LSTrAP-Cloud: A User-Friendly Cloud Computing Pipeline to Infer Coexpression Networks
Abstract
1. Introduction
2. Materials and Methods
2.1. Streaming RNA Sequencing Data
2.2. Constructing Coexpression Networks
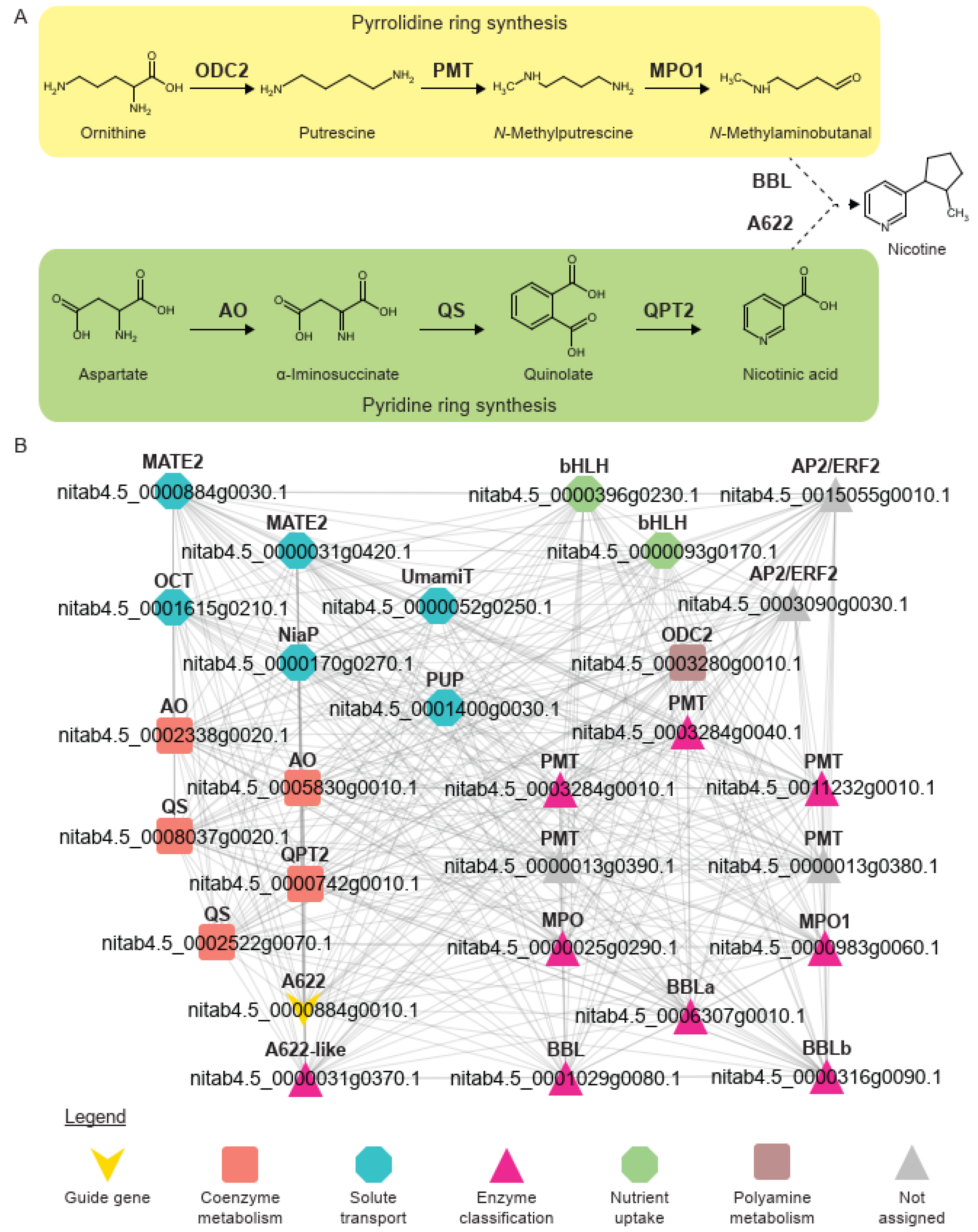
2.3. Identification of Genes Involved in Nicotine Biosynthesis
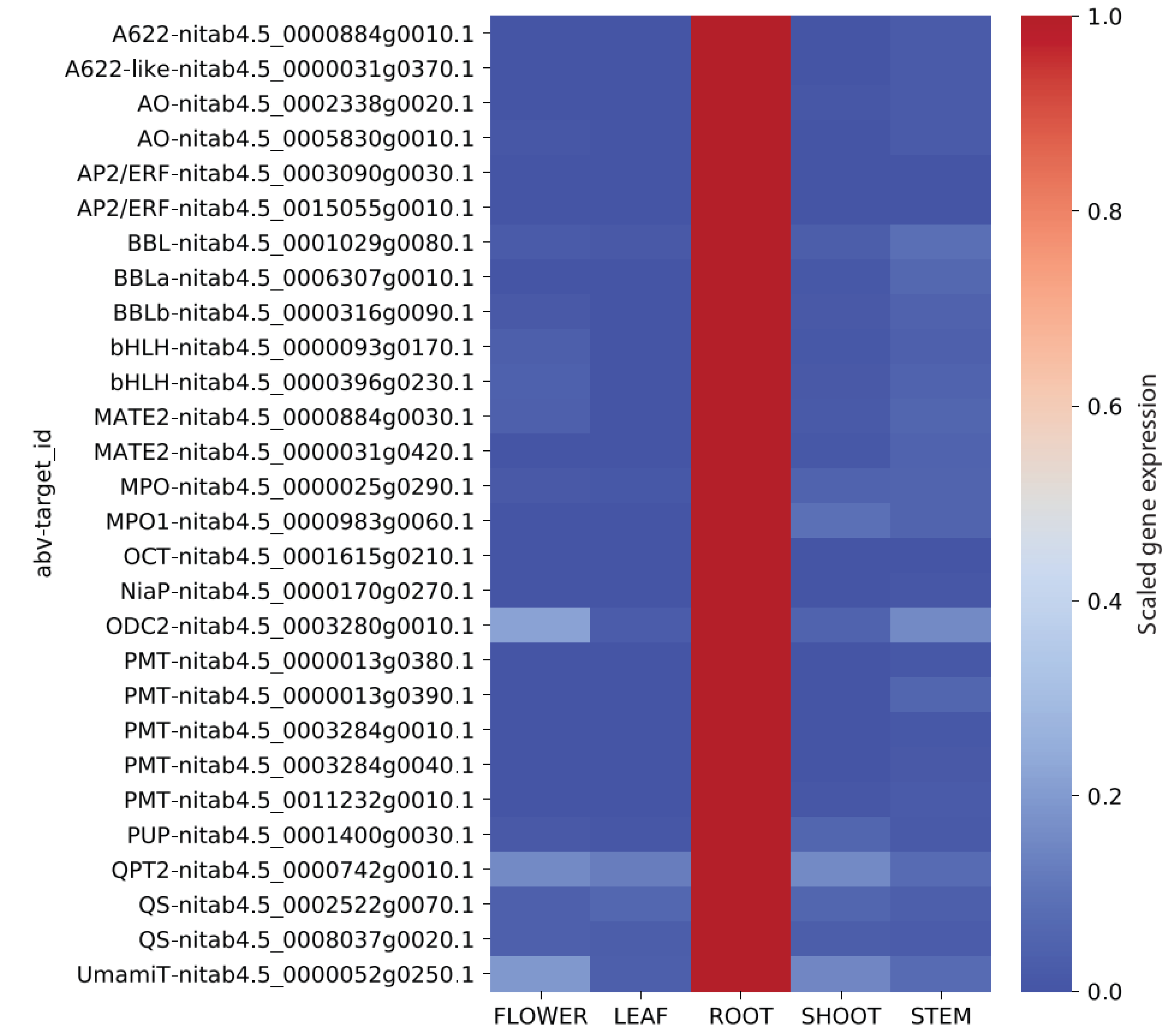
2.4. Relative Expression of Nicotine Biosynthesis Genes in 5 Major Organs
3. Results
3.1. Implementation of Gene Coexpression Pipeline on Google Colaboratory
3.2. Investigating the Nicotine Biosynthesis Coexpression Network
3.3. Expression Analysis of Genes Related to Nicotine Biosynthesis in Flower, Leaf, Root, Shoot, and Stem
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Senol Cali, D.; Kim, J.S.; Ghose, S.; Alkan, C.; Mutlu, O. Nanopore sequencing technology and tools for genome assembly: Computational analysis of the current state, bottlenecks and future directions. Brief. Bioinform. 2019, 20, 1542–1559. [Google Scholar] [CrossRef]
- Hansen, B.O.; Meyer, E.H.; Ferrari, C.; Vaid, N.; Movahedi, S.; Vandepoele, K.; Nikoloski, Z.; Mutwil, M. Ensemble gene function prediction database reveals genes important for complex I formation in Arabidopsis thaliana. New Phytol. 2018, 217, 1521–1534. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.Y.; Mutwil, M. Towards revealing the functions of all genes in plants. Trends Plant Sci. 2014, 19, 212–221. [Google Scholar] [CrossRef]
- Bouché, N.; Bouché, D. Arabidopsis gene knockout: Phenotypes wanted. Curr. Opin. Plant Biol. 2001, 4, 111–117. [Google Scholar] [CrossRef]
- Zhang, X.; Henriques, R.; Lin, S.S.; Niu, Q.W.; Chua, N.H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef]
- Ruprecht, C.; Vaid, N.; Proost, S.; Persson, S.; Mutwil, M. Beyond Genomics: Studying Evolution with Gene Coexpression Networks. Trends Plant Sci. 2017, 22, 298–307. [Google Scholar] [CrossRef]
- Ruprecht, C.; Mendrinna, A.; Tohge, T.; Sampathkumar, A.; Klie, S.; Fernie, A.R.; Nikoloski, Z.; Persson, S.; Mutwil, M. FamNet: A framework to identify multiplied modules driving pathway diversification in plants. Plant Physiol. 2016, 170, 1878–1894. [Google Scholar] [CrossRef]
