Robust Cytonuclear Coordination of Transcription in Nascent Arabidopsis thaliana Autopolyploids
Abstract
:1. Introduction
- To compensate for increased nuclear dosage relative to organelle dosage, nuclear genes whose protein products are targeted to organelles will be down-regulated in autopolyploids relative to diploids, and/or organellar genes of the autopolyploids will be up-regulated.
- To maintain proper stoichiometry with interacting proteins encoded by organelles, nuclear genes whose products are targeted to organelles will exhibit less variation in transcriptional response to genome doubling than do other nuclear genes.
2. Materials and Methods
2.1. Plant Material
2.2. RNA Extraction and RNA-Seq
2.3. RNA-Seq Data Analysis
- java -jar trimmomatic-0.39.jar SE -phred33 {input filename}.fastq.gz {output filename}.fastq.gz ILLUMINACLIP:{path to adapter directory}/TruSeq3-SE.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15
- hisat2-build -f Athaliana_447_TAIR10.fa Athaliana_index
- hisat2 -p 8 -x Athaliana_index -q {input filename}.fastq -S {output filename}.mapped.sam -t
2.4. Comparison of Poly(A)-Selected RNA-Seq Libraries to Ribosome-Depleted RNA-Seq Libraries
3. Results
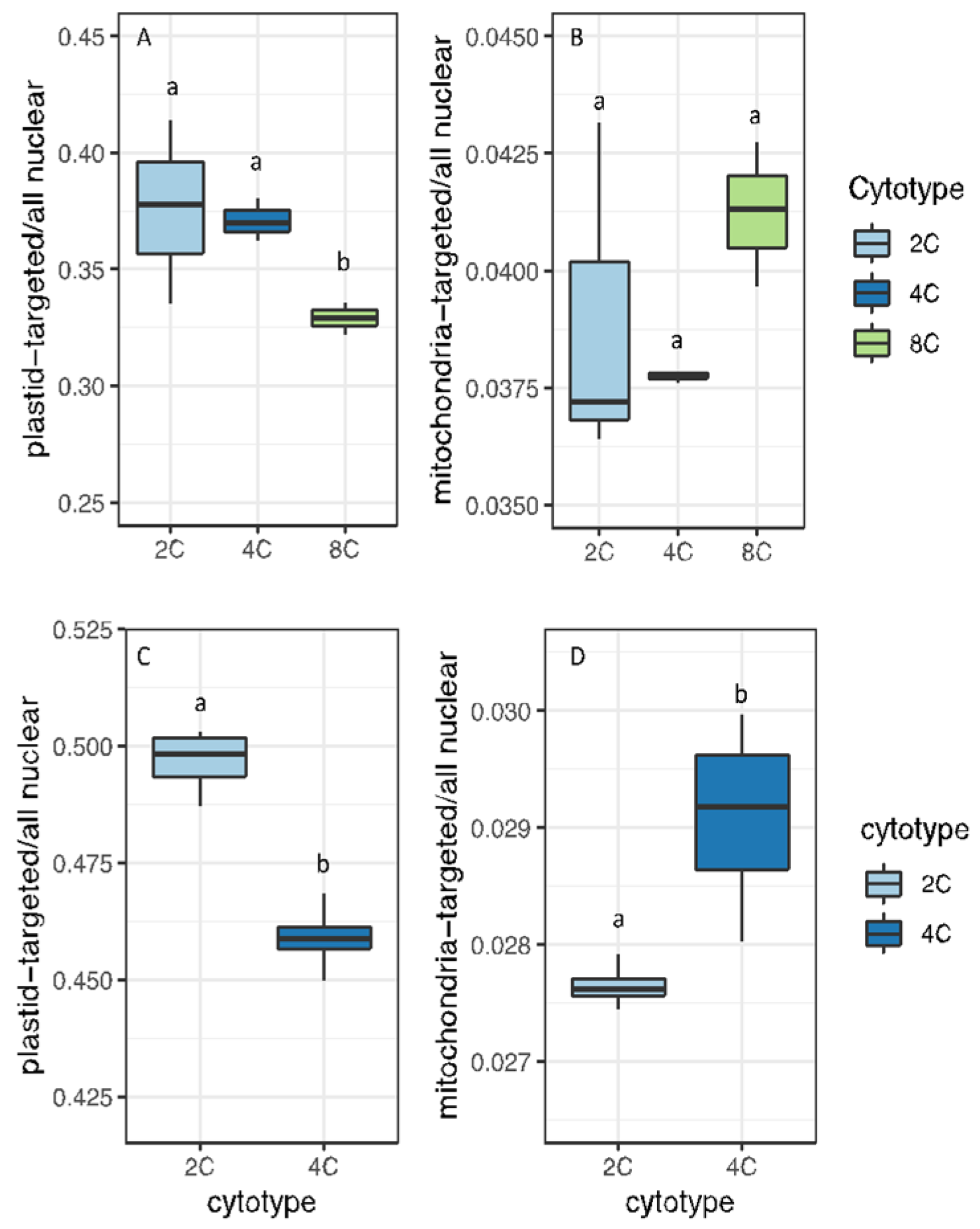
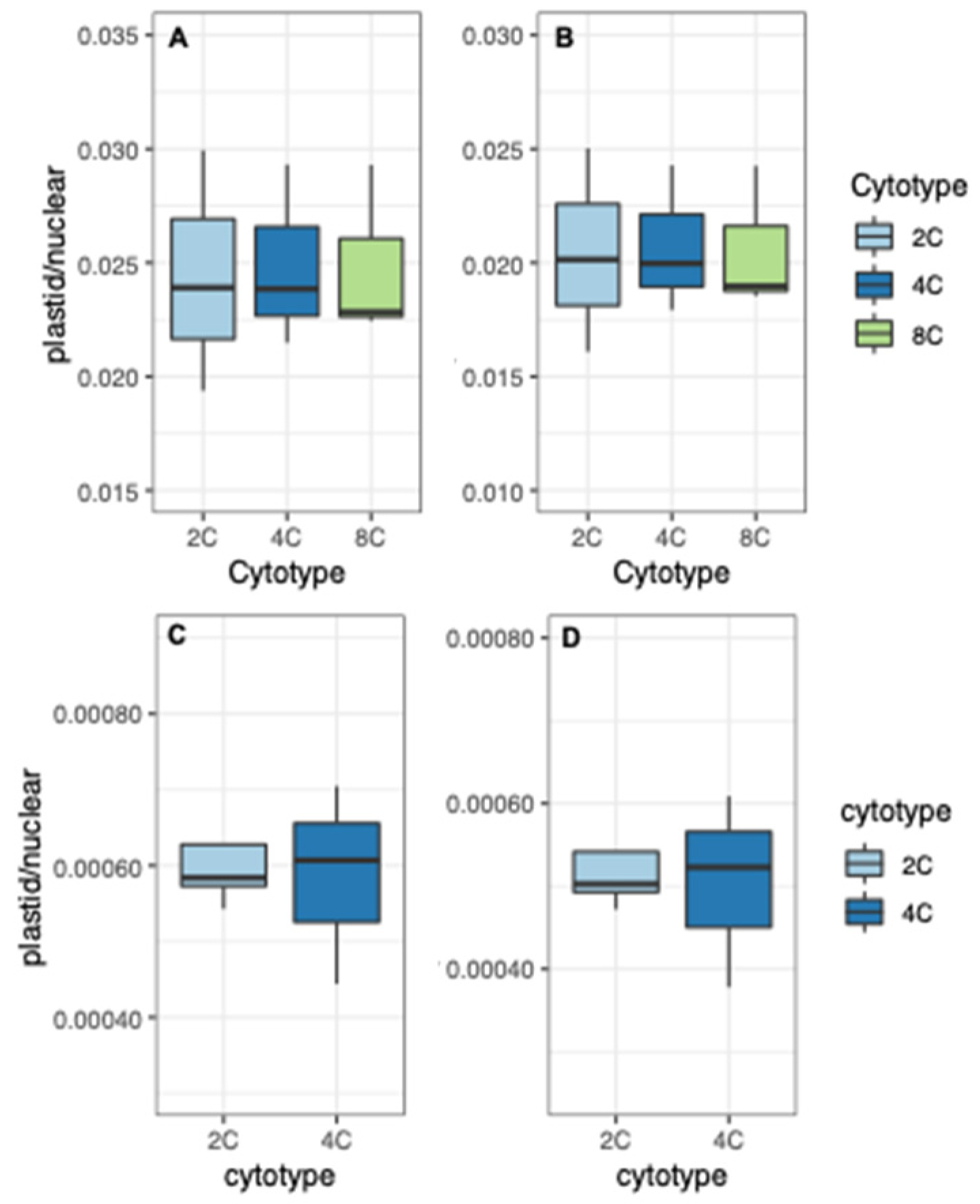
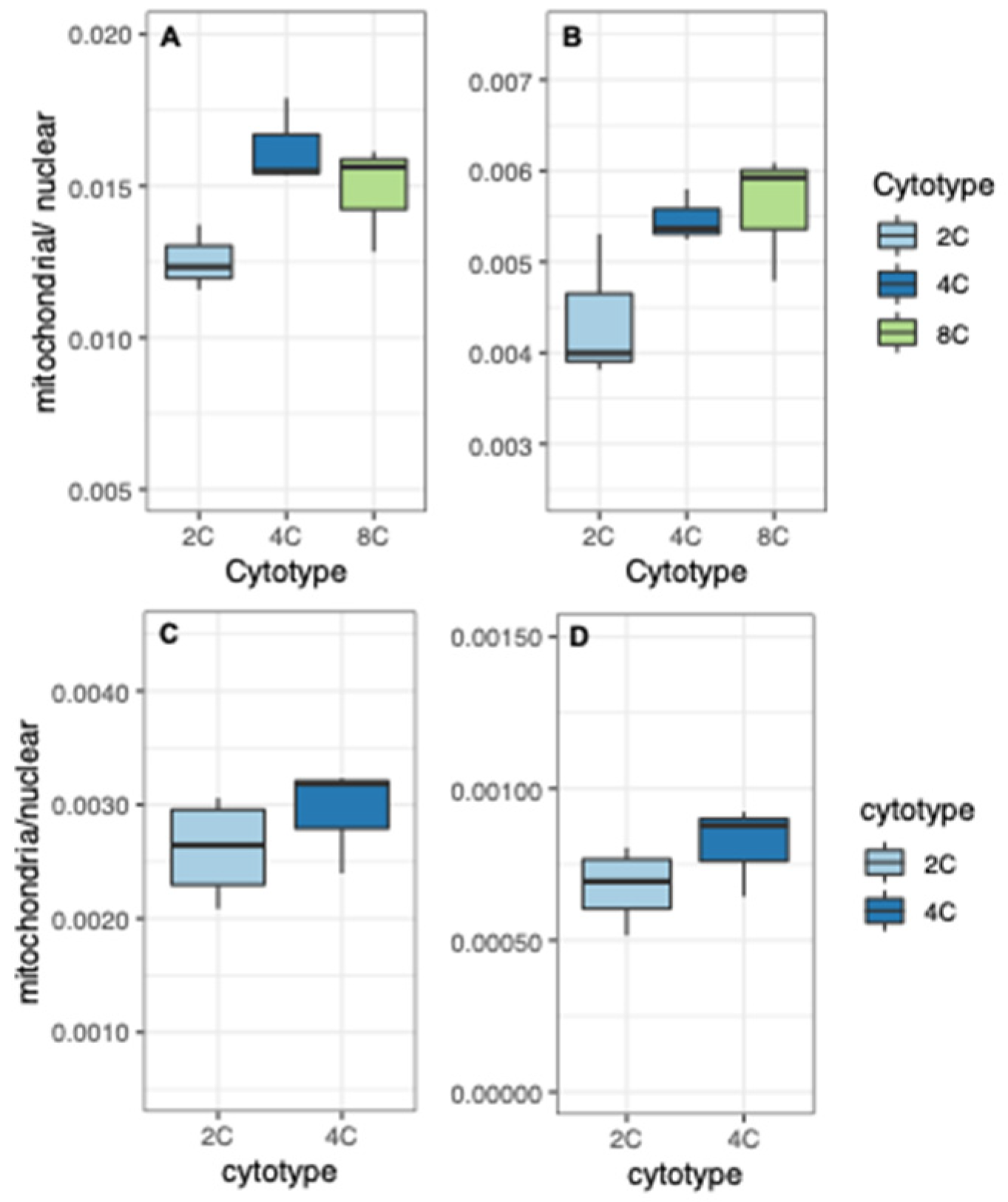
3.1. Organelle Copy Number per Nuclear Genome
3.2. Nuclear Gene Expression by Subcellular Targeting
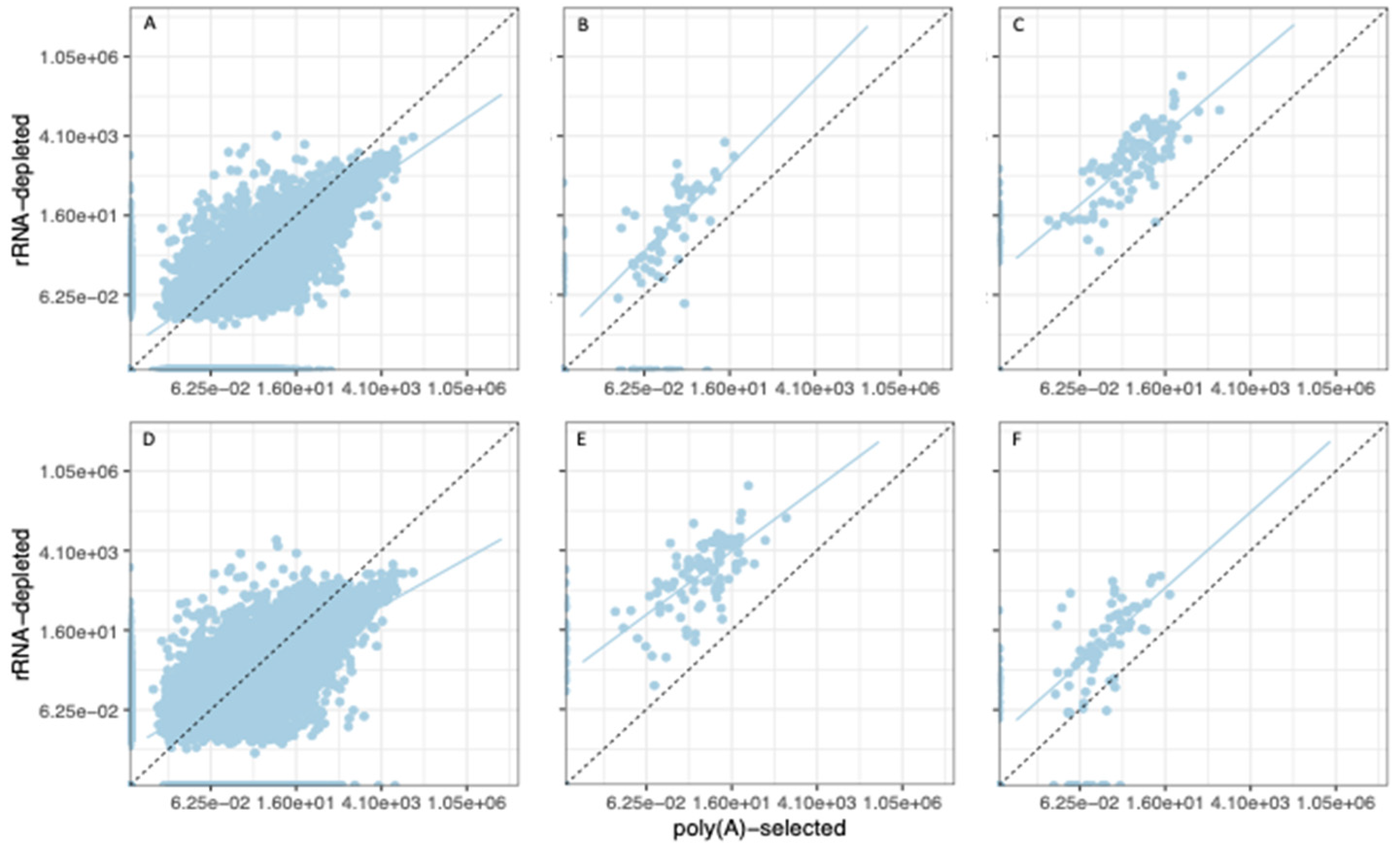
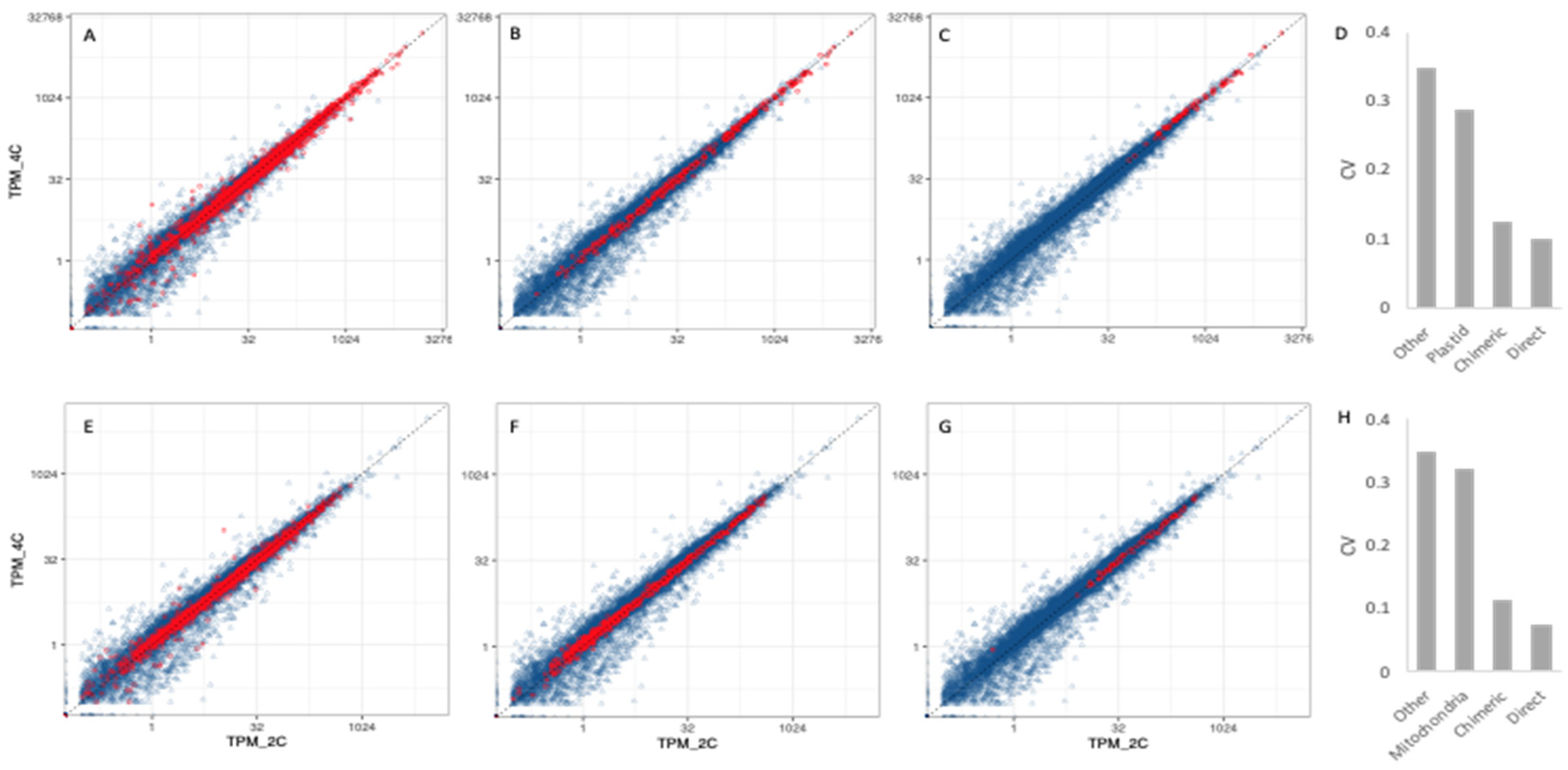
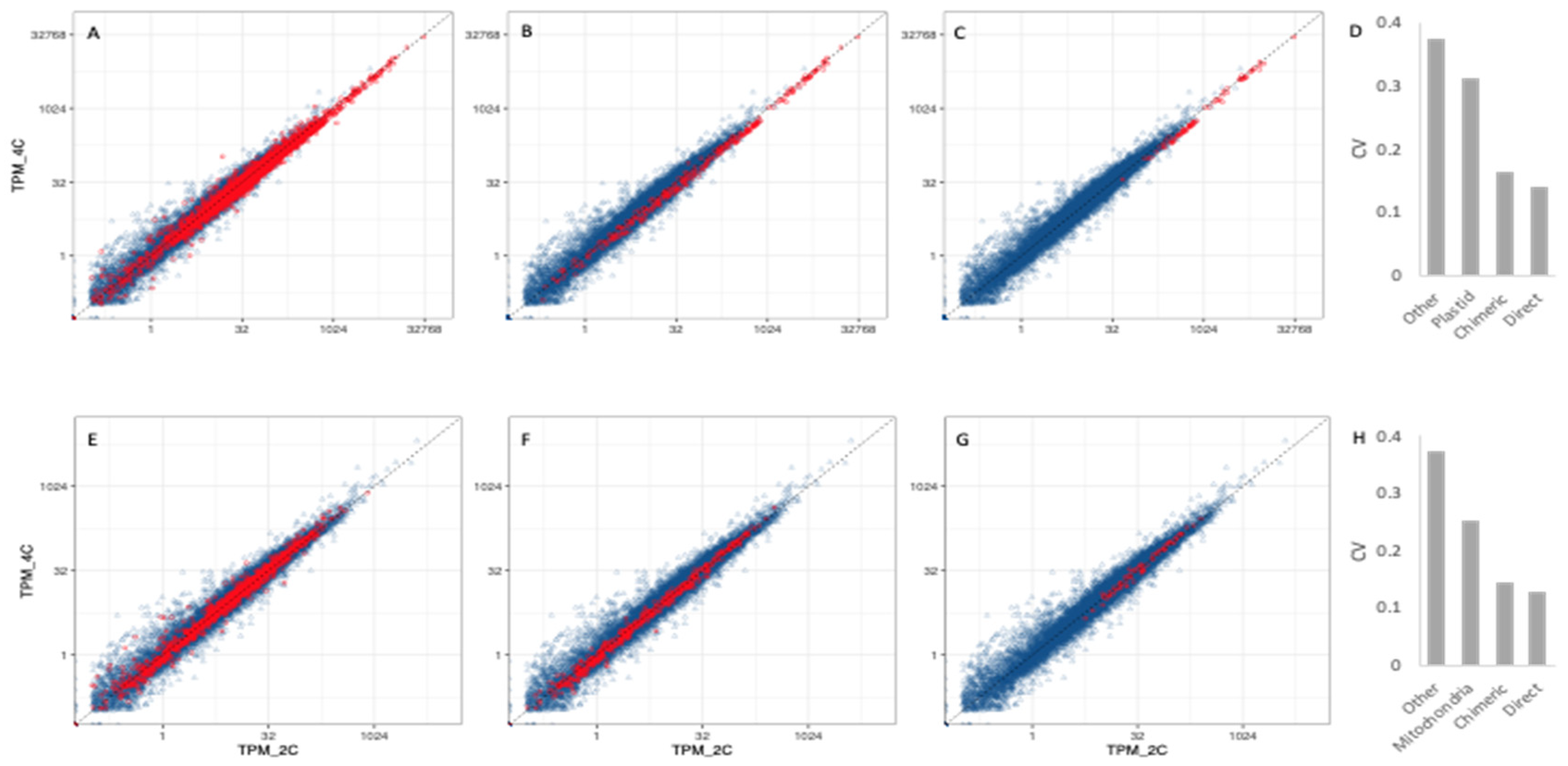
3.3. Organelle Expression Profiles Are Correlated between Poly(A)-Selected and rRNA-Depleted Libraries
