Telomere Dynamics Throughout Spermatogenesis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Germ Cell Separation
2.3. Telomere Measurement
2.4. Statistical Analysis
3. Results and Discussion
3.1. Telomere Dynamics Show Rat Strain Specificity Between Brown Norway and Sprague Dawley Rats
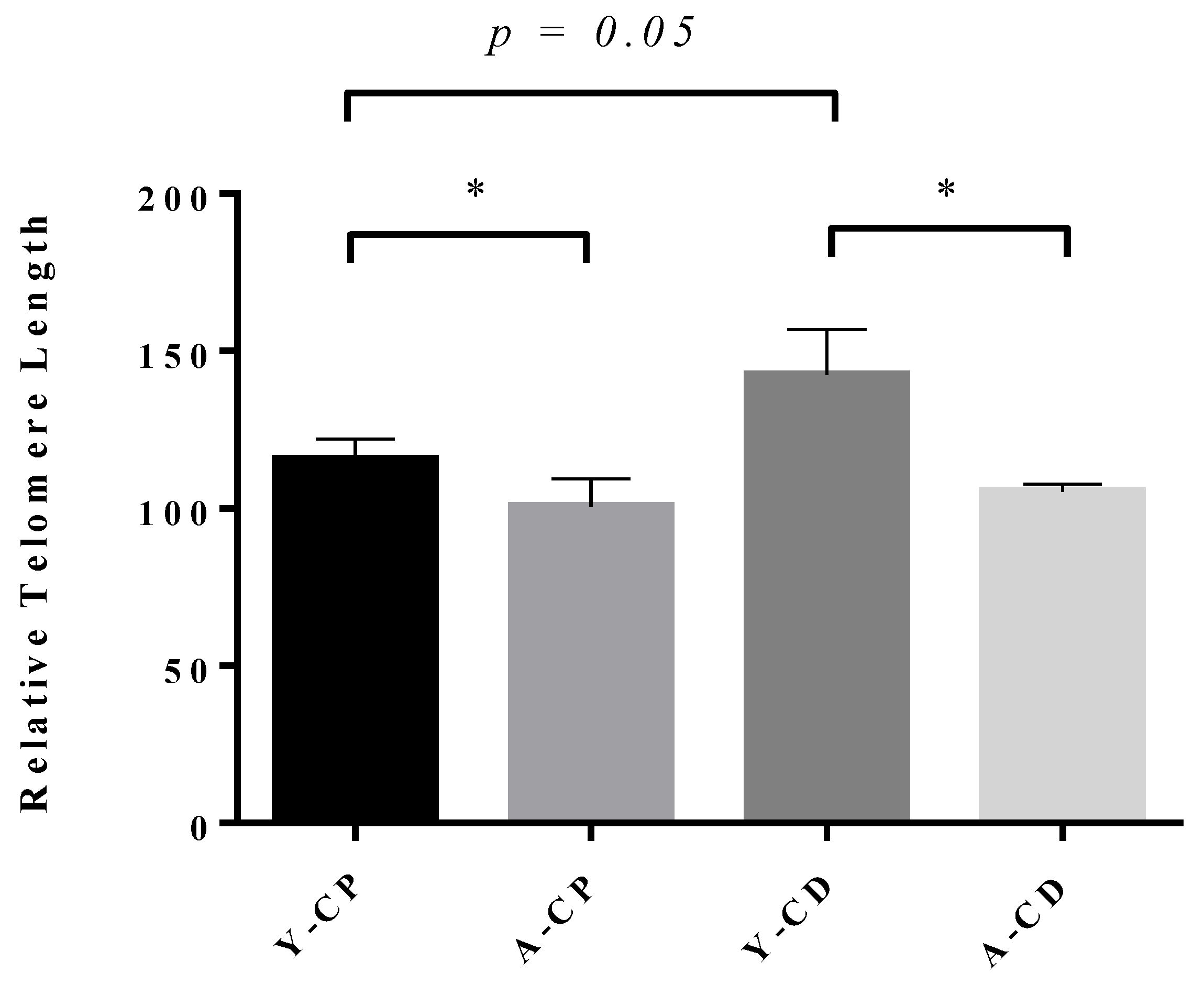
3.2. Telomere Lengths During Spermatogenesis in Brown Norway Rats
3.3. Age-Dependent Decrease in Sperm Telomere Length
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
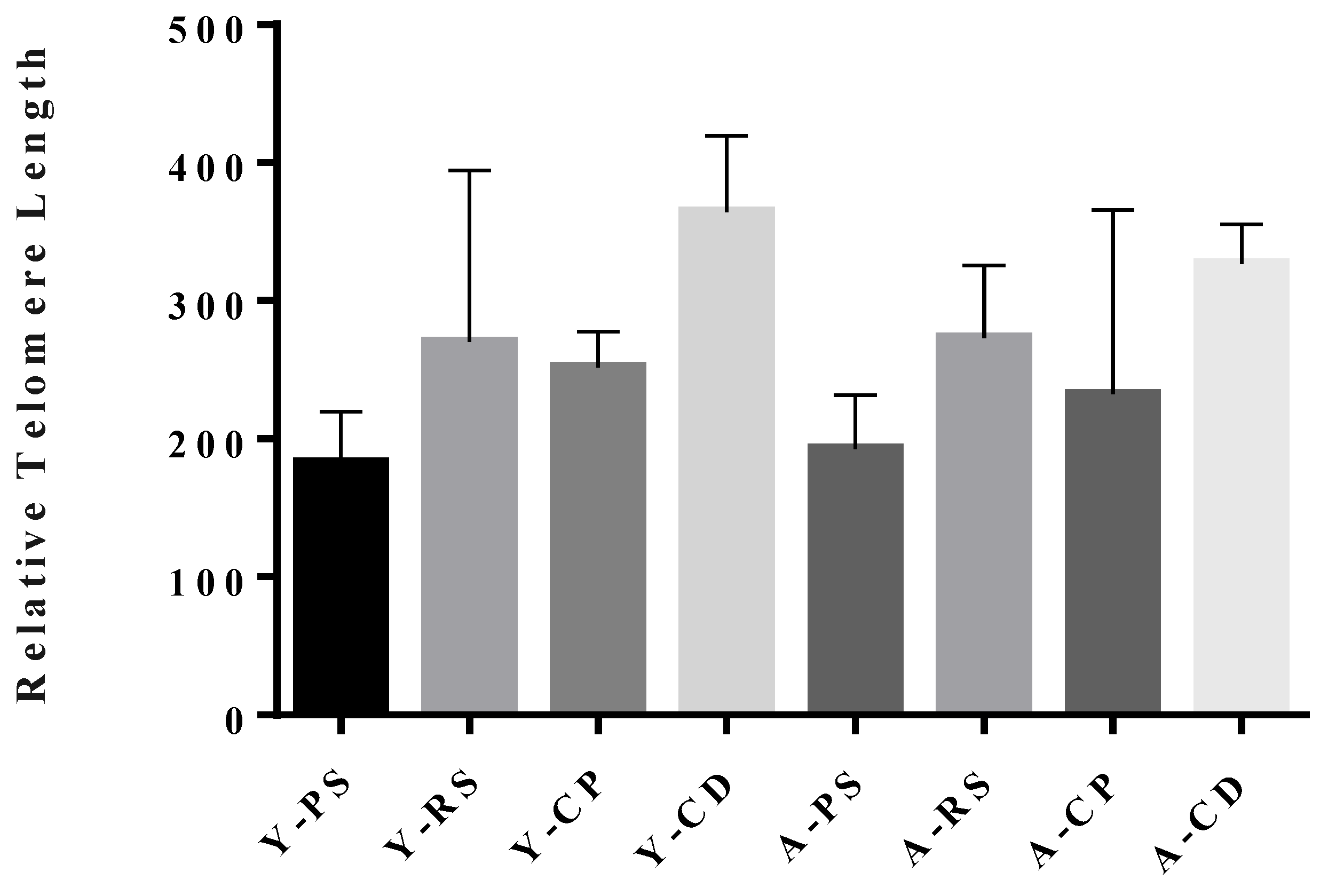
References
- Chan, S.R.W.L.; Blackburn, E.H. Telomeres and Telomerase. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004, 359, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Cesare, A.J.; Reddel, R.R. Alternative Lengthening of Telomeres: Models, Mechanisms and Implications. Nat. Rev. Genet. 2010, 11, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Rocca, M.S.; Foresta, C.; Ferlin, A. Telomere Length: Lights and Shadows on Their Role in Human Reproduction. Biol. Reprod. 2018, 100, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Achi, M.V.; Ravindranath, N.; Dym, M. Telomere Length in Male Germ Cells Is Inversely Correlated with Telomerase Activity. Biol. Reprod. 2005, 63, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Zalensky, A.O.; Tomilin, N.V.; Zalenskaya, I.A.; Teplitz, R.L.; Bradbury, E.M. Telomere-Telomere Interactions and Candidate Telomere Binding Protein(s) in Mammalian Sperm Cells. Exp. Cell Res. 1997, 232, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Zalenskaya, I.A.; Zalensky, A.O. Non-Random Positioning of Chromosomes in Human Sperm Nuclei. Chromosome Res. 2004, 12, 163–173. [Google Scholar] [CrossRef]
- Solov’eva, L.; Svetlova, M.; Bodinski, D.; Zalensky, A.O. Nature of Telomere Dimers and Chromosome Looping in Human Spermatozoa. Chromosome Res. 2005, 12, 817–823. [Google Scholar] [CrossRef]
- Ward, W.; Coffey, D. DNA Packaging and Organization in Mammalian Spermatozoa: Comparison with Somatic Cells. Biol. Reprod. 1991, 44, 569–574. [Google Scholar] [CrossRef]
- Steger, K. Transcriptional and Translational Regulation of Gene Expression in Haploid Spermatids. Anat. Embryol. 1999, 199, 471–487. [Google Scholar] [CrossRef]
- Carrell, D.T.; Emery, B.R.; Hammoud, S. Altered Protamine Expression and Diminished Spermatogenesis: What Is the Link? Hum. Reprod. Update 2007, 13, 313–327. [Google Scholar] [CrossRef]
