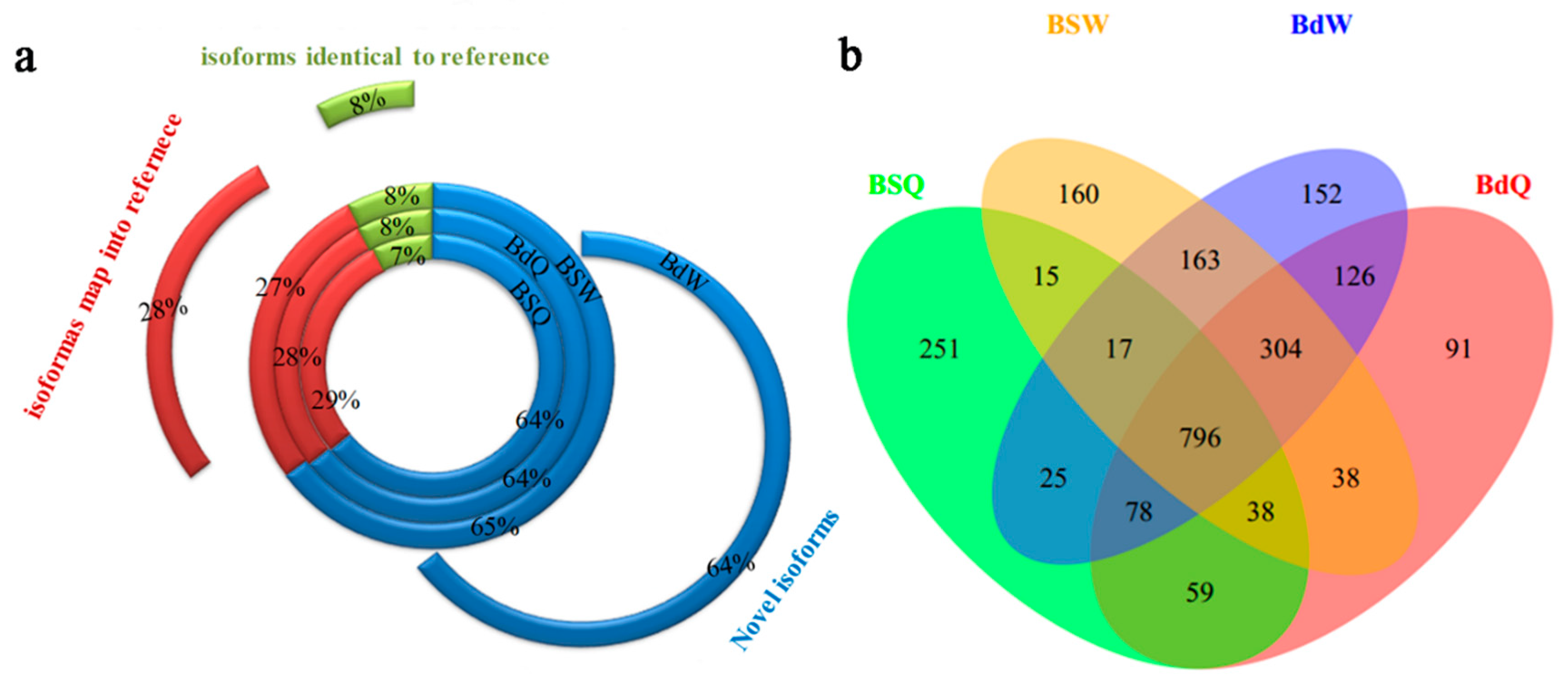
Figure 1.
The proportion of assembled transcripts and number of novel genes in the root, old leaves and juvenile leaves of B. napus cultivars Qingyou10 (QY10) and Westar10 (W10) under boron (B) sufficient and deficient conditions: (a) the proportion of novel isoforms, the isoforms mapped to the reference, and the isoforms identical to the reference; and (b) Venn diagram of the overlap of the novel genes. Q, QY, Qingyou10; W, W10, Westar10; Bs, B sufficient condition; Bd, B deficient condition.
Figure 1.
The proportion of assembled transcripts and number of novel genes in the root, old leaves and juvenile leaves of B. napus cultivars Qingyou10 (QY10) and Westar10 (W10) under boron (B) sufficient and deficient conditions: (a) the proportion of novel isoforms, the isoforms mapped to the reference, and the isoforms identical to the reference; and (b) Venn diagram of the overlap of the novel genes. Q, QY, Qingyou10; W, W10, Westar10; Bs, B sufficient condition; Bd, B deficient condition.
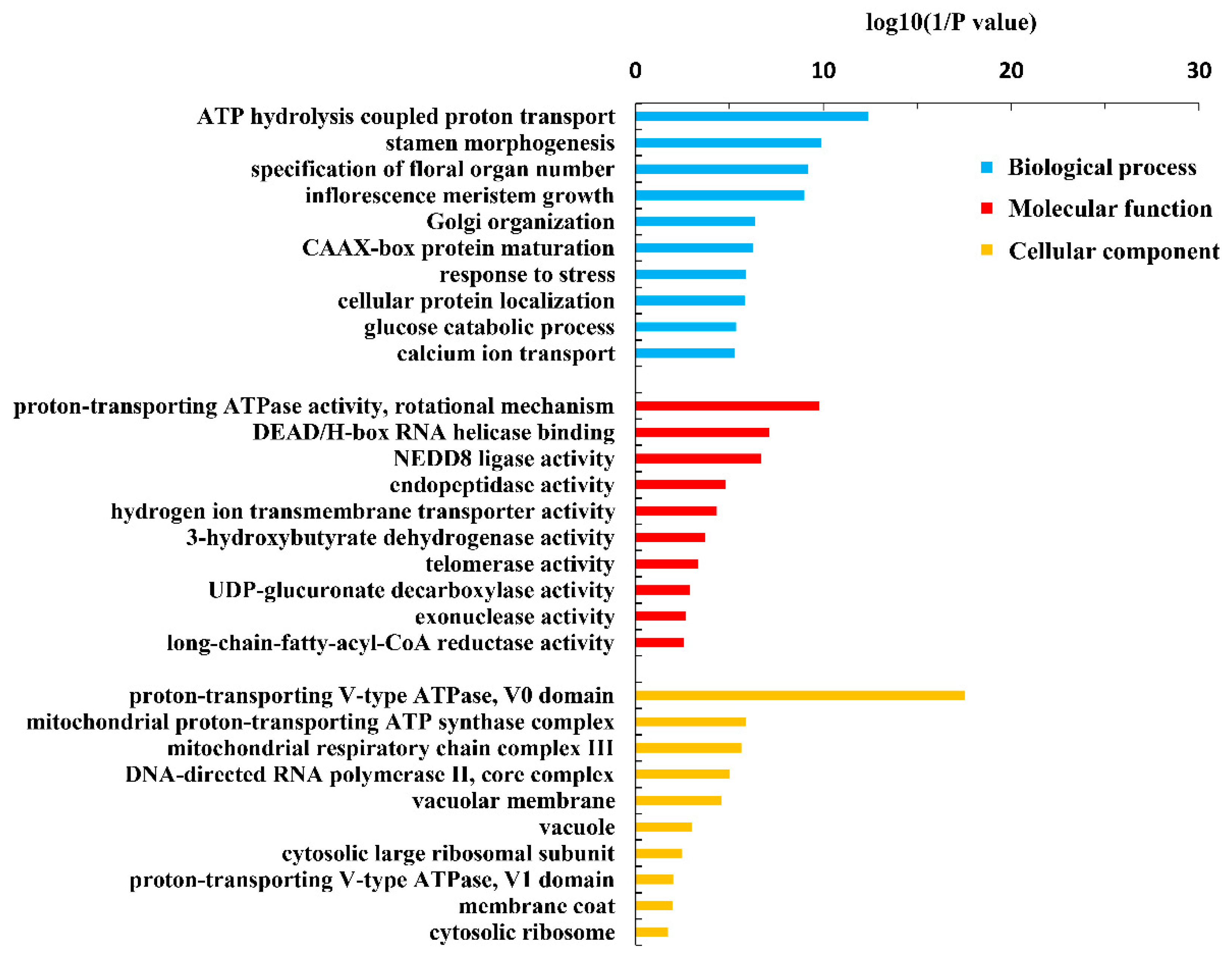
Figure 2.