3.4. Organellar Gene Expression
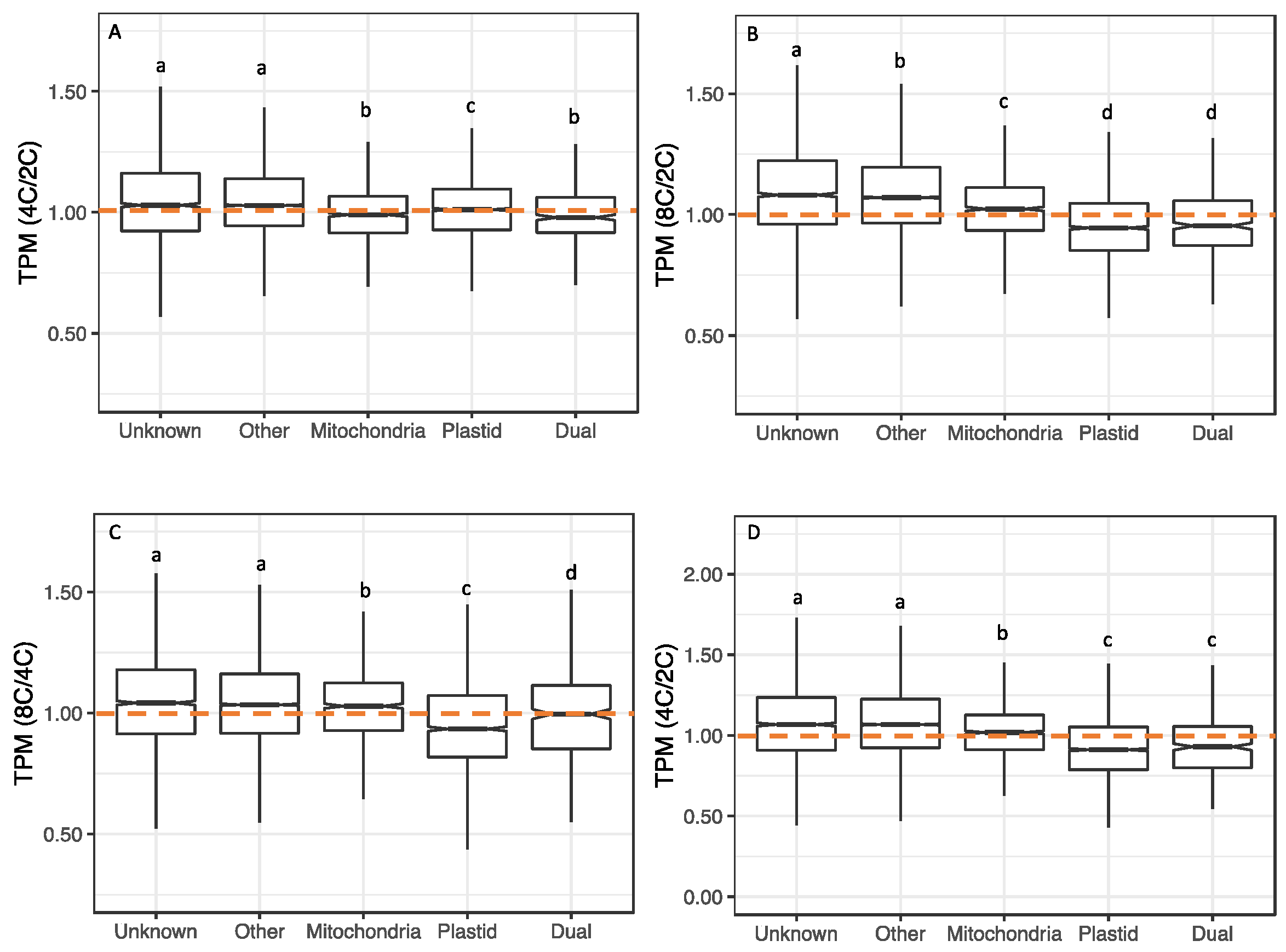
3.5. Transcriptional Responses to WGD in Nuclear Genes Encoding Organelle-Targeted Proteins
3.6. Transcriptional Responses per Cell to WGD
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rochaix, J.-D. Genetics of the Biogenesis and Dynamics of the Photosynthetic Machinery in Eukaryotes. Plant Cell 2004, 16, 1650–1660. [Google Scholar] [CrossRef]
- Woodson, J.D.; Chory, J. Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 2008, 9, 383–395. [Google Scholar] [CrossRef] [PubMed]
- Roux, F.; Mary-Huard, T.; Barillot, E.; Wenes, E.; Botran, L.; Durand, S.; Villoutreix, R.; Martin-Magniette, M.-L.; Camilleri, C.; Budar, F. Cytonuclear interactions affect adaptive traits of the annual plant Arabidopsis thaliana in the field. Proc. Natl. Acad. Sci. USA 2016, 113, 3687–3692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zascavage, R.R.; Planz, J.V. Admixture Effects on Coevolved Metabolic Systems. Front. Genet. 2018, 9, 634. [Google Scholar] [CrossRef] [PubMed]
- Sloan, D.B.; Warren, J.M.; Williams, A.M.; Wu, Z.; Abdel-Ghany, S.E.; Chicco, A.J.; Havird, J.C. Cytonuclear integration and co-evolution. Nat. Rev. Genet. 2018, 19, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Boussardon, C.; Martin-Magniette, M.-L.; Godin, B.; Benamar, A.; Vittrant, B.; Citerne, S.; Mary-Huard, T.; Macherel, D.; Rajjou, L.; Budar, F. Novel Cytonuclear Combinations Modify Arabidopsis thaliana Seed Physiology and Vigor. Front. Plant Sci. 2019, 10, 32. [Google Scholar] [CrossRef] [Green Version]