- De Vries, M.; Ramos, L.; Housein, Z.; De Boer, P. Chromatin Remodelling Initiation during Human Spermiogenesis. Biol. Open 2012, 1, 446–457. [Google Scholar] [CrossRef] [PubMed]
- Van Der Heijden, G.W.; Dieker, J.W.; Derijck, A.A.H.A.; Muller, S.; Berden, J.H.M.; Braat, D.D.M.; Van Der Vlag, J.; De Boer, P. Asymmetry in Histone H3 Variants and Lysine Methylation between Paternal and Maternal Chromatin of the Early Mouse Zygote. Mech. Dev. 2005, 122, 1008–1022. [Google Scholar] [CrossRef] [PubMed]
- Wright, W.E.; Piatyszek, M.A.; Rainey, W.E.; Byrd, W.; Shay, J.W. Telomerase Activity in Human Germline and Embryonic Tissues and Cells. Dev. Genet. 1996, 18, 173–179. [Google Scholar] [CrossRef]
- Turner, S.; Hartshorne, G.M. Telomere Lengths in Human Pronuclei, Oocytes and Spermatozoa. Mol. Hum. Reprod. 2013, 19, 510–518. [Google Scholar] [CrossRef] [PubMed]
- Baird, D.M.; Britt-Compton, B.; Rowson, J.; Amso, N.N.; Gregory, L.; Kipling, D. Telomere Instability in the Male Germline. Hum. Mol. Genet. 2006, 15, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M.; Cherkas, L.F.; Kato, B.S.; Demissie, S.; Hjelmborg, J.B.; Brimacombe, M.; Cupples, A.; Hunkin, J.L.; Gardner, J.P.; Lu, X.; et al. Offspring’s Leukocyte Telomere Length, Paternal Age, and Telomere Elongation in Sperm. PLoS Genet. 2008. [Google Scholar] [CrossRef] [PubMed]
- Kozik, A.; Bradbury, E.M.; Zalensky, A. Increased Telomere Size in Sperm Cells of Mammals with Long Terminal (TTAGGG)(n) Arrays. Mol. Reprod. Dev. 1998, 51, 98–104. [Google Scholar] [CrossRef]
- Pickett, H.A.; Henson, J.D.; Au, A.Y.M.; Neumann, A.A.; Reddel, R.R. Normal Mammalian Cells Negatively Regulate Telomere Length by Telomere Trimming. Hum. Mol. Genet. 2011, 20, 4684–4692. [Google Scholar] [CrossRef] [PubMed]
- Johnson, G.D.; Lalancette, C.; Linnemann, A.K.; Leduc, F.; Boissonneault, G.; Krawetz, S.A. The Sperm Nucleus: Chromatin, RNA, and the Nuclear Matrix. Reproduction 2011, 141, 21–36. [Google Scholar] [CrossRef]
- Zalenskaya, I.A.; Bradbury, E.M.; Zalensky, A.O. Chromatin Structure of Telomere Domain in Human Sperm. Biochem. Biophys. Res. Commun. 2000, 279, 213–218. [Google Scholar] [CrossRef]
- World Health Organization. WHO Laboratory Manual for the Examination and Processing of Human Semen, 5th ed.; World Health Organization: Geneva, Switzerland, 2010. [Google Scholar]
