Duplicated dnmt3aa and dnmt3ab DNA Methyltransferase Genes Play Essential and Non-Overlapped Functions on Modulating Behavioral Control in Zebrafish
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Ethics
2.2. Fish Lines and Husbandry
2.3. Morphometric Analysis
2.4. Measurement of 5-mC and 5-hmC Levels
2.5. Multiple Behavior Test
2.6. Short-Term Memory Test (Passive Avoidance Task)
2.7. Video Tracking and Data Analysis
2.8. Total Protein Extraction from Tissues
2.9. Determination of Neurotransmitter Contents in the Brain
2.10. Library Preparation and Whole-Genome Bisulfite Sequencing
2.11. DNA Extraction
2.12. Statistical Analysis
3. Results
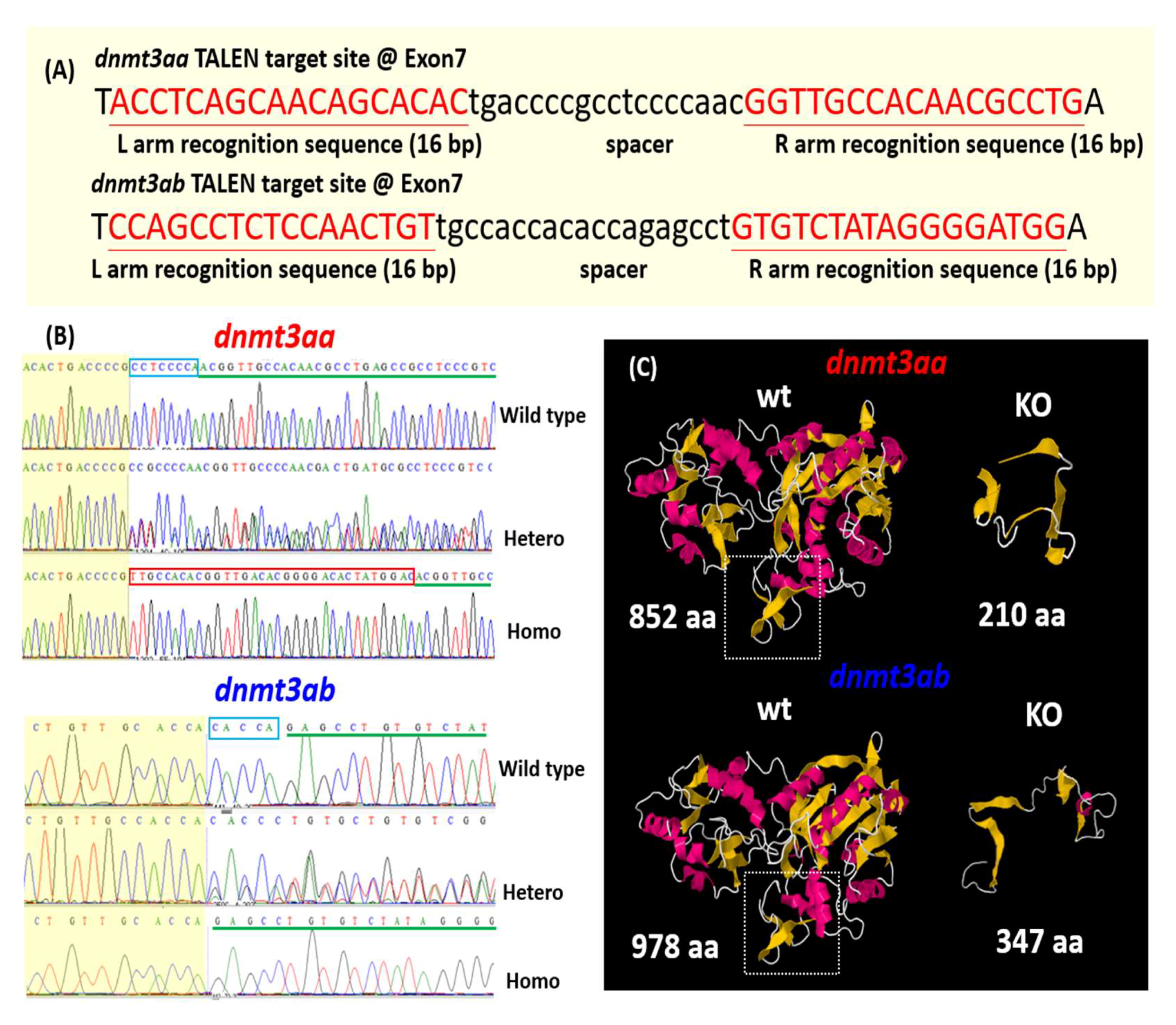
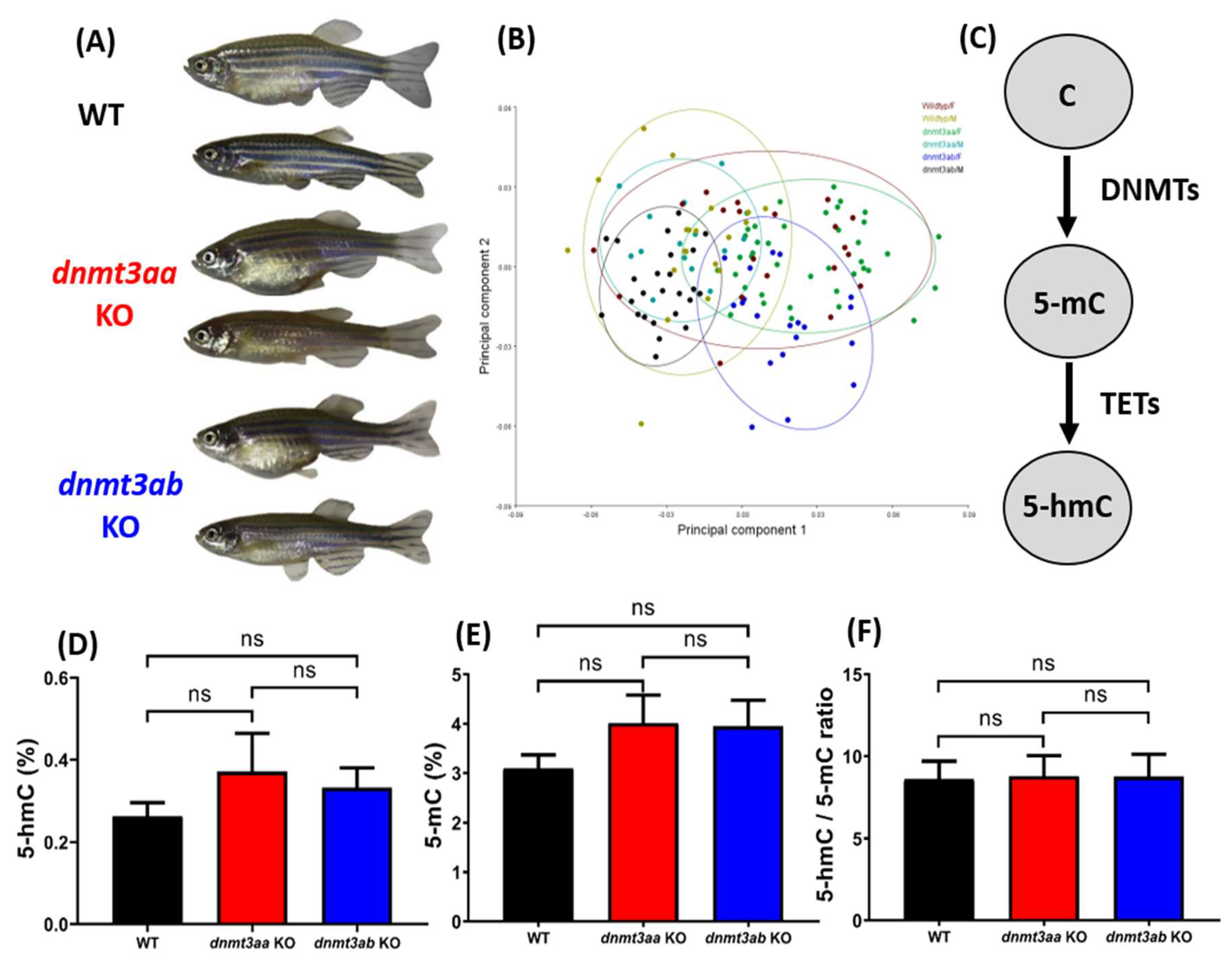
3.1. Morphometric Analysis and Detection of Global DNA Methylation Levels in dnmt3aa and dnmt3ab Mutants
3.2. Effects of dnmt3a Gene-Deficient on Zebrafish Locomotor Activity and Exploratory Behavior in Novel Tank Assay
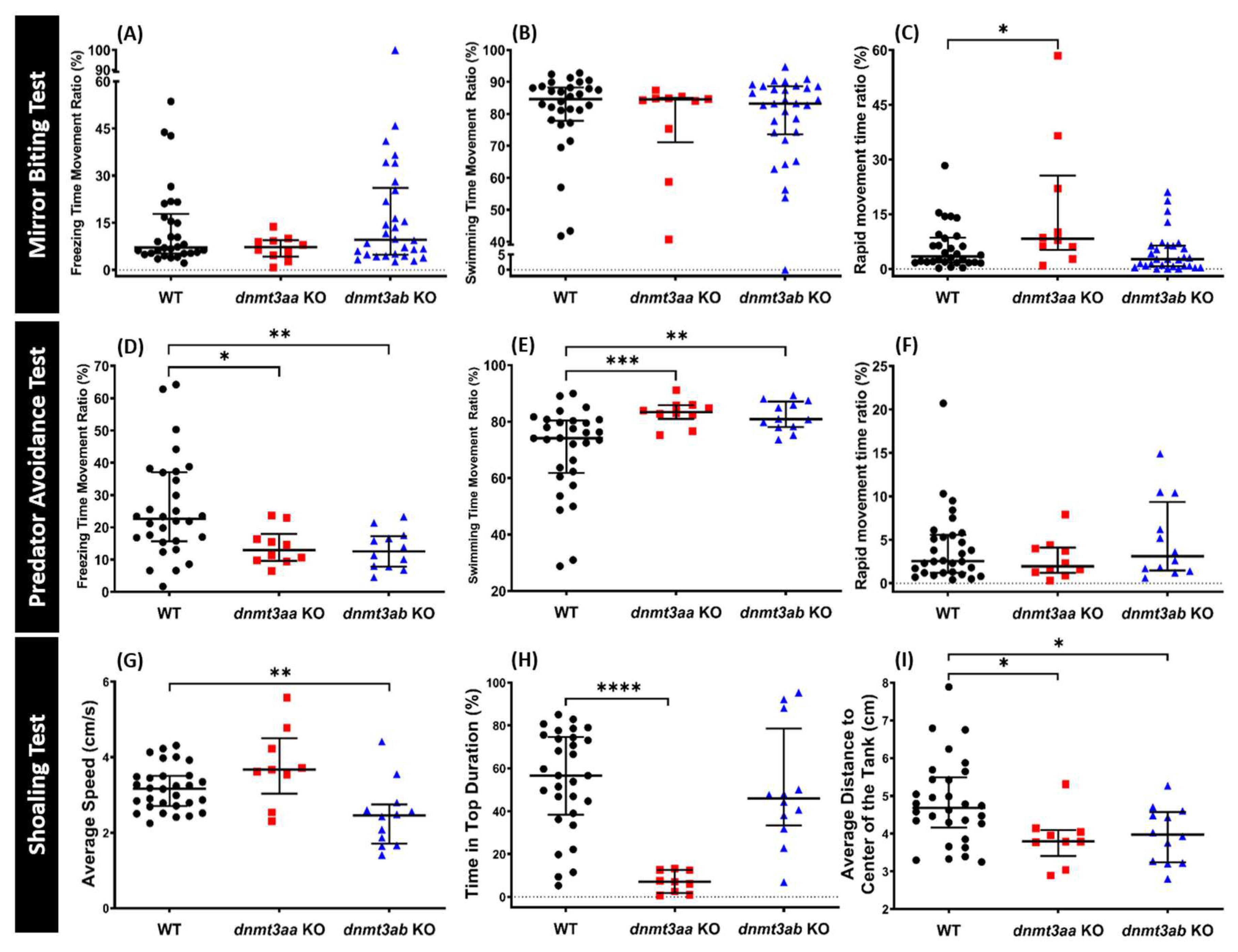
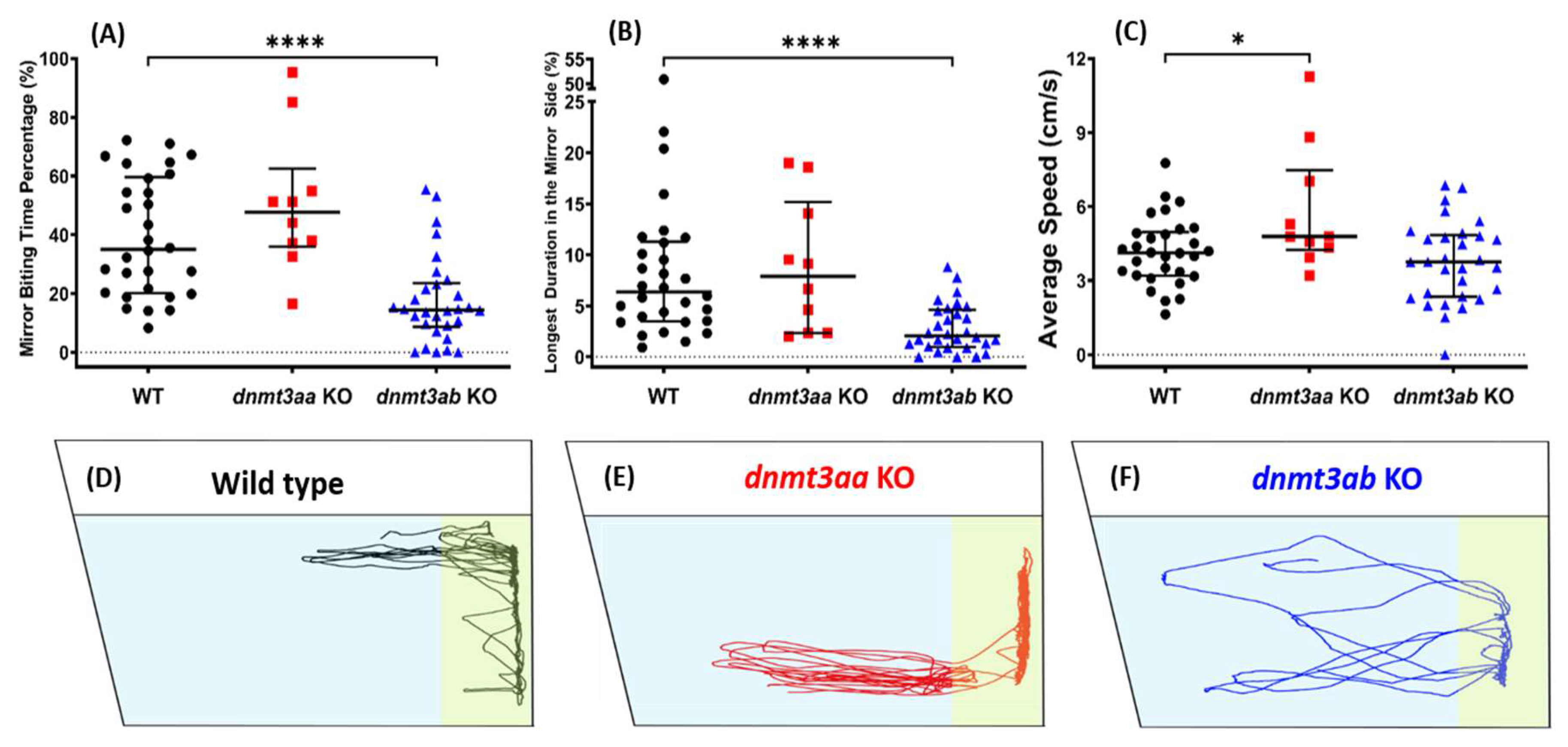
3.3. Effects of dnmt3a Gene-Deficient on Zebrafish Aggressiveness in Mirror Biting Test
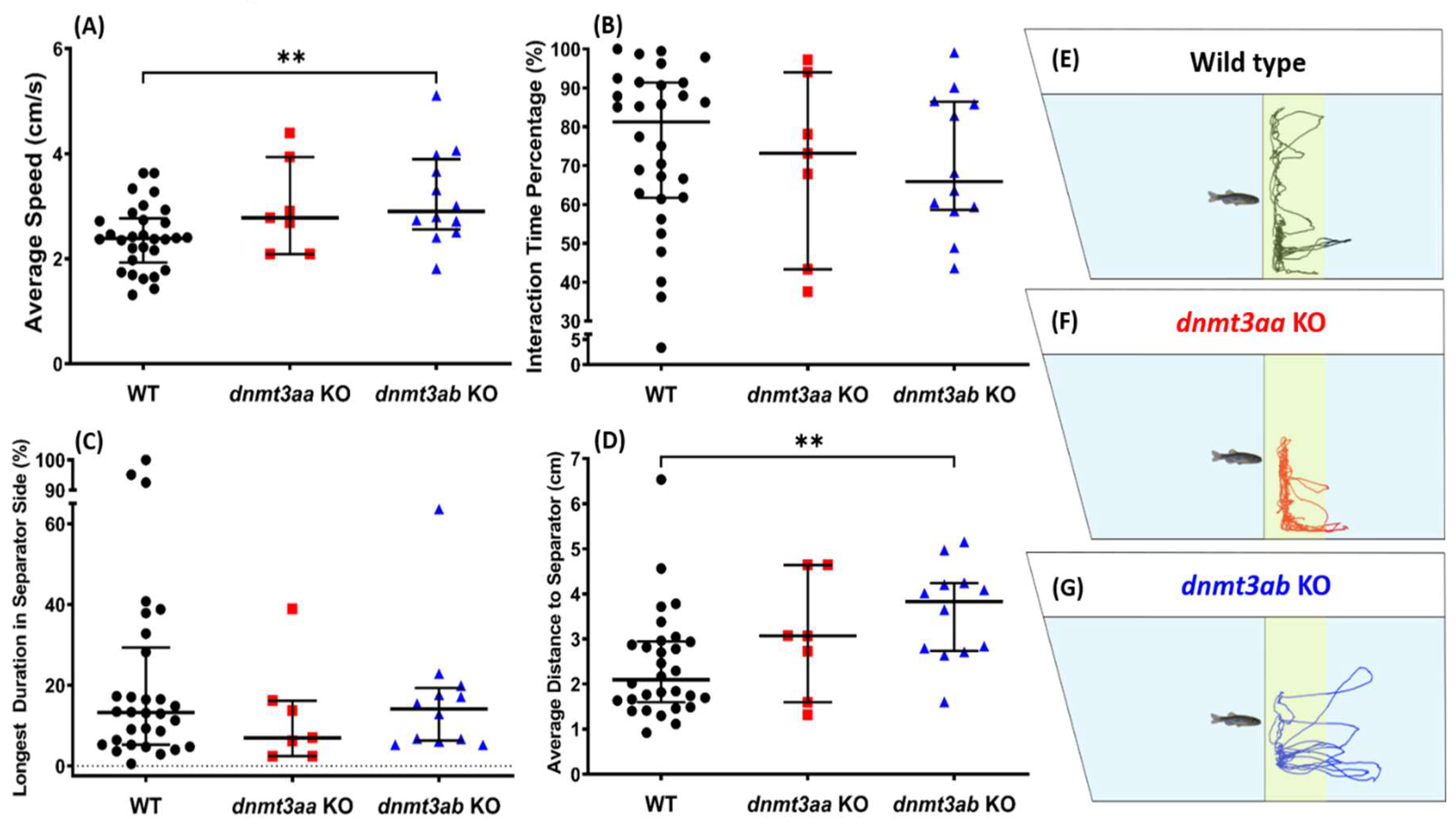
3.4. Effects of dnmt3a Gene-Deficient on Zebrafish Predator Avoidance Behavior
3.5. Effects of dnmt3a Gene-Deficient on Zebrafish Social Interaction
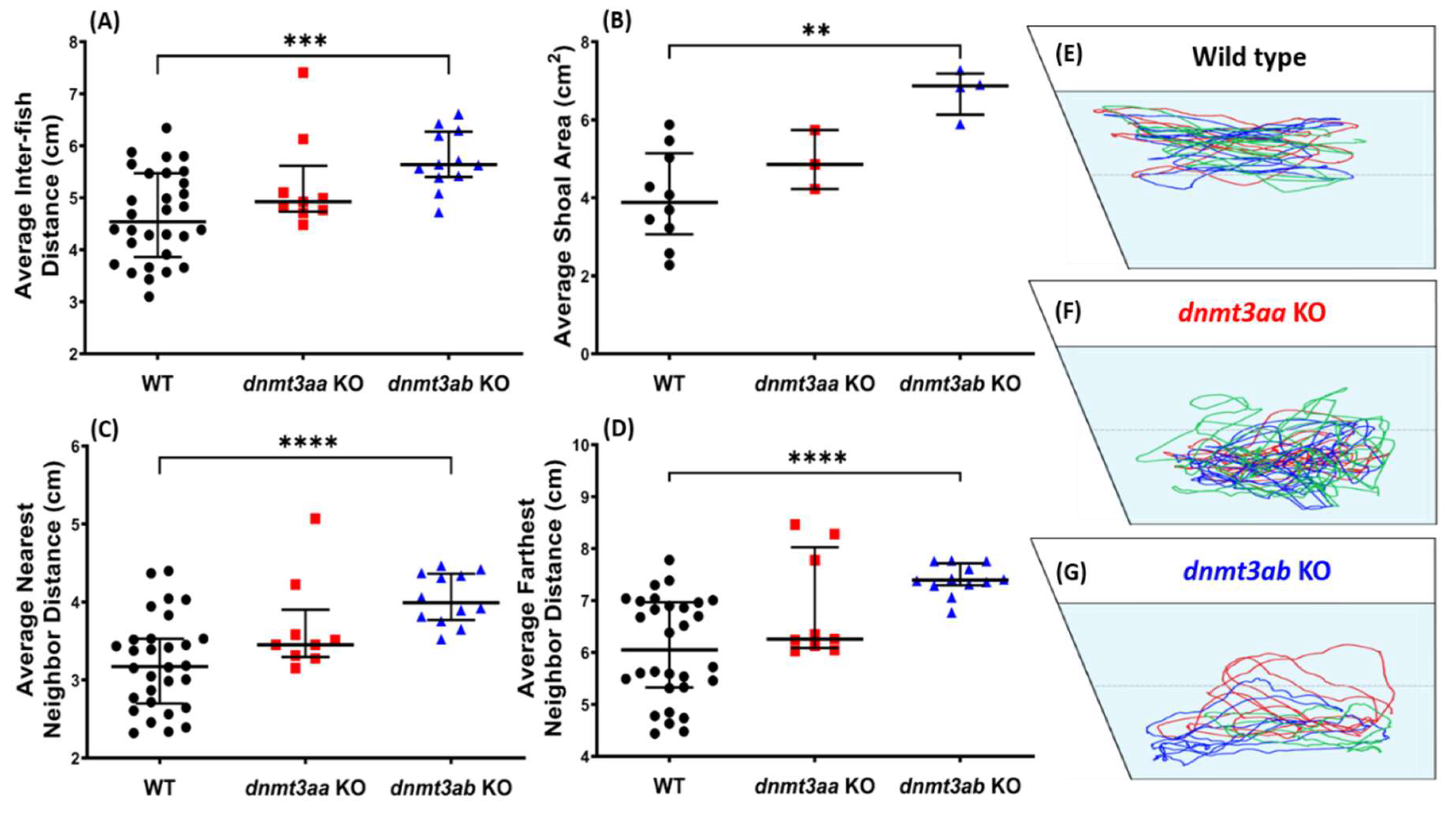
3.6. Effects of dnmt3a Gene-Deficient on Zebrafish Shoaling Formation
3.7. Effects of dnmt3a Gene-Deficient on Zebrafish Circadian Rhythm Locomotor Activity
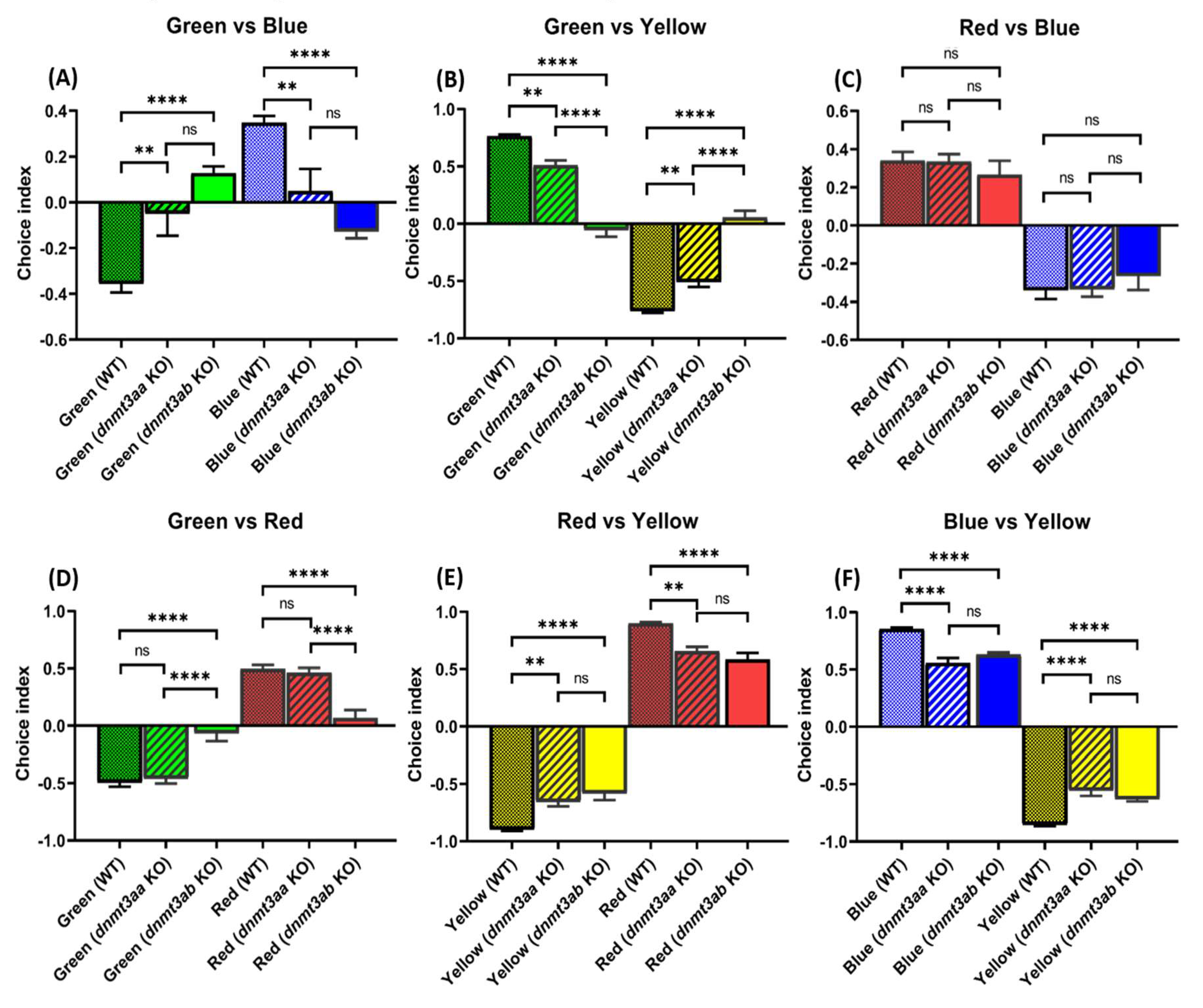
3.8. Effects of dnmt3a Gene-Deficient on Zebrafish Color Preference Ranking
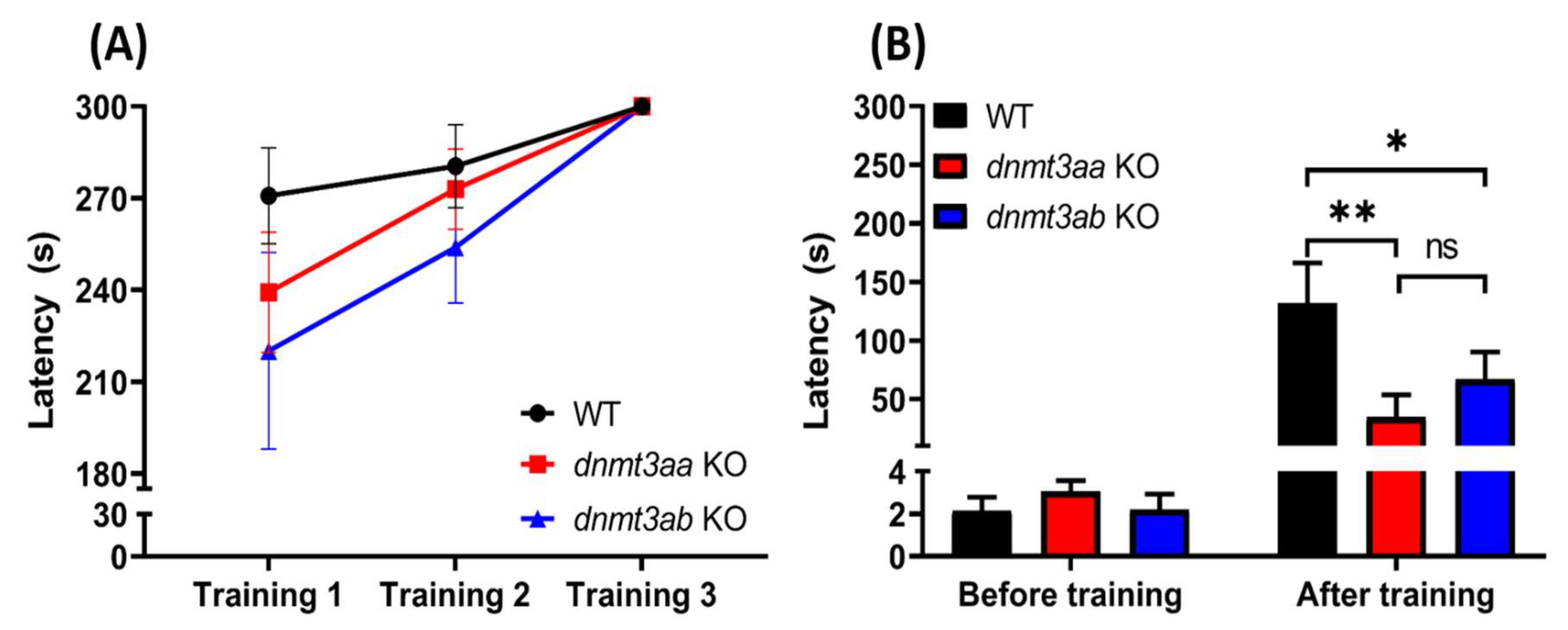
3.9. Effects of dnmt3a Gene-Deficient Zebrafish on Short-Term Memory
3.10. Biochemical Assay of dnmt3a Gene-Deficient Zebrafish
3.11. Profiling of Genome-Wide DNA Methylation Sequencing
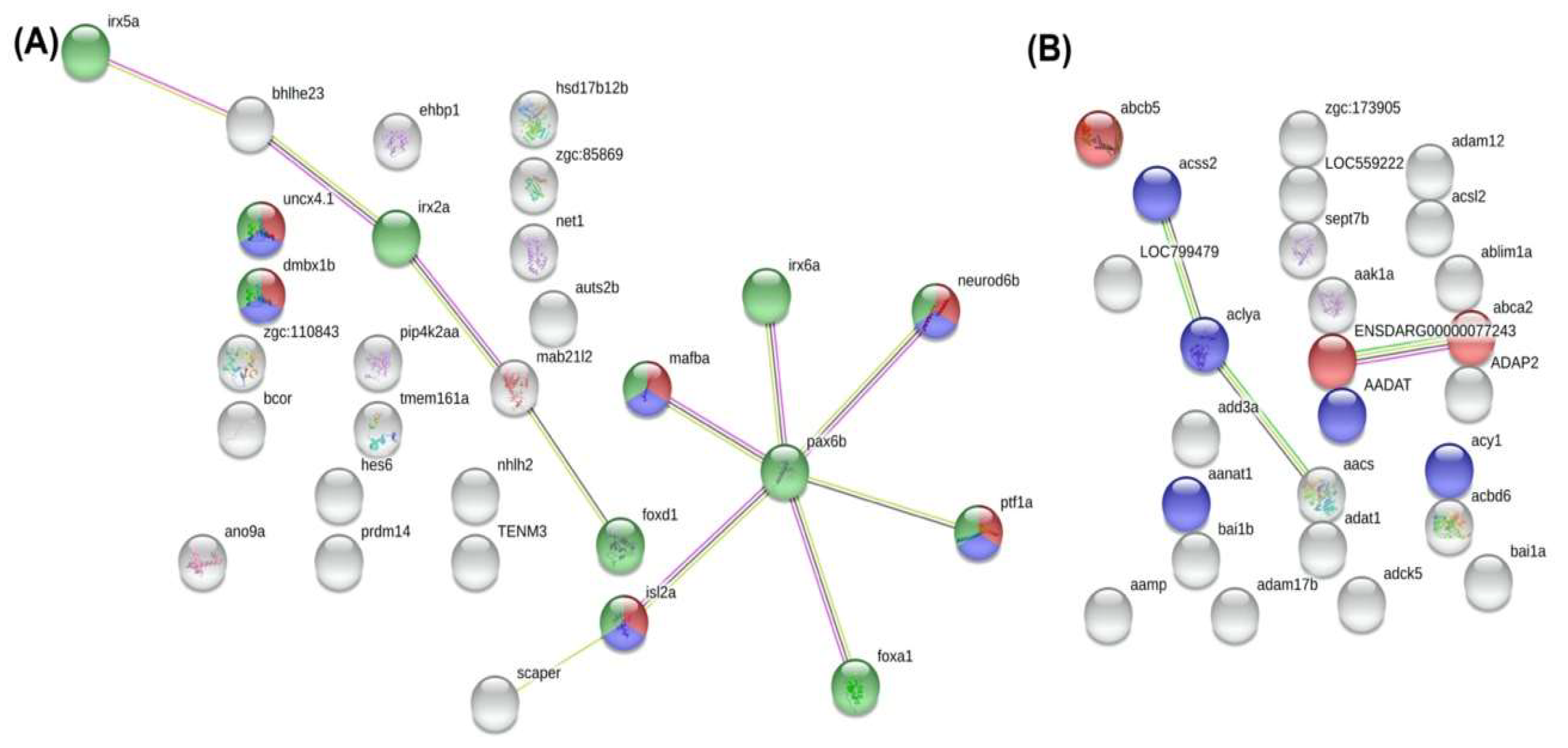
3.12. Identification of Differentially Methylated Regions (DMR) and Functional Analysis of DMR-Associated Genes
4. Discussion
4.1. Novel and Non-Overlapped Functions of dnmt3aa and dnmt3ab Genes on Modulating Behaviors in Zebrafish
4.2. The dnmt3a Gene Contributes to Zebrafish Behavior Responses to a New Environment
4.3. The dnmt3ab Gene Contributes to Zebrafish Aggressive Behavior
4.4. The dnmt3aa and dnmt3ab KO Fish Behaved Boldly in the Presence of a Predator
4.5. The dnmt3ab KO Fish Displayed Alterations in Both Social Tests
4.6. The dnmt3aa and dnmt3ab Genes Inducing the Abnormalities toward Circadian Rhythm Locomotor Activity
4.7. Distinct Preferences of dnmt3aa and dnmt3ab KO Fish toward Visual Stimuli
4.8. dnmt3a Gene is Essential for Memory Retention in Zebrafish
4.9. The Differences in Neurotransmitters and Methylation Level of dnmt3a Gene-Deficient Zebrafish
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Novel Tank Test | ||||||
|---|---|---|---|---|---|---|
| Behavior Endpoints | Fish Lines | Source of Variation | F (DFn, DFd) | p Value | Significance | |
| Average Speed | dnmt3aa KO | Column Factor | F(1, 35) = 0.02003 | 0.8883 | n.s. | |
| Row Factor | F(3.265, 114.3) = 2.265 | 0.0794 | n.s. | |||
| dnmt3ab KO | Column Factor | F(1, 58) = 0.0001992 | 0.9888 | n.s. | ||
| Row Factor | F(4.358, 252.7) = 10.57 | <0.0001 | **** | |||
| Freezing Time Movement Ratio | dnmt3aa KO | Column Factor | F(1, 35) = 0.003813 | 0.9511 | n.s. | |
| Row Factor | F(1.745, 61.06) = 0.1133 | 0.8672 | n.s. | |||
| dnmt3ab KO | Column Factor | F(1, 58) = 1.195 | 0.2788 | n.s. | ||
| Row Factor | F(3.281, 190.3) = 4.759 | 0.0024 | ** | |||
| Time in Top Duration | dnmt3aa KO | Column Factor | F(1, 35) = 18.70 | 0.0001 | *** | |
| Row Factor | F(3.828, 134) = 1.548 | 0.1941 | n.s. | |||
| dnmt3ab KO | Column Factor | F(1, 58) = 2.380 | 0.1284 | n.s. | ||
| Row Factor | F(4.707, 273) = 10.97 | <0.0001 | **** | |||
| Number of Entries to the Top | dnmt3aa KO | Column Factor | F(1, 35) = 7.937 | 0.0079 | ** | |
| Row Factor | F(3.492, 122.2) = 2.944 | 0.0287 | * | |||
| dnmt3ab KO | Column Factor | F(1, 58) = 0.9411 | 0.3360 | n.s. | ||
| Row Factor | F(4.781, 277.3) = 6.991 | <0.0001 | **** | |||
| Latency to Enter the Top | dnmt3aa KO | Column Factor | F(1, 35) = 36.68 | <0.0001 | **** | |
| Row Factor | F(4.262, 149.2) = 7.211 | <0.0001 | **** | |||
| dnmt3ab KO | Column Factor | F(1, 58) = 2.980 | 0.0896 | n.s. | ||
| Row Factor | F(4.675, 271.1) = 21.92 | <0.0001 | **** | |||
| Total Distance Traveled in the Top | dnmt3aa KO | Column Factor | F(1, 35) = 14.51 | 0.0005 | *** | |
| Row Factor | F(3.980, 139.3) = 1.620 | 0.1728 | n.s. | |||
| dnmt3ab KO | Column Factor | F(1, 58) = 2.387 | 0.1278 | n.s. | ||
| Row Factor | F(5.290, 306.8) = 8.278 | <0.0001 | **** | |||
| Mirror Biting Test | ||||||
| Behavior Endpoints | Fish Lines | U-Value | p Value | Significance | ||
| Mirror Biting Time Percentage | dnmt3aa KO | 110 | 0.2212 | n.s. | ||
| dnmt3ab KO | 161.5 | < 0.0001 | **** | |||
| Longest Duration in the Mirror Side | dnmt3aa KO | 144 | 0.8610 | n.s. | ||
| dnmt3ab KO | 164.5 | < 0.0001 | **** | |||
| Average Speed | dnmt3aa KO | 87 | 0.0498 | * | ||
| dnmt3ab KO | 384 | 0.3354 | n.s. | |||
| Freezing Time Movement Ratio | dnmt3aa KO | 117.5 | 0.3190 | n.s. | ||
| dnmt3ab KO | 417.5 | 0.6359 | n.s. | |||
| Swimming Time Movement Ratio | dnmt3aa KO | 117.5 | 0.3183 | n.s. | ||
| dnmt3ab KO | 416.5 | 0.6254 | n.s. | |||