- Rousseau-Gueutin, M.; Keller, J.; de Carvalho, J.F.; Aïnouche, A.; Martin, G. The Intertwined Chloroplast and Nuclear Genome Coevolution in Plants. In Plant Growth Regul. - Alter. Sustain Unfavorable Cond; Hamim, R.D., Ed.; IntechOpen Limited: London, UK, 2018. [Google Scholar]
- Oberprieler, C.; Talianova, M.; Griesenbeck, J. Effects of polyploidy on the coordination of gene expression between organellar and nuclear genomes in Leucanthemum Mill. (Compositae, Anthemideae). Ecol. Evol. 2019, 9, 9100–9110. [Google Scholar] [CrossRef] [Green Version]
- Sharbrough, J.; Conover, J.L.; Tate, J.A.; Wendel, J.F.; Sloan, D.B. Cytonuclear responses to genome doubling. Am. J. Bot. 2017, 104, 1277–1280. [Google Scholar] [CrossRef] [Green Version]
- Coate, J.E.; Doyle, J.J. Divergent evolutionary fates of major photosynthetic gene networks following gene and whole genome duplicaitons. Plant Signal. Behav. 2011, 6, 594–597. [Google Scholar] [CrossRef]
- Coate, J.E.; Doyle, J.J. Genomics and transcriptomics of photosynthesis in polyploids. In Hybrid and Polyploid Genomics; Chen, Z.J., Birchler, J.A., Eds.; Wiley: Hoboken, NJ, USA, 2013. [Google Scholar]
- Coate, J.E.; Schlueter, J.; Whaley, A.; Doyle, J. Comparative evolution of photosynthetic genes in response to polyploid and non-polyploid duplication. Plant Physiol. 2011, 155, 2081–2095. [Google Scholar] [CrossRef] [Green Version]