- Zini, A.; Albert, O.; Robaire, B. Assessing Sperm Chromatin and DNA Damage: Clinical Importance and Development of Standards. Andrology 2014, 2, 322–325. [Google Scholar] [CrossRef] [PubMed]
- Verhulst, S.; Susser, E.; Factor-litvak, P.R.; Simons, M.J.P.; Benetos, A.; Steenstrup, T.; Kark, J.D.; Aviv, A.; Lorraine, D.; Bank, D.; et al. Commentary: The Reliability of Telomere Length Measurements. Int. J. Epidemiol. 2015, 44, 4–7. [Google Scholar] [CrossRef] [PubMed]
- Dan, F.; Eisenberg, T.A. Letters to the Editor Telomere Length Measurement Validity: The Coefficient of Variation Is Invalid and Cannot Be Used to Compare Quantitative Polymerase Chain Reaction and Southern Blot Telomere Length Measurement Techniques. Int. J. Epidemiol. 2016, 45, 1295–1298. [Google Scholar] [CrossRef]
- Ribas-Maynou, J.; Amengual, M.J.; Garcia-Peiró, A.; Lafuente, R.; Alvarez, J.; Bosch-Rue, E.; Brassesco, C.; Brassesco, M.; Benet, J. Sperm Telomere Length in Motile Sperm Selection Techniques: A qFISH Approach. Andrologia 2017, 50, e12840. [Google Scholar] [CrossRef]
- Boniewska-Bernacka, E.; Pańczyszyn, A.; Cybulska, N. Telomeres as a Molecular Marker of Male Infertility. Hum. Fertil. 2019, 22, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Ferlin, A.; Rampazzo, E.; Rocca, M.S.; Keppel, S.; Frigo, A.C.; De Rossi, A.; Foresta, C. In Young Men Sperm Telomere Length Is Related to Sperm Number and Parental Age. Hum. Reprod. 2013, 28, 3370–3376. [Google Scholar] [CrossRef] [PubMed]
- Cariati, F.; Jaroudi, S.; Alfarawati, S.; Raberi, A.; Alviggi, C.; Pivonello, R.; Wells, D. Investigation of Sperm Telomere Length as a Potential Marker of Paternal Genome Integrity and Semen Quality. Reprod. Biomed. Online 2016, 33, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Thilagavathi, J.; Kumar, M.; Mishra, S.S.; Venkatesh, S.; Kumar, R.; Dada, R. Analysis of Sperm Telomere Length in Men with Idiopathic Infertility. Arch. Gynecol. Obstet. 2013, 287, 803–807. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Mishra, S.; Kumar, R.; Dada, R.; Malhotra, N. Mild Oxidative Stress Is Beneficial for Sperm Telomere Length Maintenance. World J. Methodol. 2016, 6, 163. [Google Scholar] [CrossRef]
- Garolla, A.; Foresta, C.; Speltra, E.; Ferlin, A.; Rocca, M.S.; Menegazzo, M. Sperm Telomere Length as a Parameter of Sperm Quality in Normozoospermic Men. Hum. Reprod. 2016, 31, 1158–1163. [Google Scholar] [CrossRef]