| Rapid Time Movement Ratio | dnmt3aa KO | 85 | 0.0417 | * | ||
| dnmt3ab KO | 369 | 0.2343 | n.s. | |||
| Predator Avoidance Test | ||||||
| Behavior Endpoints | Fish Lines | U-Value | p Value | Significance | ||
| Approaching Predator Time | dnmt3aa KO | 53 | 0.0016 | ** | ||
| dnmt3ab KO | 42 | < 0.0001 | **** | |||
| Average Distance to Separator | dnmt3aa KO | 19 | < 0.0001 | **** | ||
| dnmt3ab KO | 29 | < 0.0001 | **** | |||
| Average Speed | dnmt3aa KO | 119 | 0.3464 | n.s. | ||
| dnmt3ab KO | 98 | 0.0218 | * | |||
| Freezing Time Movement Ratio | dnmt3aa KO | 71 | 0.0123 | * | ||
| dnmt3ab KO | 78.50 | 0.0038 | ** | |||
| Swimming Time Movement Ratio | dnmt3aa KO | 48 | 0.0009 | *** | ||
| dnmt3ab KO | 79.50 | 0.0042 | ** | |||
| Rapid Time Movement Ratio | dnmt3aa KO | 124 | 0.4271 | n.s. | ||
| dnmt3ab KO | 156.5 | 0.5224 | n.s. | |||
| Social Interaction Test | ||||||
| Behavior Endpoints | Fish Lines | U-Value | p Value | Significance | ||
| Average Speed | dnmt3aa KO | 66 | 0.1379 | n.s. | ||
| dnmt3ab KO | 78 | 0.0037 | ** | |||
| Interaction Time Percentage | dnmt3aa KO | 95 | 0.7186 | n.s. | ||
| dnmt3ab KO | 146 | 0.3559 | n.s. | |||
| Longest Duration in Separator Side | dnmt3aa KO | 80 | 0.3500 | n.s. | ||
| dnmt3ab KO | 174 | 0.8800 | n.s. | |||
| Average Distance to Separator | dnmt3aa KO | 73 | 0.2274 | n.s. | ||
| dnmt3ab KO | 79 | 0.0041 | ** | |||
| Shoaling Test | ||||||
| Behavior Endpoints | Fish Lines | U-Value | p Value | Significance | ||
| Average Inter-Fish Distance | dnmt3aa KO | 88 | 0.1226 | n.s. | ||
| dnmt3ab KO | 60 | 0.0005 | *** | |||
| Average Shoal Area | dnmt3aa KO | 8 | 0.2867 | n.s. | ||
| dnmt3ab KO | 0 | 0.0020 | ** | |||
| Average Nearest Neighbor Distance | dnmt3aa KO | 80 | 0.0687 | n.s. | ||
| dnmt3ab KO | 43 | < 0.0001 | **** | |||
| Average Farthest Neighbor Distance | dnmt3aa KO | 91 | 0.1494 | n.s. | ||
| dnmt3ab KO | 29 | < 0.0001 | **** | |||
| Average Speed | dnmt3aa KO | 79 | 0.0636 | n.s. | ||
| dnmt3ab KO | 81 | 0.0050 | ** | |||
| Time in Top Duration | dnmt3aa KO | 12 | < 0.0001 | **** | ||
| dnmt3ab KO | 158 | 0.5544 | n.s. | |||
| Average Distance to Center of the Tank | dnmt3aa KO | 60 | 0.0111 | * | ||
| dnmt3ab KO | 95 | 0.0171 | * | |||
| Circadian Rhythm Locomotor Activity Test | ||||||
| Cycles | Behavior Endpoints | Fish Lines | U-Value | p Value | Significance | |
| Day | Average Speed | dnmt3aa KO | 49,862 | 0.0620 | n.s. | |
| dnmt3ab KO | 35,242 | 0.5673 | n.s. | |||
| Average Angular Velocity | dnmt3aa KO | 31,207 | 0.0055 | ** | ||
| dnmt3ab KO | 13,886 | 0.0002 | *** | |||
| Meandering | dnmt3aa KO | 17,567 | 0.6189 | n.s. | ||
| dnmt3ab KO | 32,656 | 0.0470 | * | |||
| Night | Average Speed | dnmt3aa KO | 16,195 | < 0.0001 | **** | |
| dnmt3ab KO | 4077 | < 0.0001 | **** | |||
| Average Angular Velocity | dnmt3aa KO | 31,267 | < 0.0001 | **** | ||
| dnmt3ab KO | 27,105 | < 0.0001 | **** | |||
| Meandering | dnmt3aa KO | 26,506 | < 0.0001 | **** | ||
| dnmt3ab KO | 11,843 | < 0.0001 | **** | |||
| Color Preference Test | ||||||
| Color Combinations | Fish Line Comparisons | F (DFn, DFd) | Colors | p Value | Significance | |
| Green-Blue | WT vs. dnmt3aa KO | F(5, 124) = 22.88 | Green | 0.0016 | ** | |
| WT vs. dnmt3ab KO | < 0.0001 | **** | ||||
| dnmt3aa KO vs. dnmt3ab KO | > 0.9999 | n.s. | ||||
| WT vs. dnmt3aa KO | Blue | 0.0024 | ** | |||
| WT vs. dnmt3ab KO | < 0.0001 | **** | ||||
| dnmt3aa KO vs. dnmt3ab KO | 0.2146 | n.s. | ||||
| Green-Yellow | WT vs. dnmt3aa KO | F(5, 124) = 182.4 | Green | 0.0016 | ** | |
| WT vs. dnmt3ab KO | < 0.0001 | **** | ||||
| dnmt3aa KO vs. dnmt3ab KO | < 0.0001 | **** | ||||
| WT vs. dnmt3aa KO | Yellow | 0.0016 | ** | |||
| WT vs. dnmt3ab KO | < 0.0001 | **** | ||||
| dnmt3aa KO vs. dnmt3ab KO | < 0.0001 | **** | ||||
| Red-Blue | WT vs. dnmt3aa KO | F(5, 124) = 35.28 | Red | > 0.9999 | n.s. | |
| WT vs. dnmt3ab KO | 0.9273 | n.s. | ||||
| dnmt3aa KO vs. dnmt3ab KO | 0.9658 | n.s. | ||||
| WT vs. dnmt3aa KO | Blue | > 0.9999 | n.s. | |||
| WT vs. dnmt3ab KO | 0.9274 | n.s. | ||||
| dnmt3aa KO vs. dnmt3ab KO | 0.9658 | n.s. | ||||
| Green-Red | WT vs. dnmt3aa KO | F(5, 124) = 62.59 | Green | 0.9984 | n.s. | |
| WT vs. dnmt3ab KO | < 0.0001 | **** | ||||
| dnmt3aa KO vs. dnmt3ab KO | < 0.0001 | **** | ||||
| WT vs. dnmt3aa KO | Red | 0.9984 | n.s. | |||
| WT vs. dnmt3ab KO | < 0.0001 | **** | ||||
| dnmt3aa KO vs. dnmt3ab KO | < 0.0001 | **** | ||||
| Red-Yellow | WT vs. dnmt3aa KO | F(5, 124) = 381.1 | Red | 0.0015 | ** | |
| WT vs. dnmt3ab KO | < 0.0001 | **** | ||||
| dnmt3aa KO vs. dnmt3ab KO | 0.8337 | n.s. | ||||
| WT vs. dnmt3aa KO | Yellow | 0.0015 | ** | |||
| WT vs. dnmt3ab KO | < 0.0001 | **** | ||||
| dnmt3aa KO vs. dnmt3ab KO | 0.8337 | n.s. | ||||
| Blue-Yellow | WT vs. dnmt3aa KO | F(5, 124) = 934.6 | Blue | < 0.0001 | **** | |
| WT vs. dnmt3ab KO | < 0.0001 | **** | ||||
| dnmt3aa KO vs. dnmt3ab KO | 0.3531 | n.s. | ||||
| WT vs. dnmt3aa KO | Yellow | < 0.0001 | **** | |||
| WT vs. dnmt3ab KO | < 0.0001 | **** | ||||
| dnmt3aa KO vs. dnmt3ab KO | 0.3531 | n.s. | ||||
| Short-Term Memory Test | ||||||
| Behavior Endpoints | Fish Line Comparisons | F (DFn, DFd) | p Value | Significance | ||
| Latency After 24 h | Before Training | WT vs. dnmt3aa KO | Column Factor F (2, 86) = 3.487 p = 0.0350 * Row Factor F (1, 86) = 24.59 p < 0.0001 **** | 0.9993 | n.s. | |