- Gong, L.; Salmon, A.; Yoo, M.-J.; Grupp, K.K.; Wang, Z.; Paterson, A.H.; Wendel, J.F. The Cytonuclear Dimension of Allopolyploid Evolution: An Example from Cotton Using Rubisco. Mol. Biol. Evol. 2012, 29, 3023–3036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, L.; Olson, M.; Wendel, J.F. Cytonuclear Evolution of Rubisco in Four Allopolyploid Lineages. Mol. Biol. Evol. 2014, 31, 2624–2636. [Google Scholar] [CrossRef] [PubMed]
- Sehrish, T.; Symonds, V.V.; Soltis, D.E.; Soltis, P.S.; Tate, J.A. Cytonuclear Coordination Is Not Immediate upon Allopolyploid Formation in Tragopogon miscellus (Asteraceae) Allopolyploids. PLoS ONE 2015, 10, e0144339. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Dong, Q.; Li, X.; Yuliang, A.; Yu, Y.; Li, N.; Liu, B.; Gong, L. Cytonuclear Variation of Rubisco in Synthesized Rice Hybrids and Allotetraploids. Plant Genome 2017, 10. [Google Scholar] [CrossRef]
- Forsythe, E.S.; Sharbrough, J.; Havird, J.C.; Warren, J.M.; Sloan, D.B. CyMIRA: The Cytonuclear Molecular Interactions Reference for Arabidopsis. Genome Biol. Evol. 2019, 11, 2194–2202. [Google Scholar] [CrossRef] [Green Version]
- Smet, R.D.; Adams, K.L.; Vandepoele, K.; Montagu, M.C.E.V.; Maere, S.; Peer, Y.V. de Convergent gene loss following gene and genome duplications creates single-copy families in flowering plants. Proc. Natl. Acad. Sci. USA 2013, 110, 2898–2903. [Google Scholar] [CrossRef] [Green Version]
- Oberprieler, C.; Greiner, R.; Konowalik, K.; Vogt, R. The reticulate evolutionary history of the polyploid NW Iberian Leucanthemum pluriflorum clan (Compositae, Anthemideae) as inferred from nrDNA ETS sequence diversity and eco-climatological niche-modelling. Mol. Phylogenet. Evol. 2014, 70, 478–491. [Google Scholar] [CrossRef]