GO terms of the functional categorization of the same novel genes in the root, old leaves and juvenile leaves of B. napus cultivars Qingyou10 (QY10) or Westar10 (W10) under boron sufficient and deficient conditions.
Figure 2.
GO terms of the functional categorization of the same novel genes in the root, old leaves and juvenile leaves of B. napus cultivars Qingyou10 (QY10) or Westar10 (W10) under boron sufficient and deficient conditions.
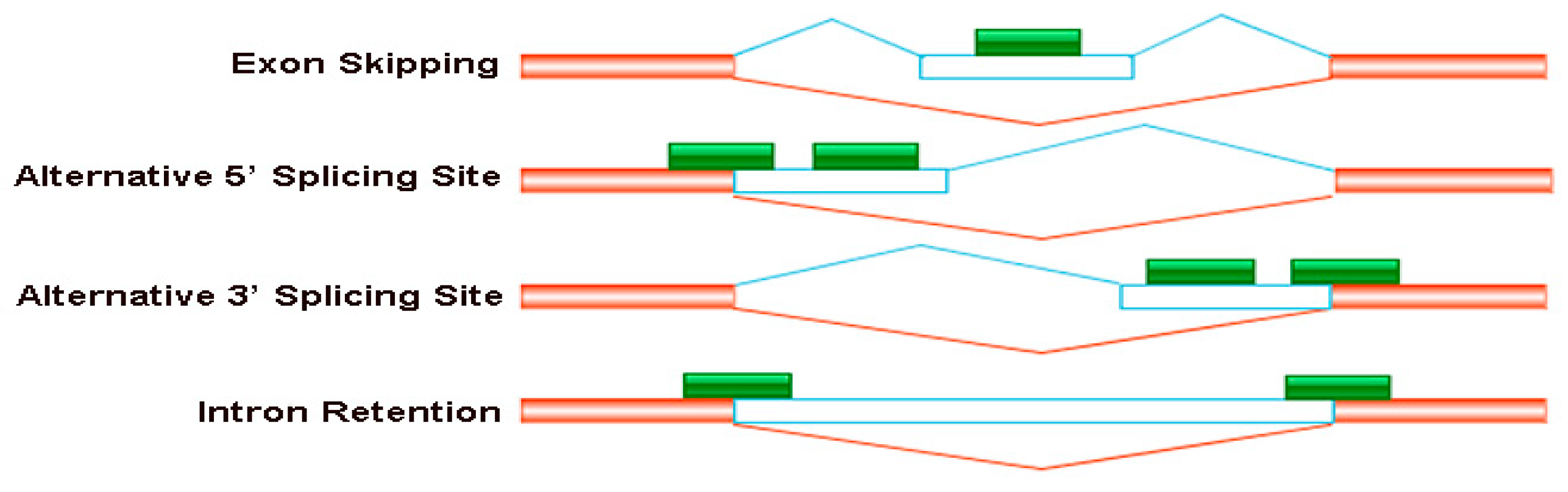
Figure 3.
Four major types of the alternative splicing events occurred in B. napus cultivars Qingyou10 and Westar10. Exon skipping (ES), alternative 5′ splice site (A5SS), alternative 3′ splice site (A3SS), and intron retention (RI) are shown schematically. The orange/transparent boxes represent exons, green boxes represent sequencing reads and lines represent introns.
Figure 3.
Four major types of the alternative splicing events occurred in B. napus cultivars Qingyou10 and Westar10. Exon skipping (ES), alternative 5′ splice site (A5SS), alternative 3′ splice site (A3SS), and intron retention (RI) are shown schematically. The orange/transparent boxes represent exons, green boxes represent sequencing reads and lines represent introns.