| WT vs. dnmt3ab KO | > 0.9999 | n.s. | ||||
| dnmt3aa KO vs. dnmt3ab KO | 0.9994 | n.s. | ||||
| After Training | WT vs. dnmt3aa KO | 0.0011 | ** | |||
| WT vs. dnmt3ab KO | 0.0443 | * | ||||
| dnmt3aa KO vs. dnmt3ab KO | 0.4340 | n.s. | ||||
| Global DNA Methylation Level Test | ||||||
| Parameters | Fish Line Comparisons | F (DFn, DFd) | p Value | Significance | ||
| 5-hmC | WT vs. dnmt3aa KO | F (2, 12) = 0.9873 | 0.2012 | n.s. | ||
| WT vs. dnmt3ab KO | 0.3745 | n.s. | ||||
| dnmt3aa KO vs. dnmt3ab KO | 0.6478 | n.s. | ||||
| 5-mC | WT vs. dnmt3aa KO | F (2, 12) = 1.439 | 0.1761 | n.s. | ||
| WT vs. dnmt3ab KO | 0.1788 | n.s. | ||||
| dnmt3aa KO vs. dnmt3ab KO | 0.9263 | n.s. | ||||
| Ratio | WT vs. dnmt3aa KO | F (2, 12) = 0.006109 | 0.9247 | n.s. | ||
| WT vs. dnmt3ab KO | 0.9299 | n.s. | ||||
| dnmt3aa KO vs. dnmt3ab KO | 0.9908 | n.s. | ||||
References
- Fernandez, A.F.; Assenov, Y.; Martin-Subero, J.I.; Balint, B.; Siebert, R.; Taniguchi, H.; Yamamoto, H.; Hidalgo, M.; Tan, A.-C.; Galm, O. A DNA methylation fingerprint of 1628 human samples. Genome Res. 2012, 22, 407–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, B.; Robertson, K.D. DNA methyltransferases, DNA damage repair, and cancer. In Epigenetic Alterations in Oncogenesis; Springer: Berlin/Heidelberg, Germany, 2013; pp. 3–29. [Google Scholar]
- Rasmussen, K.D.; Helin, K. Role of tet enzymes in DNA methylation, development, and cancer. Genes Dev. 2016, 30, 733–750. [Google Scholar] [CrossRef] [PubMed]
- Robertson, K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005, 6, 597. [Google Scholar] [CrossRef] [PubMed]
- Bestor, T.H. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000, 9, 2395–2402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA methyltransferases dnmt3a and dnmt3b are essential for de novo methylation and mammalian development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef] [Green Version]
- Bourc’his, D.; Xu, G.-L.; Lin, C.-S.; Bollman, B.; Bestor, T.H. Dnmt3l and the establishment of maternal genomic imprints. Science 2001, 294, 2536–2539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hata, K.; Okano, M.; Lei, H.; Li, E. Dnmt3l cooperates with the dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 2002, 129, 1983–1993. [Google Scholar] [PubMed]
- Jeltsch, A.; Ehrenhofer-Murray, A.; Jurkowski, T.P.; Lyko, F.; Reuter, G.; Ankri, S.; Nellen, W.; Schaefer, M.; Helm, M. Mechanism and biological role of dnmt2 in nucleic acid methylation. RNA Biol. 2017, 14, 1108–1123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karpf, A.R.; Matsui, S.-I. Genetic disruption of cytosine DNA methyltransferase enzymes induces chromosomal instability in human cancer cells. Cancer Res. 2005, 65, 8635–8639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, L.-J.; Zhang, Y.; Lv, P.-P.; He, W.-H.; Wu, Y.-T.; Liu, A.-X.; Ding, G.-L.; Dong, M.-Y.; Qu, F.; Xu, C.-M. Insufficient maintenance DNA methylation is associated with abnormal embryonic development. BMC Med. 2012, 10, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheaffer, K.L.; Elliott, E.N.; Kaestner, K.H. DNA hypomethylation contributes to genomic instability and intestinal cancer initiation. Cancer Prev. Res. 2016, 9, 534–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taberlay, P.C.; Jones, P.A. DNA methylation and cancer. In Epigenetics and Disease; Springer: Berlin/Heidelberg, Germany, 2011; pp. 1–23. [Google Scholar]
- Wang, J.; Yu, J.-T.; Tan, M.-S.; Jiang, T.; Tan, L. Epigenetic mechanisms in alzheimer’s disease: Implications for pathogenesis and therapy. Ageing Res. Rev. 2013, 12, 1024–1041. [Google Scholar] [CrossRef]
- Ménard, C.; Hodes, G.E.; Russo, S.J. Pathogenesis of depression: Insights from human and rodent studies. Neuroscience 2016, 321, 138–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nieto, S.J.; Patriquin, M.A.; Nielsen, D.A.; Kosten, T.A. Don’t worry; be informed about the epigenetics of anxiety. Pharmacol. Biochem. Behav. 2016, 146, 60–72. [Google Scholar] [CrossRef] [Green Version]
- Grayson, D.R.; Jia, X.; Chen, Y.; Sharma, R.P.; Mitchell, C.P.; Guidotti, A.; Costa, E. Reelin promoter hypermethylation in schizophrenia. Proc. Natl. Acad. Sci. USA 2005, 102, 9341–9346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Day, J.J.; Sweatt, J.D. Cognitive neuroepigenetics: A role for epigenetic mechanisms in learning and memory. Neurobiol. Learn. Mem. 2011, 96, 2–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishii, D.; Matsuzawa, D.; Matsuda, S.; Tomizawa, H.; Sutoh, C.; Shimizu, E. Methyl donor-deficient diet during development can affect fear and anxiety in adulthood in c57bl/6j mice. PLoS ONE 2014, 9, e105750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammels, C.; Prickaerts, J.; Kenis, G.; Vanmierlo, T.; Fischer, M.; Steinbusch, H.W.; van Os, J.; van den Hove, D.L.; Rutten, B.P. Differential susceptibility to chronic social defeat stress relates to the number of dnmt3a-immunoreactive neurons in the hippocampal dentate gyrus. Psychoneuroendocrinology 2015, 51, 547–556. [Google Scholar] [CrossRef] [Green Version]
- Griffith, J.S.; Mahler, H.R. DNA ticketing theory of memory. Nature 1969, 223, 580–582. [Google Scholar] [CrossRef]
- Levenson, J.M.; Sweatt, J.D. Epigenetic mechanisms in memory formation. Nat. Rev. Neurosci. 2005, 6, 108. [Google Scholar] [CrossRef]