- Robinson, D.O.; Coate, J.E.; Singh, A.; Hong, L.; Bush, M.; Doyle, J.J.; Roeder, A.H.K. Ploidy and Size at Multiple Scales in the Arabidopsis Sepal. Plant Cell 2018, 30, 2308–2329. [Google Scholar] [CrossRef] [Green Version]
- Pignatta, D.; Dilkes, B.P.; Yoo, S.-Y.; Henry, I.M.; Madlung, A.; Doerge, R.W.; Jeffrey Chen, Z.; Comai, L. Differential sensitivity of the Arabidopsis thaliana transcriptome and enhancers to the effects of genome doubling. New Phytol. 2010, 186, 194–206. [Google Scholar] [CrossRef]
- Ravi, M.; Chan, S.W.L. Haploid plants produced by centromere-mediated genome elimination. Nature 2010, 464, 615–618. [Google Scholar] [CrossRef]
- Smyth, D.R.; Bowman, J.L.; Meyerowitz, E.M. Early flower development in Arabidopsis. Plant Cell 1990, 2, 755–767. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Nallu, S.; Hill, J.A.; Don, K.; Sahagun, C.; Zhang, W.; Meslin, C.; Snell-Rood, E.; Clark, N.L.; Morehouse, N.I.; Bergelson, J.; et al. The molecular genetic basis of herbivory between butterflies and their host plants. Nat. Ecol. Evol. 2018, 2, 1418–1427. [Google Scholar] [CrossRef]
- Rorbach, J.; Bobrowicz, A.; Pearce, S.; Minczuk, M. Polyadenylation in Bacteria and Organelles. In Polyadenylation: Methods and Protocols; Rorbach, J., Bobrowicz, A.J., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2014; pp. 211–227. ISBN 978-1-62703-971-0. [Google Scholar]