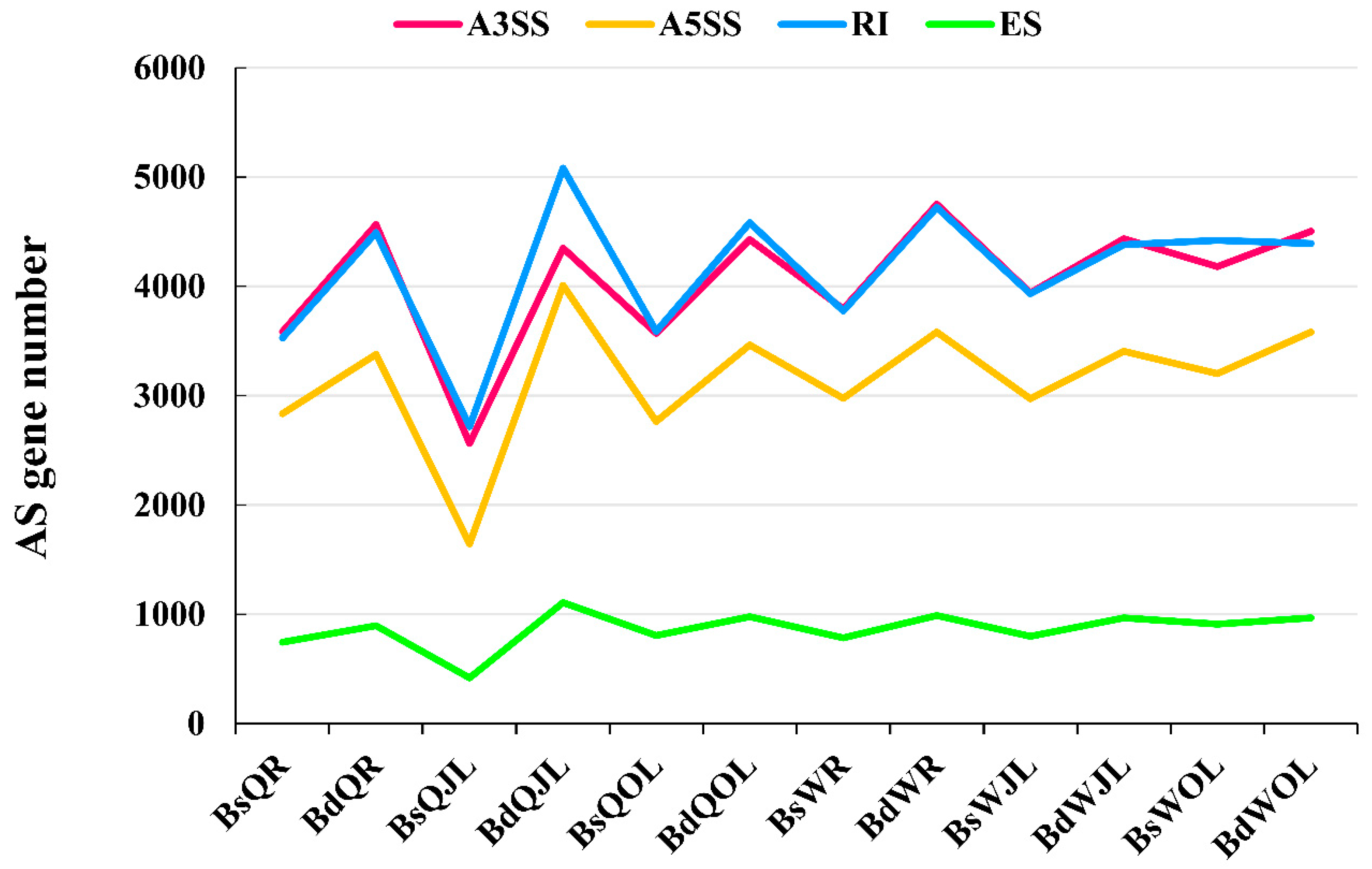
Figure 4.
The numbers of the alternative splicing (AS) events and genes in B. napus cultivars Qingyou10 (QY10) and Wesatr10 (W10) under boron (B) sufficient and deficient conditions. Q, QY10, Qingyou10; W, W10, Westar10; Bs, B sufficient condition; Bd, B deficient condition; R, root; JL, juvenile leaves; OL, old leaves.
Figure 4.
The numbers of the alternative splicing (AS) events and genes in B. napus cultivars Qingyou10 (QY10) and Wesatr10 (W10) under boron (B) sufficient and deficient conditions. Q, QY10, Qingyou10; W, W10, Westar10; Bs, B sufficient condition; Bd, B deficient condition; R, root; JL, juvenile leaves; OL, old leaves.
Figure 5.
The number of genes of four types of alternative splicing (AS) in B. napus cultivars Qingyou10 (QY10) and Wesatr10 (W10) under boron (B) sufficient and deficient conditions. Q, QY10, QY10, Qingyou10; W, W10, Westar10; Bs, B sufficient condition; Bd, B deficient condition; R, root; JL, juvenile leaves; OL, old leaves; A3SS, alternative 3′ splice site; A5SS, alternative 5′ splice site; RI, retain intron; ES, exon skip.
Figure 5.
The number of genes of four types of alternative splicing (AS) in B. napus cultivars Qingyou10 (QY10) and Wesatr10 (W10) under boron (B) sufficient and deficient conditions. Q, QY10, QY10, Qingyou10; W, W10, Westar10; Bs, B sufficient condition; Bd, B deficient condition; R, root; JL, juvenile leaves; OL, old leaves; A3SS, alternative 3′ splice site; A5SS, alternative 5′ splice site; RI, retain intron; ES, exon skip.