- Morris, M.J.; Na, E.S.; Autry, A.E.; Monteggia, L.M. Impact of dnmt1 and dnmt3a forebrain knockout on depressive-and anxiety like behavior in mice. Neurobiol. Learn. Mem. 2016, 135, 139–145. [Google Scholar] [CrossRef] [Green Version]
- Shimoda, N.; Yamakoshi, K.; Miyake, A.; Takeda, H. Identification of a gene required for de novo DNA methylation of the zebrafish no tail gene. Dev. Dyn. An Off. Publ. Am. Assoc. Anat. 2005, 233, 1509–1516. [Google Scholar]
- Aluru, N.; Kuo, E.; Helfrich, L.W.; Karchner, S.I.; Linney, E.A.; Pais, J.E.; Franks, D.G. Developmental exposure to 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin alters DNA methyltransferase (dnmt) expression in zebrafish (Danio rerio). Toxicol. Appl. Pharmacol. 2015, 284, 142–151. [Google Scholar] [CrossRef] [Green Version]
- Kalueff, A.V.; Gebhardt, M.; Stewart, A.M.; Cachat, J.M.; Brimmer, M.; Chawla, J.S.; Craddock, C.; Kyzar, E.J.; Roth, A.; Landsman, S. Towards a comprehensive catalog of zebrafish behavior 1.0 and beyond. Zebrafish 2013, 10, 70–86. [Google Scholar] [CrossRef]
- Avdesh, A.; Chen, M.; Martin-Iverson, M.T.; Mondal, A.; Ong, D.; Rainey-Smith, S.; Taddei, K.; Lardelli, M.; Groth, D.M.; Verdile, G. Regular care and maintenance of a zebrafish (Danio rerio) laboratory: An introduction. JoVE J. Vis. Exp. 2012, 69, e4196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pittman, J.T.; Lott, C.S. Startle response memory and hippocampal changes in adult zebrafish pharmacologically-induced to exhibit anxiety/depression-like behaviors. Physiol. Behav. 2014, 123, 174–179. [Google Scholar] [CrossRef]
- Parker, M.O.; Brock, A.J.; Millington, M.E.; Brennan, C.H. Behavioral phenotyping of casper mutant and 1-pheny-2-thiourea treated adult zebrafish. Zebrafish 2013, 10, 466–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fontana, B.D.; Stefanello, F.V.; Mezzomo, N.J.; Müller, T.E.; Quadros, V.A.; Parker, M.O.; Rico, E.P.; Rosemberg, D.B. Taurine modulates acute ethanol-induced social behavioral deficits and fear responses in adult zebrafish. J. Psychiatr. Res. 2018, 104, 176–182. [Google Scholar] [CrossRef] [Green Version]
- Bernardi, M.; Dias, S.; Barbosa, V. Neurotoxicity of neem commercial formulation (Azadirachta indica a. Juss) in adult zebrafish (Danio rerio). Environ. Toxicol. Pharmacol. 2013, 36, 1276–1282. [Google Scholar] [CrossRef]
- Thomas, J.; Janz, D. Dietary selenomethionine exposure in adult zebrafish alters swimming performance, energetics and the physiological stress response. Aquat. Toxicol. 2011, 102, 79–86. [Google Scholar] [CrossRef]
- Liang, S.-T.; Audira, G.; Juniardi, S.; Chen, J.-R.; Lai, Y.-H.; Du, Z.-C.; Lin, D.-S.; Hsiao, C.-D. Zebrafish carrying pycr1 gene deficiency display aging and multiple behavioral abnormalities. Cells 2019, 8, 453. [Google Scholar] [CrossRef] [Green Version]
- Audira, G.; Sampurna, B.; Juniardi, S.; Liang, S.-T.; Lai, Y.-H.; Hsiao, C.-D. A versatile setup for measuring multiple behavior endpoints in zebrafish. Inventions 2018, 3, 75. [Google Scholar] [CrossRef] [Green Version]
- Audira, G.; Sampurna, B.P.; Juniardi, S.; Liang, S.-T.; Lai, Y.-H.; Han, L.; Hsiao, C.-D. Establishing simple image-based methods and cost-effective instrument for toxicity assessment on circadian rhythm dysregulation in fish. Biol. Open 2019, 8, bio.041871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levin, E.D.; Bencan, Z.; Cerutti, D.T. Anxiolytic effects of nicotine in zebrafish. Physiol. Behav. 2007, 90, 54–58. [Google Scholar] [CrossRef] [PubMed]
- Cachat, J.; Stewart, A.; Grossman, L.; Gaikwad, S.; Kadri, F.; Chung, K.M.; Wu, N.; Wong, K.; Roy, S.; Suciu, C. Measuring behavioral and endocrine responses to novelty stress in adult zebrafish. Nat. Protoc. 2010, 5, 1786. [Google Scholar] [CrossRef]
- Kysil, E.V.; Meshalkina, D.A.; Frick, E.E.; Echevarria, D.J.; Rosemberg, D.B.; Maximino, C.; Lima, M.G.; Abreu, M.S.; Giacomini, A.C.; Barcellos, L.J. Comparative analyses of zebrafish anxiety-like behavior using conflict-based novelty tests. Zebrafish 2017, 14, 197–208. [Google Scholar] [CrossRef]
- Kalueff, A.V.; Stewart, A.M.; Gerlai, R. Zebrafish as an emerging model for studying complex brain disorders. Trends Pharmacol. Sci. 2014, 35, 63–75. [Google Scholar] [CrossRef] [Green Version]
- Siregar, P.; Juniardi, S.; Audira, G.; Lai, Y.-H.; Huang, J.-C.; Chen, K.H.-C.; Chen, J.-R.; Hsiao, C.-D. Method standardization for conducting innate color preference studies in different zebrafish strains. Biomedicines 2020, 8, 271. [Google Scholar] [CrossRef]
- Kassa, J.; Bajgar, J.; Kuča, K.; Jun, D. Behavioral toxicity of nerve agents. In Handbook of Toxicology of Chemical Warfare Agents; Elsevier: Amsterdam, The Netherlands, 2020; pp. 499–513. [Google Scholar]
- Kim, Y.-H.; Lee, Y.; Kim, D.; Jung, M.W.; Lee, C.-J. Scopolamine-induced learning impairment reversed by physostigmine in zebrafish. Neurosci. Res. 2010, 67, 156–161. [Google Scholar] [CrossRef]
- Overmier, J.B.; Flood, N.B. Passive avoidance in forebrain ablated teleost fish, carassius auratus. Physiol. Behav. 1969, 4, 791–794. [Google Scholar] [CrossRef]