- Lange, H.; Sement, F.M.; Canaday, J.; Gagliardi, D. Polyadenylation-assisted RNA degradation processes in plants. Trends Plant Sci. 2009, 14, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Schuster, G.; Stern, D. Chapter 10 RNA Polyadenylation and Decay in Mitochondria and Chloroplasts. In Progress in Molecular Biology and Translational Science; Molecular Biology of RNA Processing and Decay in Prokaryotes; Academic Press: Cambridge, MA, USA, 2009; Volume 85, pp. 393–422. [Google Scholar]
- Visger, C.J.; Wong, G.K.; Zhang., Y.; Soltis, P.S.; Soltis, D.E. Divergent Gene Expression Levels between Diploid and Autotetraploid Tolmiea (Saxifragaceae) Relative to the Total Transcriptome, the Cell, and Biomass. Am. J. Bot. 2019, 106, 280–291. [Google Scholar] [CrossRef]
- Coate, J.E.; Doyle, J.J. Quantifying Whole Transcriptome Size, a Prerequisite for Understanding Transcriptome Evolution Across Species: An Example from a Plant Allopolyploid. Genome Biol. Evol. 2010, 2, 534–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coate, J.; Doyle, J. Variation in transcriptome size: are we getting the message? Chromosoma 2014, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Lovén, J.; Orlando, D.A.; Sigova, A.A.; Lin, C.Y.; Rahl, P.B.; Burge, C.B.; Levens, D.L.; Lee, T.I.; Young, R.A. Revisiting Global Gene Expression Analysis. Cell 2014, 151, 476–482. [Google Scholar] [CrossRef] [Green Version]
- Doyle, J.J.; Coate, J.E. Polyploidy, the Nucleotype, and Novelty: The Impact of Genome Doubling on the Biology of the Cell. Int. J. Plant Sci. 2019, 180, 1–52. [Google Scholar] [CrossRef]
- Song, M.J.; Potter, B.; Doyle, J.J.; Coate, J.E. Gene balance predicts transcriptional responses immediately following ploidy change in Arabidopsis thaliana. Plant Cell 2020. in revision. [Google Scholar]