- Durairajanayagam, D. Lifestyle Causes of Male Infertility. Arab J. Urol. 2018, 16, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Liochev, S.I. Reactive Oxygen Species and the Free Radical Theory of Aging. Free Radic. Biol. Med. 2013, 60, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Aitken, R.J.; Smith, T.B.; Jobling, M.S.; Baker, M.A.; De Iuliis, G.N. Oxidative Stress and Male Reproductive Health. Asian J. Androl. 2014, 16, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Coluzzi, E.; Colamartino, M.; Cozzi, R.; Leone, S.; Meneghini, C.; O’Callaghan, N.; Sgura, A. Oxidative Stress Induces Persistent Telomeric DNA Damage Responsible for Nuclear Morphology Change in Mammalian Cells. PLoS ONE 2014, 9, e110963. [Google Scholar] [CrossRef] [PubMed]
- Richter, T.; von Zglinicki, T. A Continuous Correlation between Oxidative Stress and Telomere Shortening in Fibroblasts. Exp. Gerontol. 2007, 42, 1039–1042. [Google Scholar] [CrossRef]
- Moskovtsev, S.I.; Willis, J.; White, J.; Mullen, J.B.M. Disruption of Telomeretelomere Interactions Associated with DNA Damage in Human Spermatozoa. Syst. Biol. Reprod. Med. 2010, 56, 407–412. [Google Scholar] [CrossRef]
- De Frutos, C.; Ló Pez-Cardona, A.P.; Balvís, N.F.; Laguna-Barraza, R.; Rizos, D.; Gutierrez-Adán, A.; Bermejo-Lvarez, P. Spermatozoa Telomeres Determine Telomere Length in Early Embryos and Offspring. Reproduction 2016, 151, 1–7. [Google Scholar] [CrossRef]
- Jørgensen, P.B.; Fedder, J.; Koelvraa, S.; Graakjaer, J. Age-Dependence of Relative Telomere Length Profiles during Spermatogenesis in Man. Maturitas 2013, 75, 380–385. [Google Scholar] [CrossRef]
- Wallenfang, M.R.; Nayak, R.; DiNardo, S. Dynamics of the Male Germline Stem Cell Population during Aging of Drosophila Melanogaster. Aging Cell 2006, 5, 297–304. [Google Scholar] [CrossRef]
- Siderakis, M.; Tarsounas, M. Telomere Regulation and Function during Meiosis. Chromosom. Res. 2008, 15, 667–679. [Google Scholar] [CrossRef]
- Nordfjäll, K.; Larefalk, A.; Lindgren, P.; Holmberg, D.; Roos, G. Telomere Length and Heredity: Indications of Paternal Inheritance. Proc. Natl. Acad. Sci. USA 2005, 102, 16374–16378. [Google Scholar] [CrossRef] [PubMed]