- Day, J.J.; Kennedy, A.J.; Sweatt, J.D. DNA methylation and its implications and accessibility for neuropsychiatric therapeutics. Annu. Rev. Pharmacol. Toxicol. 2015, 55, 591–611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bui Thi, N.H.; Nguyen Thi, N.A.; Audira, G.; Siregar, P.; Liang, S.-T.; Huang, J.-C.; Hsiao, C.-D. Chronic exposure to low concentration lead chloride-induced anxiety and loss of aggression and memory in zebrafish. Int. J. Mol. Sci. 2020, 21, 1844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Escudero, A.; Vicente-Page, J.; Hinz, R.C.; Arganda, S.; de Polavieja, G.G. Idtracker: Tracking individuals in a group by automatic identification of unmarked animals. Nat. Methods 2014, 11, 743. [Google Scholar] [CrossRef]
- Audira, G.; Sampurna, B.P.; Juniardi, S.; Liang, S.-T.; Lai, Y.-H.; Hsiao, C.-D. A simple setup to perform 3D locomotion tracking in zebrafish by using a single camera. Inventions 2018, 3, 11. [Google Scholar] [CrossRef] [Green Version]
- Picciotto, M.R.; Higley, M.J.; Mineur, Y.S. Acetylcholine as a neuromodulator: Cholinergic signaling shapes nervous system function and behavior. Neuron 2012, 76, 116–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawrence, J.J. Cholinergic control of gaba release: Emerging parallels between neocortex and hippocampus. Trends Neurosci. 2008, 31, 317–327. [Google Scholar] [CrossRef]
- Fernstrom, J.D.; Fernstrom, M.H. Tyrosine, phenylalanine, and catecholamine synthesis and function in the brain. J. Nutr. 2007, 137, 1539S–1547S. [Google Scholar] [CrossRef]
- Li, N.; Xia, T.; Nel, A.E. The role of oxidative stress in ambient particulate matter-induced lung diseases and its implications in the toxicity of engineered nanoparticles. Free Radic. Biol. Med. 2008, 44, 1689–1699. [Google Scholar] [CrossRef] [Green Version]
- Tekkök, S.B.; Ye, Z.; Ransom, B.R. Excitotoxic mechanisms of ischemic injury in myelinated white matter. J. Cereb. Blood Flow Metab. 2007, 27, 1540–1552. [Google Scholar] [CrossRef] [Green Version]
- Feng, J.; Zhou, Y.; Campbell, S.L.; Le, T.; Li, E.; Sweatt, J.D.; Silva, A.J.; Fan, G. Dnmt1 and dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 2010, 13, 423. [Google Scholar] [CrossRef]
- Barba-Escobedo, P.A.; Gould, G.G. Visual social preferences of lone zebrafish in a novel environment: Strain and anxiolytic effects. Genes Brain Behav. 2012, 11, 366–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engeszer, R.E.; Ryan, M.J.; Parichy, D.M. Learned social preference in zebrafish. Curr. Biol. 2004, 14, 881–884. [Google Scholar] [CrossRef] [Green Version]
- Pham, M.; Raymond, J.; Hester, J.; Kyzar, E.; Gaikwad, S.; Bruce, I.; Fryar, C.; Chanin, S.; Enriquez, J.; Bagawandoss, S. Assessing social behavior phenotypes in adult zebrafish: Shoaling, social preference, and mirror biting tests. In Zebrafish Protocols for Neurobehavioral Research; Springer: Berlin/Heidelberg, Germany, 2012; pp. 231–246. [Google Scholar]
- Gerlai, R.; Lee, V.; Blaser, R. Effects of acute and chronic ethanol exposure on the behavior of adult zebrafish (Danio rerio). Pharmacol. Biochem. Behav. 2006, 85, 752–761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magalhaes, F.E.A.; Batista, F.L.A.; Lima, L.M.G.; Abrante, I.D.A.; Batista, F.L.A.; Abrante, I.D.A.; de Araújo, J.I.F.; Santos, S.A.A.R.; de Oliveira, B.A.; Raposo, R.D.S. Adult zebrafish (Danio rerio) as a model for the study of corneal antinociceptive compounds. Zebrafish 2018, 15, 566–574. [Google Scholar] [CrossRef] [PubMed]
- Menezes, F.P.; da Silva, R.S. The influence of temperature on adult zebrafish sensitivity to pentylenetetrazole. Epilepsy Res. 2017, 135, 14–18. [Google Scholar] [CrossRef]
- Fontana, B.D.; Cleal, M.; Parker, M.O. Female adult zebrafish (Danio rerio) show higher levels of anxiety-like behavior than males, but do not differ in learning and memory capacity. Eur. J. Neurosci. 2019, 52, 2604–2613. [Google Scholar] [CrossRef]
- Rambo, C.L.; Mocelin, R.; Marcon, M.; Villanova, D.; Koakoski, G.; de Abreu, M.S.; Oliveira, T.A.; Barcellos, L.J.; Piato, A.L.; Bonan, C.D. Gender differences in aggression and cortisol levels in zebrafish subjected to unpredictable chronic stress. Physiol. Behav. 2017, 171, 50–54. [Google Scholar] [CrossRef]
- Genario, R.; Giacomini, A.C.; de Abreu, M.S.; Marcon, L.; Demin, K.A.; Kalueff, A.V. Sex differences in adult zebrafish anxiolytic-like responses to diazepam and melatonin. Neurosci. Lett. 2020, 714, 134548. [Google Scholar] [CrossRef]
- Wendelaar Bonga, S.E. The stress response in fish. Physiol. Rev. 1997, 77, 591–625. [Google Scholar] [CrossRef] [PubMed]
- Weis, J.S.; Smith, G.; Zhou, T.; Santiago-Bass, C.; Weis, P. Effects of contaminants on behavior: Biochemical mechanisms and ecological consequences: Killifish from a contaminated site are slow to capture prey and escape predators; altered neurotransmitters and thyroid may be responsible for this behavior, which may produce population changes in the fish and their major prey, the grass shrimp. Bioscience 2001, 51, 209–217. [Google Scholar]
- Rao, J.V.; Begum, G.; Pallela, R.; Usman, P.; Rao, R.N. Changes in behavior and brain acetylcholinesterase activity in mosquito fish, gambusia affinis in response to the sub-lethal exposure to chlorpyrifos. Int. J. Environ. Res. Public Health 2005, 2, 478–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Won, Y.K.; Liu, J.; Olivier, K., Jr.; Zheng, Q.; Pope, C.N. Age-related effects of chlorpyrifos on acetylcholine release in rat brain. Neurotoxicology 2001, 22, 39–48. [Google Scholar] [CrossRef]