- Fox, T.D. Mitochondrial Protein Synthesis, Import, and Assembly. Genetics 2012, 192, 1203–1234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bendich, A.J. Why do chloroplasts and mitochondria contain so many copies of their genome? BioEssays 1987, 6, 279–282. [Google Scholar] [CrossRef] [PubMed]
- das Neves, R.P.; Jones, N.S.; Andreu, L.; Gupta, R.; Enver, T.; Iborra, F.J. Connecting Variability in Global Transcription Rate to Mitochondrial Variability. PLoS Biol. 2010, 8, e1000560. [Google Scholar] [CrossRef] [Green Version]
- Butterfass, T. A nucleotypic control of chloroplast reproduction. Protoplasma 1983, 118, 71–74. [Google Scholar] [CrossRef]
- Warner, D.A.; Edwards, G.E. Effects of Polyploidy on Photosynthesis. Photosynth. Res. 1993, 35, 135–147. [Google Scholar] [CrossRef]
- Kawade, K.; Horiguchi, G.; Ishikawa, N.; Hirai, M.Y.; Tsukaya, H. Promotion of chloroplast proliferation upon enhanced post-mitotic cell expansion in leaves. BMC Plant Biol. 2013, 13, 143. [Google Scholar] [CrossRef] [Green Version]
- Coate, J.E.; Luciano, A.K.; Seralathan, V.; Minchew, K.J.; Owens, T.G.; Doyle, J.J. Anatomical, biochemical, and photosynthetic responses to recent allopolyploidy in Glycine dolichocarpa (Fabaceae). Am. J. Bot. 2012, 99, 55–67. [Google Scholar] [CrossRef]
- Preuten, T.; Cincu, E.; Fuchs, J.; Zoschke, R.; Liere, K.; Börner, T. Fewer genes than organelles: extremely low and variable gene copy numbers in mitochondria of somatic plant cells: Gene copy numbers in mitochondria. Plant J. 2010, 64, 948–959. [Google Scholar] [CrossRef]