- Broer, L.; Codd, V.; Nyholt, D.R.; Deelen, J.; Mangino, M.; Willemsen, G.; Albrecht, E.; Amin, N.; Beekman, M.; De Geus, E.J.C.; et al. Meta-Analysis of Telomere Length in 19 713 Subjects Reveals High Heritability, Stronger Maternal Inheritance and a Paternal Age Effect. Eur. J. Hum. Genet. 2013, 21, 1163–1168. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, D.T.A.; Tackney, J.; Cawthon, R.M.; Theresa, C.; Kristen, C. Paternal and Grandpaternal Ages at Conception and Descendant Telomere Lengths in Chimpanzees and Humans. Am. J. Phys. Anthropol. 2017, 162, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Bauch, C.; Boonekamp, J.J.; Korsten, P.; Mulder, E.; Verhulst, S. Epigenetic Inheritance of Telomere Length in Wild Birds. PLoS Genet. 2019, 15, e1007827. [Google Scholar] [CrossRef] [PubMed]
- Paul, C.; Robaire, B. Ageing of the Male Germ Line. Nat. Rev. Urol. 2013, 10, 227. [Google Scholar] [CrossRef]
- Bryant, J.M.; Meyer-ficca, M.L.; Dang, V.M.; Berger, S.L.; Meyer, R.G. Separation of Spermatogenic Cell Types Using STA-PUT Velocity Sedimentation. JoVE 2013, 80, e50648. [Google Scholar] [CrossRef]
- Callaghan, N.J.O.; Fenech, M. A Quantitative PCR Method for Measuring Absolute Telomere Length. Biol. Proced. Online 2011, 13, 3. [Google Scholar] [CrossRef]
- Bebbington, K.; Spurgin, L.G.; Fairfield, E.A.; Dugdale, H.L.; Komdeur, J.; Burke, T.; Richardson, D.S. Telomere Length Reveals Cumulative Individual and Transgenerational Inbreeding Effects in a Passerine Bird. Mol. Ecol. 2016, 25, 2949–2960. [Google Scholar] [CrossRef]
- Fujita, T.; Yuno, M.; Okuzaki, D.; Ohki, R.; Fujii, H. Identification of Non-Coding RNAs Associated with Telomeres Using a Combination of enChIP and RNA Sequencing. PLoS ONE 2015, 10, e0123387. [Google Scholar] [CrossRef]
- Sullivan, R. Epididymosomes: A Heterogeneous Population of Microvesicles with Multiple Functions in Sperm Maturation and Storage. Asian J. Androl. 2015, 17, 726. [Google Scholar] [CrossRef]
- Schoeftner, S.; Blasco, M.A. Chromatin Regulation and Non-Coding RNAs at Mammalian Telomeres. Semin. Cell Dev. Biol. 2010, 21, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Azzalin, C.M.; Reichenbach, P.; Khoriauli, L.; Giulotto, E.; Lingner, J. Telomeric Repeat-Containing RNA and RNA Surveillance Factors at Mammalian Chromosome Ends. Science 2007, 318, 798–802. [Google Scholar] [CrossRef] [PubMed]
- Luke, B.; Panza, A.; Redon, S.; Iglesias, N.; Li, Z.; Lingner, J. The Rat1p 5′ to 3′ Exonuclease Degrades Telomeric Repeat-Containing RNA and Promotes Telomere Elongation in Saccharomyces Cerevisiae. Mol. Cell 2008, 32, 465–477. [Google Scholar] [CrossRef] [PubMed]