- Gogos, J.A.; Morgan, M.; Luine, V.; Santha, M.; Ogawa, S.; Pfaff, D.; Karayiorgou, M. Catechol-o-methyltransferase-deficient mice exhibit sexually dimorphic changes in catecholamine levels and behavior. Proc. Natl. Acad. Sci. USA 1998, 95, 9991–9996. [Google Scholar] [CrossRef] [Green Version]
- Halappa, R.; David, M. Behavioral responses of the freshwater fish, Cyprinus carpio (Linnaeus) following sublethal exposure to chlorpyrifos. Turk. J. Fish. Aquat. Sci. 2009, 9, 233–238. [Google Scholar] [CrossRef]
- Oliveira, R.F.; Silva, J.F.; Simoes, J.M. Fighting zebrafish: Characterization of aggressive behavior and winner–loser effects. Zebrafish 2011, 8, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Audira, G.; Sarasamma, S.; Chen, J.-R.; Juniardi, S.; Sampurna, B.; Liang, S.-T.; Lai, Y.-H.; Lin, G.-M.; Hsieh, M.-C.; Hsiao, C.-D. Zebrafish mutants carrying leptin a (lepa) gene deficiency display obesity, anxiety, less aggression and fear, and circadian rhythm and color preference dysregulation. Int. J. Mol. Sci. 2018, 19, 4038. [Google Scholar] [CrossRef] [Green Version]
- Miczek, K.A.; Fish, E.W.; Joseph, F.; de Almeida, R.M. Social and neural determinants of aggressive behavior: Pharmacotherapeutic targets at serotonin, dopamine and γ-aminobutyric acid systems. Psychopharmacology 2002, 163, 434–458. [Google Scholar] [CrossRef] [PubMed]
- Miczek, K.A.; Maxson, S.C.; Fish, E.W.; Faccidomo, S. Aggressive behavioral phenotypes in mice. Behav. Brain Res. 2001, 125, 167–181. [Google Scholar] [CrossRef]
- De Almeida, R.M.; Ferrari, P.F.; Parmigiani, S.; Miczek, K.A. Escalated aggressive behavior: Dopamine, serotonin and gaba. Eur. J. Pharmacol. 2005, 526, 51–64. [Google Scholar] [CrossRef]
- Thomas, S.A.; Palmiter, R.D. Impaired maternal behavior in mice lacking norepinephrine and epinephrine. Cell 1997, 91, 583–592. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; Gainetdinov, R.R.; Wetsel, W.C.; Jones, S.R.; Bohn, L.M.; Miller, G.W.; Wang, Y.-M.; Caron, M.G. Mice lacking the norepinephrine transporter are supersensitive to psychostimulants. Nat. Neurosci. 2000, 3, 465. [Google Scholar] [CrossRef]
- Ellison, G.D. Behavior and the balance between norepinephrine and serotonin. Acta Neurobiol. Exp. 1975, 35, 499–515. [Google Scholar]
- Ahmed, O.; Seguin, D.; Gerlai, R. An automated predator avoidance task in zebrafish. Behav. Brain Res. 2011, 216, 166–171. [Google Scholar] [CrossRef] [Green Version]
- Saif, M.; Chatterjee, D.; Buske, C.; Gerlai, R. Sight of conspecific images induces changes in neurochemistry in zebrafish. Behav. Brain Res. 2013, 243, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Larson, E.T.; Summers, C.H. Serotonin reverses dominant social status. Behav. Brain Res. 2001, 121, 95–102. [Google Scholar] [CrossRef]
- Di Giusto, E.; Cairncross, K.; King, M. Hormonal influences on fear-motivated responses. Psychol. Bull. 1971, 75, 432. [Google Scholar] [CrossRef]
- Pezze, M.A.; Feldon, J. Mesolimbic dopaminergic pathways in fear conditioning. Prog. Neurobiol. 2004, 74, 301–320. [Google Scholar] [CrossRef]
- Shekhar, A.; Katner, J.; Rusche, W.; Sajdyk, T.; Simon, J. Fear-potentiated startle elevates catecholamine levels in the dorsomedial hypothalamus of rats. Pharmacol. Biochem. Behav. Brain Funct. 1994, 48, 525–529. [Google Scholar] [CrossRef]
- Shams, S.; Chatterjee, D.; Gerlai, R. Chronic social isolation affects thigmotaxis and whole-brain serotonin levels in adult zebrafish. Behav. Brain Res. 2015, 292, 283–287. [Google Scholar] [CrossRef]
- Buske, C.; Gerlai, R. Early embryonic ethanol exposure impairs shoaling and the dopaminergic and serotoninergic systems in adult zebrafish. Neurotoxicol. Teratol. 2011, 33, 698–707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edwards, D.H.; Kravitz, E.A. Serotonin, social status and aggression. Curr. Opin. Neurobiol. 1997, 7, 812–819. [Google Scholar] [CrossRef]
- Sánchez-Vázquez, F.J.; López-Olmeda, J.F.; Vera, L.M.; Migaud, H.; López-Patiño, M.A.; Míguez, J.M. Environmental cycles, melatonin, and circadian control of stress response in fish. Front. Endocrinol. 2019, 10, 279. [Google Scholar] [CrossRef] [PubMed]
- Falcon, J.; Migaud, H.; Munoz-Cueto, J.A.; Carrillo, M. Current knowledge on the melatonin system in teleost fish. Gen. Comp. Endocrinol. 2010, 165, 469–482. [Google Scholar] [CrossRef] [Green Version]
- Iuvone, P.M.; Tosini, G.; Pozdeyev, N.; Haque, R.; Klein, D.C.; Chaurasia, S.S. Circadian clocks, clock networks, arylalkylamine n-acetyltransferase, and melatonin in the retina. Prog. Retin. Eye Res. 2005, 24, 433–456. [Google Scholar] [CrossRef]
- Cahill, G.M. Circadian melatonin rhythms in cultured zebrafish pineals are not affected by catecholamine receptor agonists. Gen. Comp. Endocrinol. 1997, 105, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-S.; Ryu, J.-H.; Choi, T.-I.; Bae, Y.-K.; Lee, S.; Kang, H.J.; Kim, C.-H. Innate color preference of zebrafish and its use in behavioral analyses. Mol. Cells 2016, 39, 750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Liu, X.; Sun, M.; Zhang, Q.; Li, T.; Li, X.; Xu, J.; Zhao, X.; Chen, D.; Feng, X. Reversal of reserpine-induced depression and cognitive disorder in zebrafish by sertraline and traditional chinese medicine (tcm). Behav. Brain Funct. 2018, 14, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.-B.; Yao, Y.-Y.; Zhang, H.-F.; Kawakami, K.; Du, J.-L. Left habenula mediates light-preference behavior in zebrafish via an asymmetrical visual pathway. Neuron 2017, 93, 914–928. [Google Scholar] [CrossRef] [Green Version]