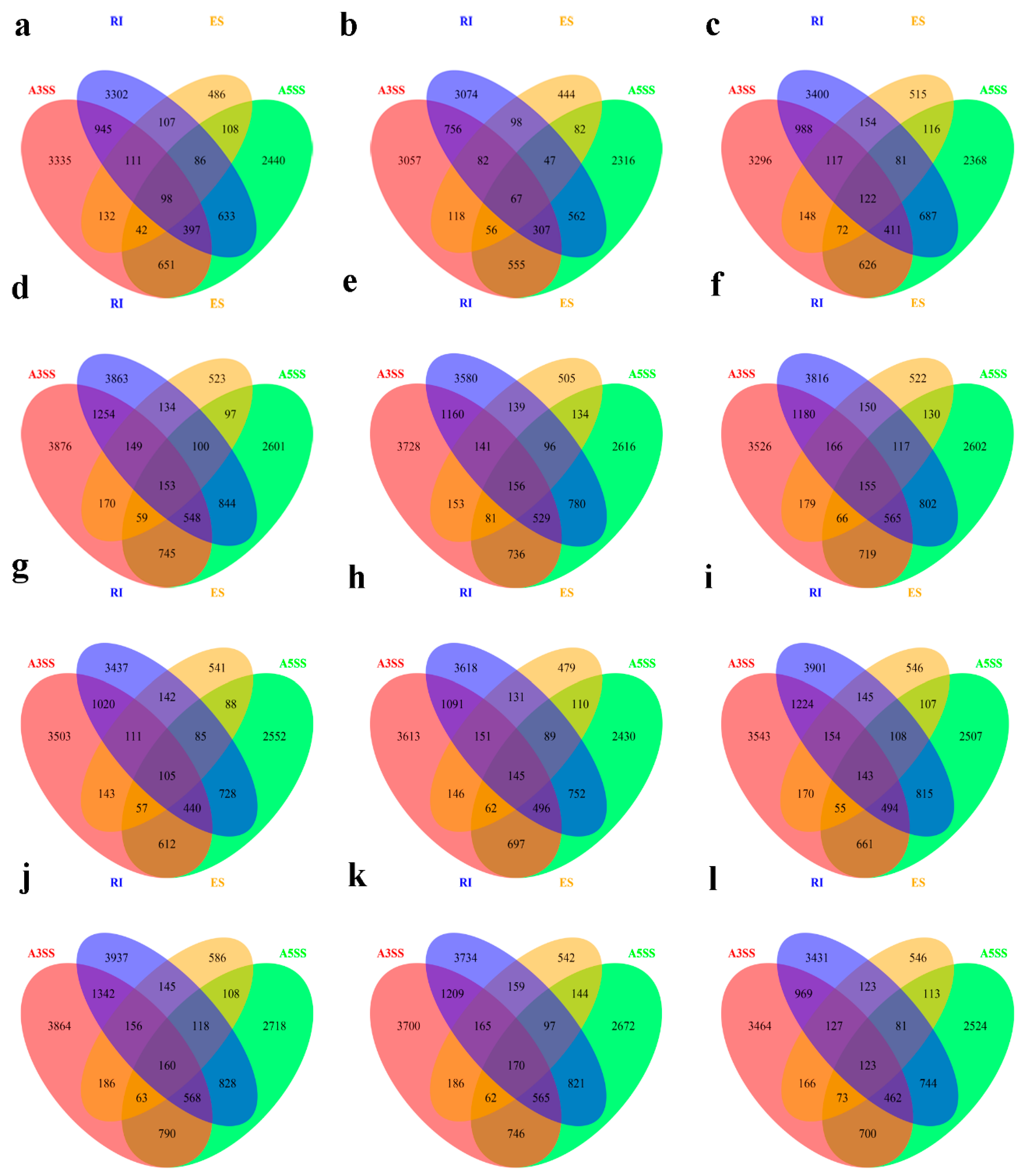
Figure 6.
Venn diagram of the overlap of the four types of AS genes in B. napus cultivars Qingyou10 (QY10) and Westar10 (W10) under boron (B) sufficient and deficient conditions: (a) BsQR; (b) BsQJL; (c) BsQOL; (d) BdQR; (e) BdQJL; (f) BdQOL; (g) BsWR; (h) BsWJL; (i) BsWOL; (j) BdWR; (k) BdWJL; and (l) BdWOL. Q, QY10, Qingyou10; W, W10, Westar10; R, root; JL, juvenile leaves; OL, old leaves; Bs, B sufficient condition; Bd, B deficient condition.
Figure 6.
Venn diagram of the overlap of the four types of AS genes in B. napus cultivars Qingyou10 (QY10) and Westar10 (W10) under boron (B) sufficient and deficient conditions: (a) BsQR; (b) BsQJL; (c) BsQOL; (d) BdQR; (e) BdQJL; (f) BdQOL; (g) BsWR; (h) BsWJL; (i) BsWOL; (j) BdWR; (k) BdWJL; and (l) BdWOL. Q, QY10, Qingyou10; W, W10, Westar10; R, root; JL, juvenile leaves; OL, old leaves; Bs, B sufficient condition; Bd, B deficient condition.
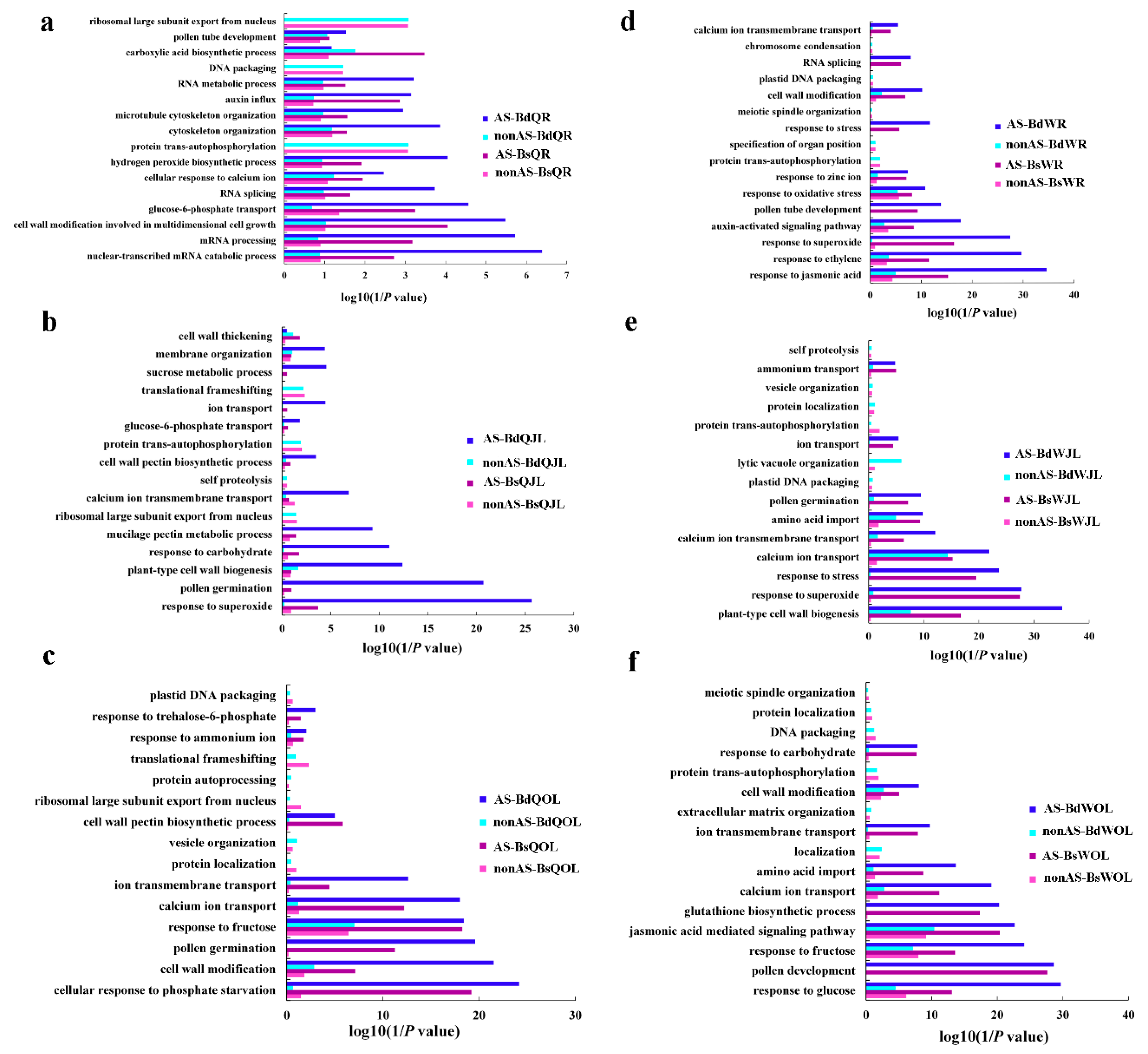
Figure 7.
