signal 2004, 16, 769–779. [Google Scholar] [CrossRef] [PubMed]
- Purwana, I.N.; Kanasaki, H.; Mijiddorj, T.; Oride, A.; Miyazaki, K. Induction of dual-specificity phosphatase 1 (DUSP1) by pulsatile gonadotropin-releasing hormone stimulation: Role for gonadotropin subunit expression in mouse pituitary LbetaT2 cells. Biol. Reprod. 2011, 84, 996–1004. [Google Scholar] [CrossRef]
- Yang, J.; Sun, L.; Han, J.; Zheng, W.; Peng, W. DUSP1/MKP-1 regulates proliferation and apoptosis in keratinocytes through the ERK/Elk-1/Egr-1 signaling pathway. Life Sci. 2019, 223, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Ji, D.; Yang, B.; Li, Y.; Cai, M.; Zhang, W.; Cheng, G.; Guo, H. Transcriptomic inspection revealed a possible pathway regulating the formation of the high-quality brush hair in Chinese Haimen goat (Capra hircus). R. Soc. Open Sci. 2018, 5, 170907. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Wang, J.; Feng, Y.; Zhang, L.; Hu, H.; Wang, Q.; Chu, C.; Qu, J.; Wang, Y.; Li, Y. Silencing MAP3K1 expression inhibits the proliferation of goat hair follicle stem cells. Vitr. Cell. Dev. Biol. Anim. 2021, 57, 428–437. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, S.; Chu, Z.; Dang, Y.; Zhu, J.; Su, X. MicroRNA-101 in the ventrolateral orbital cortex (VLO) modulates depressive-like behaviors in rats and targets dual-specificity phosphatase 1 (DUSP1). Brain Res. 2017, 1669, 55–62. [Google Scholar] [CrossRef]
- Xin, Y.; Tang, L.; Chen, J.; Chen, D.; Wen, W.; Han, F. Inhibition of miR-101-3p protects against sepsis-induced myocardial injury by inhibiting MAPK and NF-κB pathway activation via the upregulation of DUSP1. Int. J. Mol. Med. 2021, 47, 20. [Google Scholar] [CrossRef]
- Wei, X.; Tang, C.; Lu, X.; Liu, R.; Zhou, M.; He, D.; Zheng, D.; Sun, C.; Wu, Z. MiR-101 targets DUSP1 to regulate the TGF-β secretion in sorafenib inhibits macrophage-induced growth of hepatocarcinoma. Oncotarget 2015, 6, 18389–18405. [Google Scholar] [CrossRef]
- Ye, Y.; Bao, C.; Fan, W. Overexpression of miR-101 May Target DUSP1 to Promote the Cartilage Degradation in Rheumatoid Arthritis. J. Comput. Biol. 2019, 26, 1067–1079. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).