- Usadel, B.; Obayashi, T.; Mutwil, M.; Giorgi, F.M.; Bassel, G.W.; Tanimoto, M.; Chow, A.; Steinhauser, D.; Persson, S.; Provart, N.J. Co-expression tools for plant biology: Opportunities for hypothesis generation and caveats. Plant Cell Environ. 2009, 32, 1633–1651. [Google Scholar] [CrossRef]
- Lee, I.; Ambaru, B.; Thakkar, P.; Marcotte, E.M.; Rhee, S.Y. Rational association of genes with traits using a genome-scale gene network for Arabidopsis thaliana. Nat. Biotechnol. 2010, 28, 149–156. [Google Scholar] [CrossRef]
- Hansen, B.O.; Vaid, N.; Musialak-Lange, M.; Janowski, M.; Mutwil, M. Elucidating gene function and function evolution through comparison of co-expression networks of plants. Front. Plant Sci. 2014, 5, 1–9. [Google Scholar] [CrossRef]
- Proost, S.; Mutwil, M. Tools of the trade: Studying molecular networks in plants. Curr. Opin. Plant Biol. 2016, 30, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Takabayashi, A.; Ishikawa, N.; Obayashi, T.; Ishida, S.; Obokata, J.; Endo, T.; Sato, F. Three novel subunits of Arabidopsis chloroplastic NAD(P)H dehydrogenase identified by bioinformatic and reverse genetic approaches. Plant J. 2009, 57, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Lammens, T.; Boudolf, V.; Maes, S.; Yoshizumi, T.; De Jaeger, G.; Witters, E.; Inzé, D.; De Veylder, L. The DNA replication checkpoint aids survival of plants deficient in the novel replisome factor ETG1. EMBO J. 2008, 27, 1840–1851. [Google Scholar] [CrossRef] [PubMed]
- Stuart, J.M.; Segal, E.; Koller, D.; Kim, S.K. A Gene-Coexpression Network for Global Discovery of Conserved Genetic Modules. Science 2003, 302, 249–255. [Google Scholar] [CrossRef]
- Yu, H.; Luscombe, N.M.; Qian, J.; Gerstein, M. Genomic analysis of gene expression relationships in transcriptional regulatory networks. Trends Genet. 2003, 19, 422–427. [Google Scholar] [CrossRef]
- Jiménez-Gómez, J.M.; Wallace, A.D.; Maloof, J.N. Network analysis identifies ELF3 as a QTL for the shade avoidance response in arabidopsis. PLoS Genet. 2010, 6, e1001100. [Google Scholar] [CrossRef]
- Persson, S.; Wei, H.; Milne, J.; Page, G.P.; Somerville, C.R. Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc. Natl. Acad. Sci. USA 2005, 102, 8633–8638. [Google Scholar] [CrossRef]