GO terms of the functional categorization of AS and non-alternative splicing (nonAS) genes (biological process) in the root, juvenile leaves and old leaves of B. napus cultivars Qingyou10 (QY10) and Westar10 (W10) under boron (B) sufficient and deficient conditions: (a) root of QY10; (b) juvenile leaves of QY10; (c) old leaves of QY10; (d) root of W10; (e) juvenile leaves of W10; and (f) old leaves of W10. AS, alternative splicing; nonAS, non-alternative splicing; Bs, B sufficient condition; Bd, B deficient condition; QR, root of QY10; QOL, old leaves of QY10; QJL, juvenile leaves of QY10; WR, root of W10; WOL, old leaves of W10; WJL, juvenile leaves of W10.
Figure 7.
GO terms of the functional categorization of AS and non-alternative splicing (nonAS) genes (biological process) in the root, juvenile leaves and old leaves of B. napus cultivars Qingyou10 (QY10) and Westar10 (W10) under boron (B) sufficient and deficient conditions: (a) root of QY10; (b) juvenile leaves of QY10; (c) old leaves of QY10; (d) root of W10; (e) juvenile leaves of W10; and (f) old leaves of W10. AS, alternative splicing; nonAS, non-alternative splicing; Bs, B sufficient condition; Bd, B deficient condition; QR, root of QY10; QOL, old leaves of QY10; QJL, juvenile leaves of QY10; WR, root of W10; WOL, old leaves of W10; WJL, juvenile leaves of W10.
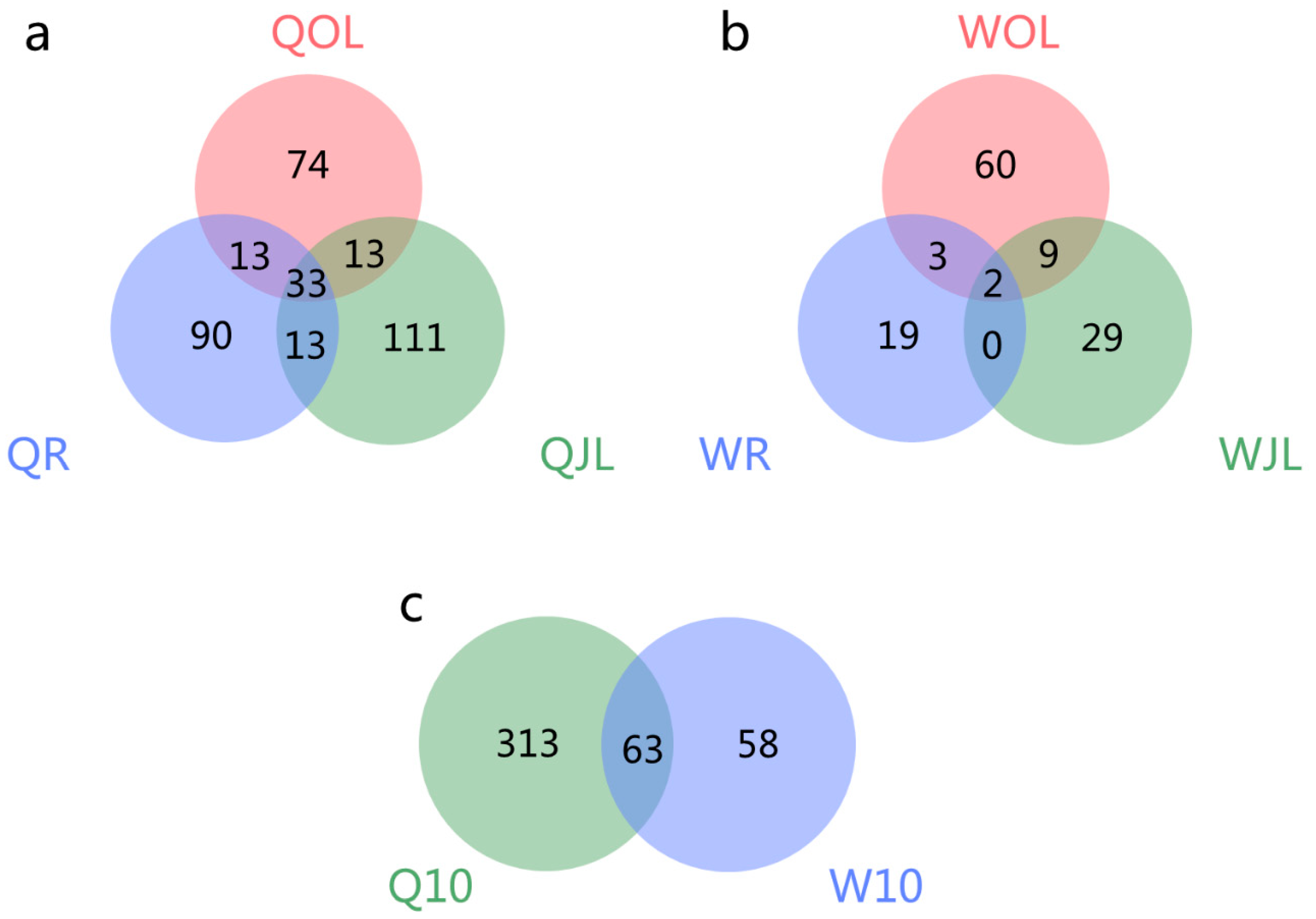
Figure 8.
Venn diagram of the overlap of differential alternative splicing (DAS) genes in the root, juvenile leaves and old leaves of B. napus cultivars Qingyou10 (QY10) and Westar10 (W10) under boron deficient conditions: (a) QY10; (b)W10; and (c) QY10 and W10 under B deficient conditions; QR, root of QY10; QOL, old leaves of QY10; QJL, juvenile leaves of QY10; WR, root of W10; WOL, old leaves of W10; WJL, juvenile leaves of W10.