- Savitsky, M.; Kwon, D.; Georgiev, P.; Kalmykova, A.; Gvozdev, V. Telomere Elongation Is under the Control of the RNAi-Based Mechanism in the Drosophila Germline. Genes Dev. 2006, 20, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Montero, J.J.; López De Silanes, I.; Granã, O.; Blasco, M.A. Telomeric RNAs Are Essential to Maintain Telomeres. Nat. Commun. 2016, 7, 12534. [Google Scholar] [CrossRef] [PubMed]
- Montero, J.J.; López-Silanes, I.; Megías, D.; Fraga, M.; Castells-García, Á.; Blasco, M.A. TERRA Recruitment of Polycomb to Telomeres Is Essential for Histone Trymethylation Marks at Telomeric Heterochromatin. Nat. Commun. 2018, 9, 4–6. [Google Scholar] [CrossRef] [PubMed]
- Reig-Viader, R.; Vila-cejudo, M.; Vitelli, V.; Busca, R.; Ruiz-herrera, A.; Valle, C.; Uab, C.; Spallanzani, B.L. Telomeric Repeat-Containing RNA (TERRA) and Telomerase Are Components of Telomeres During Mammalian Gametogenesis. Biol. Reprod. 2014, 90, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Reig-Viader, R.; Capilla, L.; Vila-cejudo, M.; Ruiz-herrera, A. Telomere Homeostasis Is Compromised in Spermatocytes from Patients with Idiopathic Infertility. Fertil. Steril. 2014, 102. [Google Scholar] [CrossRef]

| Rat Strain | Cell Type | N | Median rTL | IQR | SEM |
|---|---|---|---|---|---|
| SD - Young | PS | 5 | 205.89 | 126.14 | 37.21 |
| RS | 4 | 180.53 | 141.46 | 124.19 | |
| CP | 6 | 230.47 | 79.17 | 25.54 | |
| CD | 10 | 396.81 | 253.57 | 54.94 | |
| SD - Aged | PS | 6 | 204.92 | 132.51 | 38.99 |
| RS | 5 | 301.42 | 122.45 | 52.70 | |
| CP | 3 | 116.93 | 205.51 | 133.00 | |
| CD | 12 | 302.82 | 132.82 | 28.49 | |
| BN - Young | PS | 5 | 143.70 | 13.35 | 11.63 |
| RS | 5 | 165.79 | 77.18 | 20.09 | |
| CP | 5 | 116.61 | 17.56 | 6.49 | |
| CD | 5 | 129.67 | 17.42 | 14.61 | |
| BN - Aged | CP | 4 | 97.47 | 13.40 | 6.35 |
| CD | 4 | 106.54 | 4.64 | 2.52 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fice, H.E.; Robaire, B. Telomere Dynamics Throughout Spermatogenesis. Genes 2019, 10, 525. https://doi.org/10.3390/genes10070525
Fice HE, Robaire B. Telomere Dynamics Throughout Spermatogenesis. Genes. 2019; 10(7):525. https://doi.org/10.3390/genes10070525
Chicago/Turabian StyleFice, Heather E., and Bernard Robaire. 2019. "Telomere Dynamics Throughout Spermatogenesis" Genes 10, no. 7: 525. https://doi.org/10.3390/genes10070525