- Giegé, P.; Sweetlove, L.J.; Cognat, V.; Leaver, C.J. Coordination of Nuclear and Mitochondrial Genome Expression during Mitochondrial Biogenesis in Arabidopsis. Plant Cell 2005, 17, 1497–1512. [Google Scholar] [CrossRef] [Green Version]
- Eberhard, S.; Drapier, D.; Wollman, F.-A. Searching limiting steps in the expression of chloroplast-encoded proteins: relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J. 2002, 31, 149–160. [Google Scholar] [CrossRef] [PubMed]






| Comparison | Genome | F | DF | p | R |
|---|---|---|---|---|---|
| Poly(A) vs. PRJNA380344 | Mitochondrion | 140.9 | 1, 150 | <0.0001 | 0.696 |
| Plastid | 4.242 | 1, 131 | 0.041 | 0.177 | |
| Nuclear | 3.1 × 104 | 1, 32907 | <0.0001 | 0.696 | |
| Poly(A) vs. PRJNA437291 | Mitochondrion | 77.86 | 1, 150 | <0.0001 | 0.585 |
| Plastid | 4.02 | 1, 131 | 0.047 | 0.172 | |
| Nuclear | 1.8 × 103 | 1, 32907 | <0.0001 | 0.229 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coate, J.E.; Schreyer, W.M.; Kum, D.; Doyle, J.J. Robust Cytonuclear Coordination of Transcription in Nascent Arabidopsis thaliana Autopolyploids. Genes 2020, 11, 134. https://doi.org/10.3390/genes11020134
Coate JE, Schreyer WM, Kum D, Doyle JJ. Robust Cytonuclear Coordination of Transcription in Nascent Arabidopsis thaliana Autopolyploids. Genes. 2020; 11(2):134. https://doi.org/10.3390/genes11020134
Chicago/Turabian StyleCoate, Jeremy E., W. Max Schreyer, David Kum, and Jeff J. Doyle. 2020. "Robust Cytonuclear Coordination of Transcription in Nascent Arabidopsis thaliana Autopolyploids" Genes 11, no. 2: 134. https://doi.org/10.3390/genes11020134



_Kum.jpg)