Figure 8.
Venn diagram of the overlap of differential alternative splicing (DAS) genes in the root, juvenile leaves and old leaves of B. napus cultivars Qingyou10 (QY10) and Westar10 (W10) under boron deficient conditions: (a) QY10; (b)W10; and (c) QY10 and W10 under B deficient conditions; QR, root of QY10; QOL, old leaves of QY10; QJL, juvenile leaves of QY10; WR, root of W10; WOL, old leaves of W10; WJL, juvenile leaves of W10.
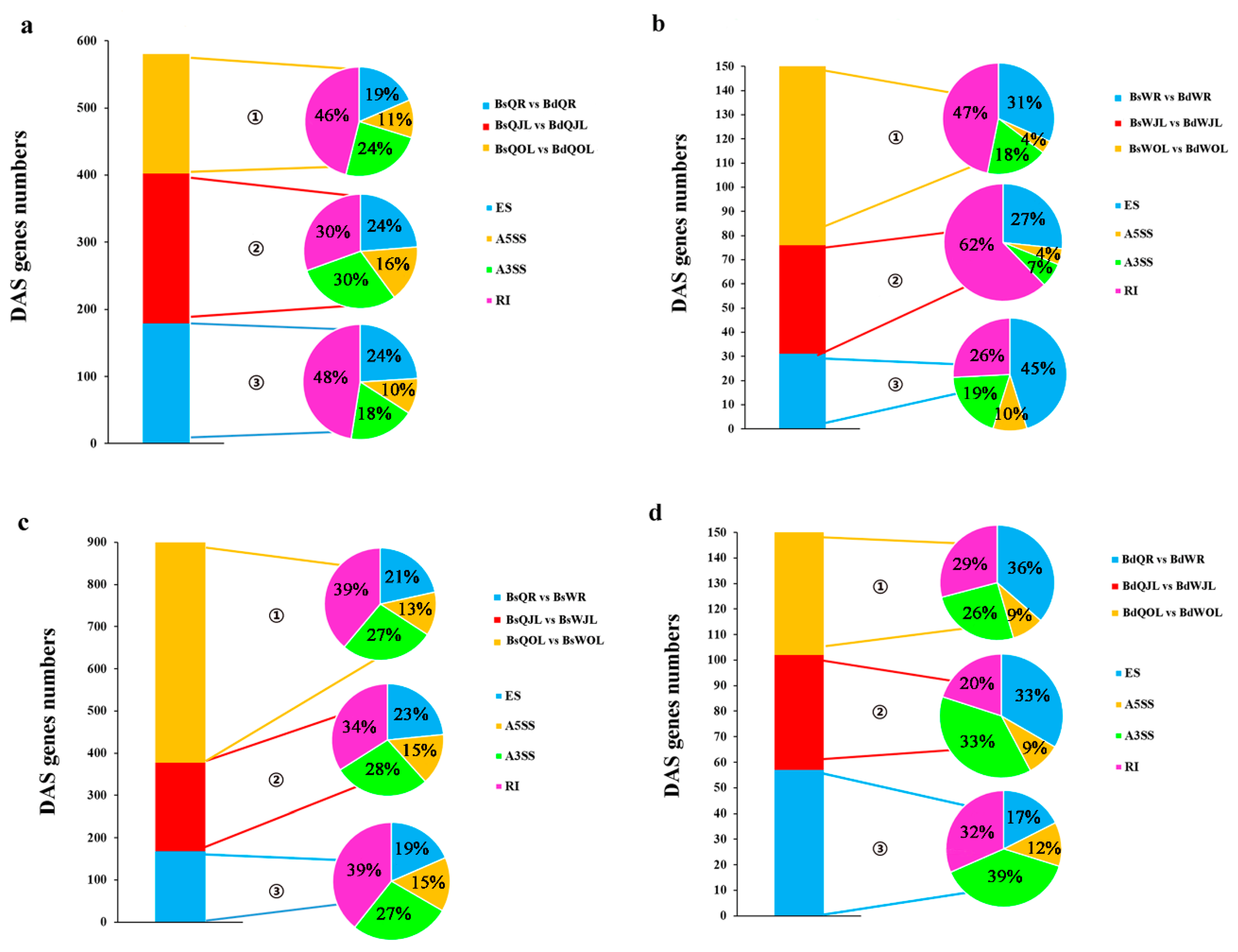
Figure 9.
The number of DAS genes in the root, juvenile leaves and old leaves of B. napus cultivars Qingyou10 (QY10) and Westar10 (W10) under boron deficient conditions: (a,b) the total number of DAS genes (control, B sufficient condition) identified in QY10 and W10, respectively; and (c,d), the total number of DAS genes in QY10 (control, W10) under boron sufficient and deficient conditions, respectively. ① Ratio of four alternative splicing patterns in old leaves; ② ratio of four alternative splicing patterns in juvenile leaves; and ③ ratio of four alternative splicing patterns in root. DAS, differential alternative splicing; Bs, boron sufficient condition; Bd, boron deficient condition; QR, root of QY10; QOL, old leaves of QY10; QJL, juvenile leaves of QY10; WR, root of W10; WOL, old leaves of W10; WJL, juvenile leaves of W10. A3SS, Alternative 3′ splice site; A5SS, alternative 5′ splice site; RI, retain intron; ES, exon skip.
Figure 9.
The number of DAS genes in the root, juvenile leaves and old leaves of B. napus cultivars Qingyou10 (QY10) and Westar10 (W10) under boron deficient conditions: (a,b) the total number of DAS genes (control, B sufficient condition) identified in QY10 and W10, respectively; and (c,d), the total number of DAS genes in QY10 (control, W10) under boron sufficient and deficient conditions, respectively. ① Ratio of four alternative splicing patterns in old leaves; ② ratio of four alternative splicing patterns in juvenile leaves; and ③ ratio of four alternative splicing patterns in root. DAS, differential alternative splicing; Bs, boron sufficient condition; Bd, boron deficient condition; QR, root of QY10; QOL, old leaves of QY10; QJL, juvenile leaves of QY10; WR, root of W10; WOL, old leaves of W10; WJL, juvenile leaves of W10. A3SS, Alternative 3′ splice site; A5SS, alternative 5′ splice site; RI, retain intron; ES, exon skip.