- Bault, Z.A.; Peterson, S.M.; Freeman, J.L. Directional and color preference in adult zebrafish: Implications in behavioral and learning assays in neurotoxicology studies. J. Appl. Toxicol. 2015, 35, 1502–1510. [Google Scholar] [CrossRef]
- Avdesh, A.; Martin-Iverson, M.T.; Mondal, A.; Chen, M.; Askraba, S.; Morgan, N.; Lardelli, M.; Groth, D.M.; Verdile, G.; Martins, R.N. Evaluation of color preference in zebrafish for learning and memory. J. Alzheimer’s Dis. 2012, 28, 459–469. [Google Scholar] [CrossRef]
- Miller, C.A.; Sweatt, J.D. Covalent modification of DNA regulates memory formation. Neuron 2007, 53, 857–869. [Google Scholar] [CrossRef] [Green Version]
- Levenson, J.M.; Qiu, S.; Weeber, E.J. The role of reelin in adult synaptic function and the genetic and epigenetic regulation of the reelin gene. Biochim. Et Biophys. Acta BBA Gene Regul. Mech. 2008, 1779, 422–431. [Google Scholar] [CrossRef]
- Morris, M.J.; Adachi, M.; Na, E.S.; Monteggia, L.M. Selective role for dnmt3a in learning and memory. Neurobiol. Learn. Mem. 2014, 115, 30–37. [Google Scholar] [CrossRef] [Green Version]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [Green Version]
- Peaston, A.E.; Whitelaw, E. Epigenetics and phenotypic variation in mammals. Mamm. Genome 2006, 17, 365–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ledón-Rettig, C.C.; Richards, C.L.; Martin, L.B. Epigenetics for behavioral ecologists. Behav. Ecol. 2013, 24, 311–324. [Google Scholar] [CrossRef] [Green Version]
- Abdolmaleky, H.M.; Smith, C.L.; Zhou, J.-R.; Thiagalingam, S. Epigenetic alterations of the dopaminergic system in major psychiatric disorders. In Pharmacogenomics in Drug Discovery and Development; Springer: Berlin/Heidelberg, Germany, 2008; pp. 187–212. [Google Scholar]
- Ntziachristos, P.; Mullenders, J.; Trimarchi, T.; Aifantis, I. Mechanisms of epigenetic regulation of leukemia onset and progression. In Advances in Immunology; Elsevier: Amsterdam, The Netherlands, 2013; Volume 117, pp. 1–38. [Google Scholar]
- Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.-F.; Chan, W.-Y. The de novo DNA methyltransferase dnmt3a in development and cancer. Epigenetics 2014, 9, 669–677. [Google Scholar] [CrossRef] [Green Version]
- Avecilla, A.; Doke, M.; Jovellanos, J.; Avecilla, V. Contribution of inhibitor of differentiation and estrogenic endocrine disruptors to neurocognitive disorders. Med. Sci. 2018, 6, 61. [Google Scholar] [CrossRef] [Green Version]
- Katrancha, S.M.; Shaw, J.E.; Zhao, A.Y.; Myers, S.A.; Cocco, A.R.; Jeng, A.T.; Zhu, M.; Pittenger, C.; Greer, C.A.; Carr, S.A.; et al. Trio haploinsufficiency causes neurodevelopmental disease-associated deficits. Cell Rep. 2019, 26, 2805–2817. [Google Scholar] [CrossRef] [Green Version]
- Borromeo, M.D.; Meredith, D.M.; Castro, D.S.; Chang, J.C.; Tung, K.C.; Guillemot, F.; Johnson, J.E. Correction: A transcription factor network specifying inhibitory versus excitatory neurons in the dorsal spinal cord. Development 2017, 144, 2539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takamiya, M.; Weger, B.D.; Schindler, S.; Beil, T.; Yang, L.; Armant, O.; Ferg, M.; Schlunck, G.; Reinhard, T.; Dickmeis, T.; et al. Molecular description of eye defects in the zebrafish pax6b mutant, sunrise, reveals a pax6b-dependent genetic network in the developing anterior chamber. PLoS ONE 2015, 10, e0117645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moosajee, M.; Hingorani, M.; Moore, A.T. Pax6-related aniridia. In Genereviews ((r)); Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Stephens, K., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- De Roeck, A.; van Broeckhoven, C.; Sleegers, K. The role of abca7 in Alzheimer’s disease: Evidence from genomics, transcriptomics and methylomics. Acta Neuropathol. 2019, 138, 201–220. [Google Scholar] [CrossRef] [Green Version]
- Szutowicz, A.; Bielarczyk, H.; Jankowska-Kulawy, A.; Pawelczyk, T.; Ronowska, A. Acetyl-coa the key factor for survival or death of cholinergic neurons in course of neurodegenerative diseases. Neurochem. Res. 2013, 38, 1523–1542. [Google Scholar] [CrossRef] [Green Version]
- Currais, A.; Huang, L.; Goldberg, J.; Petrascheck, M.; Ates, G.; Pinto-Duarte, A.; Shokhirev, M.N.; Schubert, D.; Maher, P. Elevating acetyl-coa levels reduces aspects of brain aging. eLife 2019, 8, e47866. [Google Scholar] [CrossRef] [PubMed]



| Biomarkers | WT | dnmt3aa KO | dnmt3ab KO | Unit | F (DFn, DFd) | ANOVA | ||
|---|---|---|---|---|---|---|---|---|
| Concentration | Concentration | p-Value | Concentration | p-Value | Significance | |||
| Dopamine | 1.7380 ± 0.2852 | 1.9060 ± 0.3808 | 0.7410 | 1.7550 ± 0.3898 | 0.9737 | pg/μg | F (2, 27) = 0.06767 | NO |
| GABA | 0.0093 ± 0.0017 | 0.0099 ± 0.0023 | 0.8345 | 0.0091 ± 0.0020 | 0.9445 | nmol/μg | F (2, 27) = 0.04286 | NO |
| Serotonin (5-HT) | 0.0290 ± 0.0058 | 0.0331 ± 0.0087 | 0.7020 | 0.0289 ± 0.0077 | 0.9925 | ng/μg | F (2, 27) = 0.1022 | NO |
| Norepinephrine | 0.1711 ± 0.0333 | 0.1749 ± 0.0379 | 0.9417 | 0.1499 ± 0.0378 | 0.6837 | ng/μg | F (2, 27) = 0.1369 | NO |
| ROS | 0.5943 ± 0.0918 | 0.5890 ± 0.1273 | 0.9721 | 0.4874 ± 0.0961 | 0.4831 | U/mL | F (2, 27) = 0.3212 | NO |
| ACh | 0.7061 ± 0.1742 | 0.9756 ± 0.2387 | 0.3193 | 0.8205 ± 0.1360 | 0.6701 | ug/μg | F (2, 27) = 0.5187 | NO |