- Itkin, M.; Heinig, U.; Tzfadia, O.; Bhide, A.J.; Shinde, B.; Cardenas, P.D.; Bocobza, S.E.; Unger, T.; Malitsky, S.; Finkers, R.; et al. Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 2013, 341, 175–179. [Google Scholar] [CrossRef]
- Proost, S.; Mutwil, M. PlaNet: Comparative Co-Expression Network Analyses for Plants. In Methods in Molecular Biology; van Dijk, A.D.J., Ed.; Springer: New York, NY, USA, 2017; Volume 1533, pp. 213–227. [Google Scholar] [CrossRef]
- Sibout, R.; Proost, S.; Hansen, B.O.; Vaid, N.; Giorgi, F.M.; Ho-Yue-Kuang, S.; Legée, F.; Cézart, L.; Bouchabké-Coussa, O.; Soulhat, C.; et al. Expression atlas and comparative coexpression network analyses reveal important genes involved in the formation of lignified cell wall in Brachypodium distachyon. New Phytol. 2017, 215, 1009–1025. [Google Scholar] [CrossRef]
- Alejandro, S.; Lee, Y.Y.; Tohge, T.; Sudre, D.; Osorio, S.; Park, J.; Bovet, L.; Lee, Y.Y.; Geldner, N.; Fernie, A.R.; et al. AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Curr. Biol. 2012, 22, 1207–1212. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Tan, Q.W.; Mutwil, M. Inferring biosynthetic and gene regulatory networks from Artemisia annua RNA sequencing data on a credit card-sized ARM computer. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 194429. [Google Scholar] [CrossRef]
- Kohen, R.; Barlev, J.; Hornung, G.; Stelzer, G.; Feldmesser, E.; Kogan, K.; Safran, M.; Leshkowitz, D. UTAP: User-friendly Transcriptome Analysis Pipeline. BMC Bioinform. 2019, 20, 154. [Google Scholar] [CrossRef]
- Proost, S.; Krawczyk, A.; Mutwil, M. LSTrAP: Efficiently combining RNA sequencing data into co-expression networks. BMC Bioinform. 2017, 18, 444. [Google Scholar] [CrossRef]
- Melsted, P.; Booeshaghi, A.S.; Gao, F.; Beltrame, E.; Lu, L.; Hjorleifsson, K.E.; Gehring, J.; Pachter, L. Modular and efficient pre-processing of single-cell RNA-seq. BioRxiv 2019, 673285. [Google Scholar] [CrossRef]
- Leinonen, R.; Akhtar, R.; Birney, E.; Bower, L.; Cerdeno-Tárraga, A.; Cheng, Y.; Cleland, I.; Faruque, N.; Goodgame, N.; Gibson, R.; et al. The European Nucleotide Archive. Nucleic Acids Res. 2011, 39, D28–D31. [Google Scholar] [CrossRef]