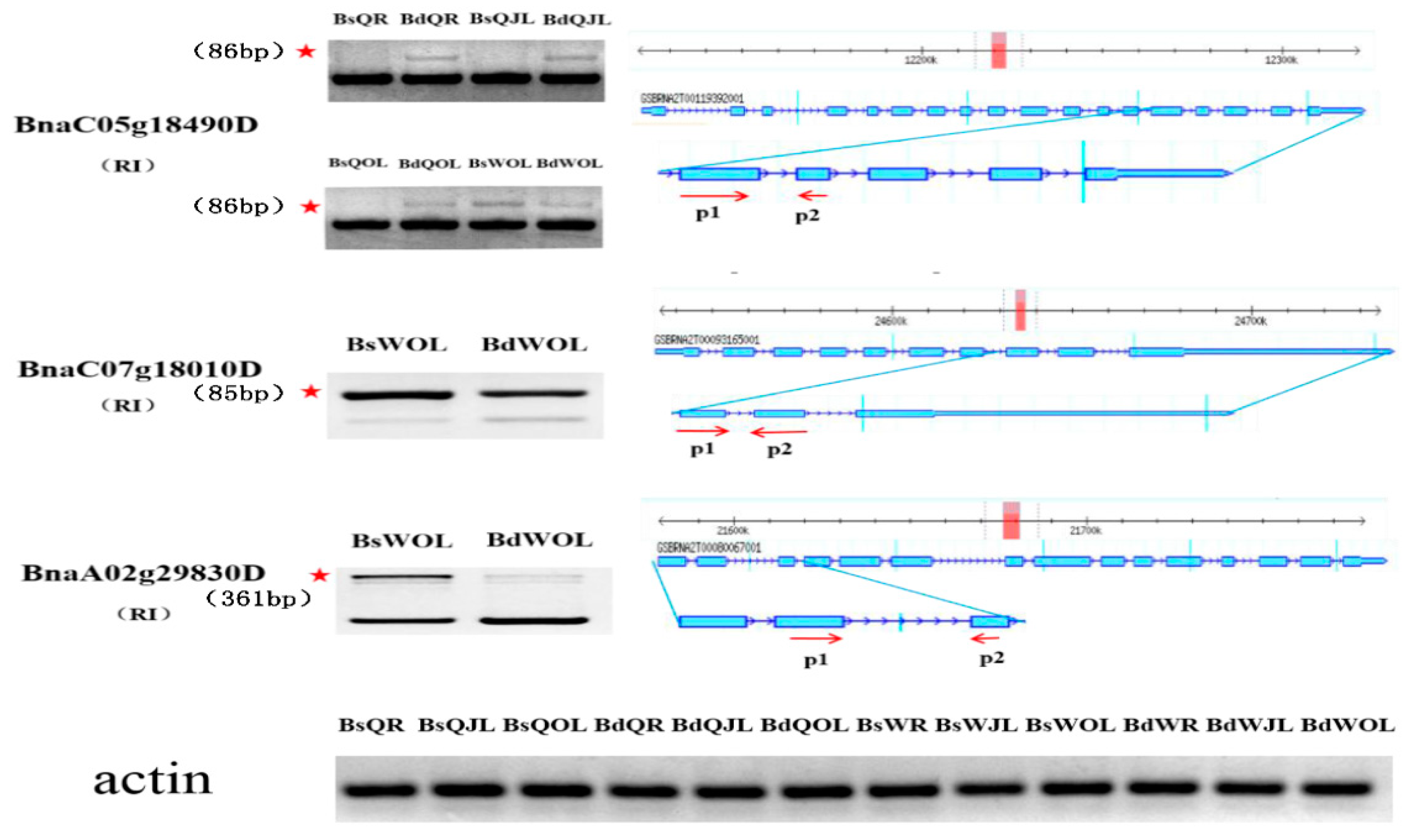
Figure 10.
Validation of DAS events in the root, juvenile leaves and old leaves of B. napus cultivars Qingyou10 (QY10) and Westar10 (W10) under boron deficient conditions. The band with the red asterisk showed the fragment generated by the alternative splicing of the gene, which was determined according to the size of the retained intron fragment. The forward and reverse primers, P1 and P2, were designed based on the exon between the retained intron. Bs, boron sufficient condition; Bd, boron deficient condition; QR, root of QY10; QOL, old leaves of QY10; QJL, juvenile leaves of QY10; WR, root of W10; WOL, old leaves of W10; WJL, juvenile leaves of W10. RI, retain intron.
Figure 10.
Validation of DAS events in the root, juvenile leaves and old leaves of B. napus cultivars Qingyou10 (QY10) and Westar10 (W10) under boron deficient conditions. The band with the red asterisk showed the fragment generated by the alternative splicing of the gene, which was determined according to the size of the retained intron fragment. The forward and reverse primers, P1 and P2, were designed based on the exon between the retained intron. Bs, boron sufficient condition; Bd, boron deficient condition; QR, root of QY10; QOL, old leaves of QY10; QJL, juvenile leaves of QY10; WR, root of W10; WOL, old leaves of W10; WJL, juvenile leaves of W10. RI, retain intron.
Figure 11.
GO terms of the functional categorization of DAS and DE genes (biological process) in the root, juvenile leaves and old leaves of B. napus cultivars Qingyou10 (QY10) and Westar10 (W10) under B deficient conditions: (a) root of QY10; (b) juvenile leaves of QY10; (c) old leaves of QY10; (d) root of W10; (e) juvenile leaves of W10; and (f) old leaves of W10.
Figure 11.
GO terms of the functional categorization of DAS and DE genes (biological process) in the root, juvenile leaves and old leaves of B. napus cultivars Qingyou10 (QY10) and Westar10 (W10) under B deficient conditions: (a) root of QY10; (b) juvenile leaves of QY10; (c) old leaves of QY10; (d) root of W10; (e) juvenile leaves of W10; and (f) old leaves of W10.