| Cortisol | 2.2670 ± 0.3324 | 2.2670 ± 0.3272 | 0.9985 | 1.7780 ± 0.1878 | 0.2445 | pg/μg | F (2, 27) = 0.9451 | NO |
| Glutamate | 0.0262 ± 0.0044 | 0.0311 ± 0.0054 | 0.4975 | 0.0329 ± 0.0052 | 0.3554 | ug/μg | F (2, 27) = 0.4737 | NO |
| Glycine | 0.1369 ± 0.0207 | 0.1788 ± 0.0240 | 0.1911 | 0.1526 ± 0.0214 | 0.6194 | ug/μg | F (2, 27) = 0.9181 | NO |
| Histamine | 0.0170 ± 0.0049 | 0.0183 ± 0.0040 | 0.8360 | 0.0177 ± 0.0043 | 0.9112 | ng/μg | F (2, 27) = 0.02188 | NO |
| Catecholamine | 1.4080 ± 0.3270 | 1.3790 ± 0.2444 | 0.9437 | 1.3450 ± 0.2853 | 0.8792 | ng/μg | F (2, 27) = 0.01179 | NO |
| Melatonin | 0.0596 ± 0.0083 | 0.0718 ± 0.0096 | 0.5514 | 0.0782 ± 0.0213 | 0.3659 | pg/μg | F (2, 27) = 0.4366 | NO |
| Epinephrine | 0.0091 ± 0.0014 | 0.0092 ± 0.0012 | 0.9579 | 0.0099 ± 0.0013 | 0.6733 | ng/μg | F (2, 27) = 0.1079 | NO |
| Biomarkers | WT | dnmt3aa KO | dnmt3ab KO | Unit | F (DFn, DFd) | ANOVA | ||
|---|---|---|---|---|---|---|---|---|
| Concentration | Concentration | p-Value | Concentration | p-Value | Significance | |||
| Dopamine | 0.7943 ± 0.2082 | 0.5660 ± 0.1293 | 0.3937 | 1.0830 ± 0.2098 | 0.2821 | pg/μg | F (2, 27) = 1.938 | NO |
| GABA | 0.0041 ± 0.0012 | 0.0026 ± 0.0007 | 0.3083 | 0.0054 ± 0.0010 | 0.3761 | nmol/μg | F (2, 27) = 1.882 | NO |
| Serotonin (5-HT) | 0.0086 ± 0.0022 | 0.0051 ± 0.0010 | 0.2629 | 0.0138 ± 0.0027 | 0.1009 | ng/μg | F (2, 27) = 4.091 | YES |
| Norepinephrine | 0.0013 ± 0.0004 | 0.0007 ± 0.0002 | 0.2669 | 0.0033 ± 0.0004 | 0.0008 *** | ng/μg | F (2, 27) = 13.07 | YES |
| ROS | 0.1864 ± 0.0465 | 0.1204 ± 0.0196 | 0.2376 | 0.3410 ± 0.0440 | 0.0087 ** | U/mL | F (2, 27) = 8.586 | YES |
| ACh | 0.5470 ± 0.1099 | 0.3712 ± 0.0536 | 0.1991 | 0.7664 ± 0.1086 | 0.1120 | ug/μg | F (2, 27) = 4.398 | YES |
| Cortisol | 1.1900 ± 0.2340 | 0.8416 ± 0.1621 | 0.2683 | 1.5870 ± 0.2474 | 0.2082 | pg/μg | F (2, 27) = 2.932 | NO |
| Glutamate | 0.0054 ± 0.0013 | 0.0035 ± 0.0006 | 0.2452 | 0.0074 ± 0.0014 | 0.2218 | ug/μg | F (2, 27) = 2.974 | NO |
| Glycine | 0.0321 ± 0.0056 | 0.0193 ± 0.0037 | 0.0644 | 0.0349 ± 0.0046 | 0.6765 | ug/μg | F (2, 27) = 3.139 | NO |
| Histamine | 0.0129 ± 0.0038 | 0.0104 ± 0.0025 | 0.5926 | 0.0178 ± 0.0034 | 0.2979 | ng/μg | F (2, 27) = 1.33 | NO |
| Catecholamine | 0.6510 ± 0.1080 | 0.4566 ± 0.0871 | 0.2636 | 0.9893 ± 0.1557 | 0.0572 | ng/μg | F (2, 27) = 5.012 | YES |
| Melatonin | 0.0235 ± 0.0065 | 0.0155 ± 0.0034 | 0.2450 | 0.0335 ± 0.0037 | 0.1490 | pg/μg | F (2, 27) = 3.59 | YES |
| Epinephrine | 0.0044 ± 0.0013 | 0.0032 ± 0.0007 | 0.4703 | 0.0075 ± 0.0013 | 0.0693 | ng/μg | F (2, 27) = 3.667 | YES |
| Categories | DNA Methylation Counts | Differentially Methylated Regions (DMRs) | ||||
|---|---|---|---|---|---|---|
| WT | dnmt3aa KO | dnmt3ab KO | WT vs dnmt3aa KO | WT vs dnmt3ab KO | dnmt3aa KO vs dnmt3ab KO | |
| Exonic | 2,176,682 | 2,186,207 | 2,183,519 | 640 | 430 | 613 |
| Intergenic | 14,826,766 | 1,5002,735 | 14,869,769 | 6279 | 3740 | 6215 |
| Intronic | 17,219,672 | 17,423,241 | 17,313,624 | 6870 | 4035 | 6622 |
| Splicing | 6,211 | 6,276 | 6,225 | 4 | 4 | 4 |
| Upstream | 533,791 | 538,935 | 535,204 | 1038 | 651 | 963 |
| Downstream | 471,226 | 475,277 | 472,228 | 322 | 195 | 318 |
| UTR3 | 568,462 | 572,270 | 569,330 | 287 | 161 | 273 |
| UTR5 | 331,319 | 332,694 | 331,889 | 436 | 280 | 459 |
| Total | 36,166,511 | 36,570,202 | 36,314,198 | 15,962 | 9543 | 15,554 |
| Fish Lines | dnmt3aa KO Fish | dnmt3ab KO Fish |
|---|---|---|
| Morphological analysis | ||
| Morphometric analysis | unaltered | unaltered |
| Biochemical analysis | ||
| 5-hmC/5-mC ratio | unaltered | unaltered |
| Neurotransmitters in the brain | unaltered | unaltered |
| Neurotransmitters in the whole body | ↓ | ↑ |
| Behavioral analysis | ||
| Locomotor Activity in Novel Environment | unaltered | abnormal |
| Exploratory Behavior in Novel Environment | ↓ | unaltered |
| Aggressiveness | unaltered | ↓ |
| Predator Avoidance | ↓ | ↓ |
| Social Interaction | unaltered | ↓ |
| Shoaling | unaltered | loosen |
| Circadian Rhythm Locomotor Activity (Light Cycle) | unaltered | unaltered |
| Circadian Rhythm Locomotor Activity (Dark Cycle) | ↑ | ↑ |
| Color preference index ranking | unaltered | dysregulated |
| Short-term memory | ↓ | ↓ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lai, Y.-H.; Audira, G.; Liang, S.-T.; Siregar, P.; Suryanto, M.E.; Lin, H.-C.; Villalobos, O.; Villaflores, O.B.; Hao, E.; Lim, K.-H.; et al. Duplicated dnmt3aa and dnmt3ab DNA Methyltransferase Genes Play Essential and Non-Overlapped Functions on Modulating Behavioral Control in Zebrafish. Genes 2020, 11, 1322. https://doi.org/10.3390/genes11111322
Lai Y-H, Audira G, Liang S-T, Siregar P, Suryanto ME, Lin H-C, Villalobos O, Villaflores OB, Hao E, Lim K-H, et al. Duplicated dnmt3aa and dnmt3ab DNA Methyltransferase Genes Play Essential and Non-Overlapped Functions on Modulating Behavioral Control in Zebrafish. Genes. 2020; 11(11):1322. https://doi.org/10.3390/genes11111322
Chicago/Turabian StyleLai, Yu-Heng, Gilbert Audira, Sung-Tzu Liang, Petrus Siregar, Michael Edbert Suryanto, Huan-Chau Lin, Omar Villalobos, Oliver B. Villaflores, Erwei Hao, Ken-Hong Lim, and et al. 2020. "Duplicated dnmt3aa and dnmt3ab DNA Methyltransferase Genes Play Essential and Non-Overlapped Functions on Modulating Behavioral Control in Zebrafish" Genes 11, no. 11: 1322. https://doi.org/10.3390/genes11111322