- Edwards, K.D.; Fernandez-Pozo, N.; Drake-Stowe, K.; Humphry, M.; Evans, A.D.; Bombarely, A.; Allen, F.; Hurst, R.; White, B.; Kernodle, S.P.; et al. A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genom. 2017, 18, 448. [Google Scholar] [CrossRef]
- Fernandez-Pozo, N.; Menda, N.; Edwards, J.D.; Saha, S.; Tecle, I.Y.; Strickler, S.R.; Bombarely, A.; Fisher-York, T.; Pujar, A.; Foerster, H.; et al. The Sol Genomics Network (SGN)—From genotype to phenotype to breeding. Nucleic Acids Res. 2015, 43, D1036–D1041. [Google Scholar] [CrossRef]
- Franz, M.; Lopes, C.T.; Huck, G.; Dong, Y.; Sumer, O.; Bader, G.D. Cytoscape.js: A graph theory library for visualisation and analysis. Bioinformatics 2015, 32, 309–311. [Google Scholar] [CrossRef]
- Schwacke, R.; Ponce-Soto, G.Y.; Krause, K.; Bolger, A.M.; Arsova, B.; Hallab, A.; Gruden, K.; Stitt, M.; Bolger, M.E.; Usadel, B. MapMan4: A Refined Protein Classification and Annotation Framework Applicable to Multi-Omics Data Analysis. Mol. Plant 2019, 12, 879–892. [Google Scholar] [CrossRef]
- Kajikawa, M.; Sierro, N.; Kawaguchi, H.; Bakaher, N.; Ivanov, N.V.; Hashimoto, T.; Shoji, T. Genomic Insights into the Evolution of the Nicotine Biosynthesis Pathway in Tobacco. Plant Physiol. 2017, 174, 999–1011. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Brockmöller, T.; Navarro-Quezada, A.; Kuhl, H.; Gase, K.; Ling, Z.; Zhou, W.; Kreitzer, C.; Stanke, M.; Tang, H.; et al. Wild tobacco genomes reveal the evolution of nicotine biosynthesis. Proc. Natl. Acad. Sci. USA 2017, 114, 6133–6138. [Google Scholar] [CrossRef] [PubMed]
- Shoji, T.; Inai, K.; Yazaki, Y.; Sato, Y.; Takase, H.; Shitan, N.; Yazaki, K.; Goto, Y.; Toyooka, K.; Matsuoka, K.; et al. Multidrug and Toxic Compound Extrusion-Type Transporters Implicated in Vacuolar Sequestration of Nicotine in Tobacco Roots. Plant Physiol. 2009, 149, 708–718. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, I.T. An Ecologically Motivated Analysis of Plant-Herbivore Interactions in Native Tobacco. Plant Physiol. 2001, 127, 1449–1458. [Google Scholar] [CrossRef]
- Ruprecht, C.; Mutwil, M.; Saxe, F.; Eder, M.; Nikoloski, Z.; Persson, S. Large-Scale Co-Expression Approach to Dissect Secondary Cell Wall Formation Across Plant Species. Front. Plant Sci. 2011, 2, 1–13. [Google Scholar] [CrossRef]
- Liu, H.; Kotova, T.I.; Timko, M.P. Increased Leaf Nicotine Content by Targeting Transcription Factor Gene Expression in Commercial Flue-Cured Tobacco (Nicotiana tabacum L.). Genes 2019, 10, 930. [Google Scholar] [CrossRef]
- Mutwil, M.; Klie, S.; Tohge, T.; Giorgi, F.M.; Wilkins, O.; Campbell, M.M.; Fernie, A.R.; Usadel, B.; Nikoloski, Z.; Persson, S. PlaNet: Combined Sequence and Expression Comparisons across Plant Networks Derived from Seven Species. Plant Cell 2011, 23, 895–910. [Google Scholar] [CrossRef]
- Ferrari, C.; Shivhare, D.; Hansen, B.O.; Pasha, A.; Esteban, E.; Provart, N.J.; Kragler, F.; Fernie, A.R.; Tohge, T.; Mutwil, M. Expression Atlas of Selaginella moellendorffii Provides Insights into the Evolution of Vasculature, Secondary Metabolism, and Roots. Plant Cell 2020, 32, 853–870. [Google Scholar] [CrossRef]
- Friesner, J.; Assmann, S.M.; Bastow, R.; Bailey-Serres, J.; Beynon, J.; Brendel, V.; Buell, C.R.; Bucksch, A.; Busch, W.; Demura, T.; et al. The Next Generation of Training for Arabidopsis Researchers: Bioinformatics and Quantitative Biology. Plant Physiol. 2017, 175, 1499–1509. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, Q.W.; Goh, W.; Mutwil, M. LSTrAP-Cloud: A User-Friendly Cloud Computing Pipeline to Infer Coexpression Networks. Genes 2020, 11, 428. https://doi.org/10.3390/genes11040428
Tan QW, Goh W, Mutwil M. LSTrAP-Cloud: A User-Friendly Cloud Computing Pipeline to Infer Coexpression Networks. Genes. 2020; 11(4):428. https://doi.org/10.3390/genes11040428
Chicago/Turabian StyleTan, Qiao Wen, William Goh, and Marek Mutwil. 2020. "LSTrAP-Cloud: A User-Friendly Cloud Computing Pipeline to Infer Coexpression Networks" Genes 11, no. 4: 428. https://doi.org/10.3390/genes11040428
APA StyleTan, Q. W., Goh, W., & Mutwil, M. (2020). LSTrAP-Cloud: A User-Friendly Cloud Computing Pipeline to Infer Coexpression Networks. Genes, 11(4), 428. https://doi.org/10.3390/genes11040428