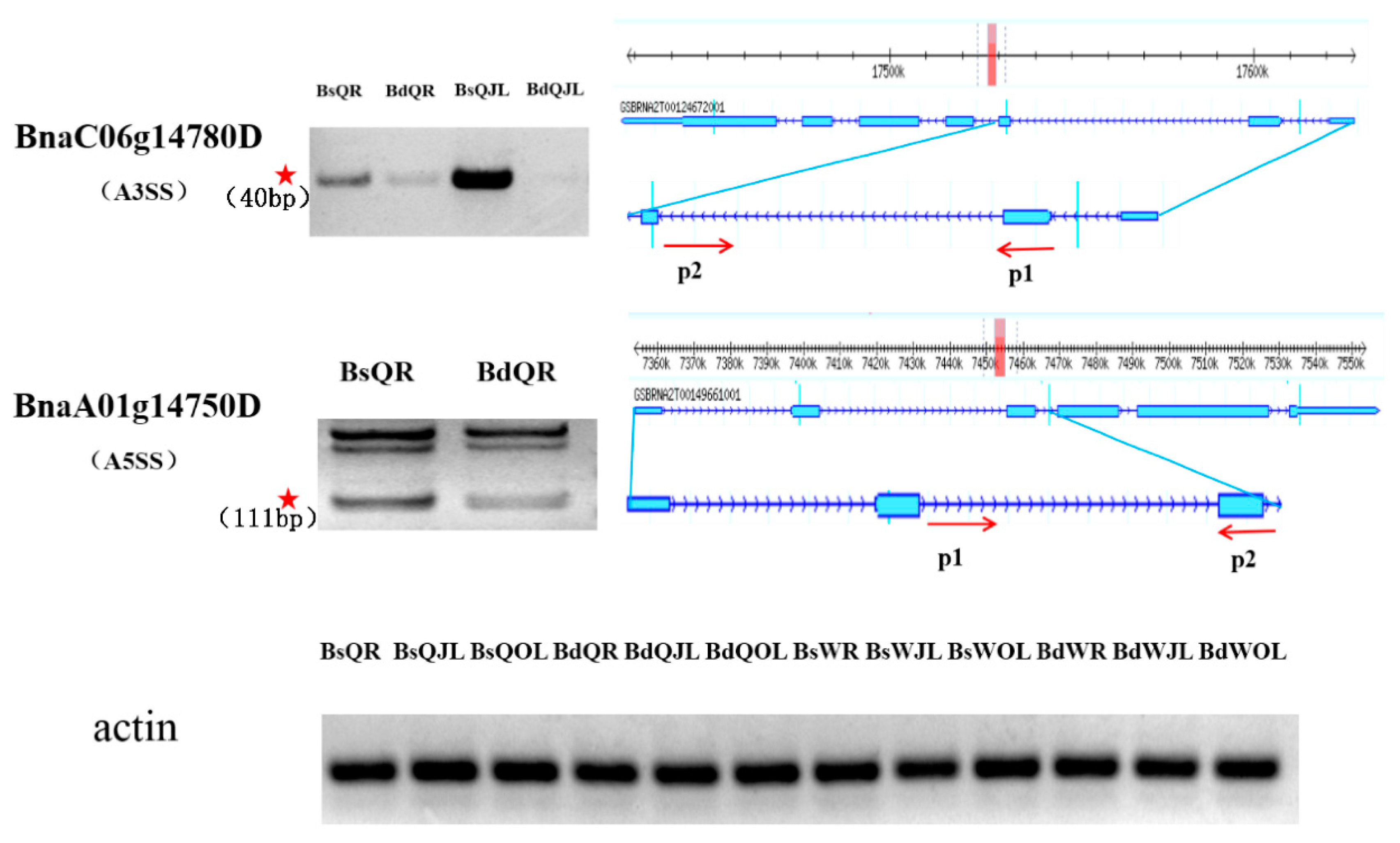
Figure 12.
Validation of differential alternative splicing factors in the root and juvenile leaves of B. napus cultivars Qingyou10 (QY10) and Westar10 (W10) under boron deficient conditions. The band with the red asterisk show the fragment generated by the alternative splicing of the gene, which was determined according to the size of the insert fragment. The forward and reverse primers, P1 and P2, were used to amplify the insert fragment and side exon. Bs, boron sufficient condition; Bd, boron deficient condition; QR, root of QY10; QOL, old leaves of QY10; QJL, juvenile leaves of QY10; WR, root of W10; WOL, old leaves of W10; WJL, juvenile leaves of W10. A3SS, alternative 3′ splicing site; A5SS, alternative 5′ splicing site.
Figure 12.
Validation of differential alternative splicing factors in the root and juvenile leaves of B. napus cultivars Qingyou10 (QY10) and Westar10 (W10) under boron deficient conditions. The band with the red asterisk show the fragment generated by the alternative splicing of the gene, which was determined according to the size of the insert fragment. The forward and reverse primers, P1 and P2, were used to amplify the insert fragment and side exon. Bs, boron sufficient condition; Bd, boron deficient condition; QR, root of QY10; QOL, old leaves of QY10; QJL, juvenile leaves of QY10; WR, root of W10; WOL, old leaves of W10; WJL, juvenile leaves of W10. A3SS, alternative 3′ splicing site; A5SS, alternative 5′ splicing site.
Table 1.
The starting reads, clean reads ratio, and mapped reads ratio against in the B. napus reference genome of c.v. Darmor-bzh.
Table 1.
The starting reads, clean reads ratio, and mapped reads ratio against in the B. napus reference genome of c.v. Darmor-bzh.
| | No. of Reads (Millions) | Reads Percentage (%) |
|---|
| Starting reads | 2078.8 | - |
| Clean reads | 1921.9 | 92.45 |
| Mapped reads | 1310.9 | 68.2 |
Table 2.
The number of novel transcripts and novel genes in the root, juvenile leaves and old leaves of Qingyou10 and Westar10 under B sufficient and deficient conditions (above 1.0 FPKM).
Table 2.
The number of novel transcripts and novel genes in the root, juvenile leaves and old leaves of Qingyou10 and Westar10 under B sufficient and deficient conditions (above 1.0 FPKM).
| Sample Name | Novel Transcripts | Reference Like Transcripts | Novel Genes | Reference Like Genes |
|---|
| BsQR1 | 32,762 | 16,656 | 1019 | 5877 |
| BsQR2 | 33,451 | 16,384 | 1136 | 5766 |
| BsQR3 | 33,662 | 16,661 | 1096 | 5841 |
| BsQJL1 | 33,763 | 15,740 | 1147 | 5539 |
| BsQJL2 | 35,091 | 15,637 | 1248 | 5742 |
| BsQJL3 | 33,274 | 15,608 | 1221 | 5400 |
| BsQOL1 | 34,228 | 14,921 | 1288 | 5033 |
| BsQOL2 | 34,501 | 14,959 | 1299 | 5074 |
| BsQOL3 | 33,610 | 14,592 | 1330 | 4927 |
| BdQR1 | 35,565 | 16,441 | 1415 | 5648 |
| BdQR2 | 35,780 | 16,570 | 1549 | 5643 |
| BdQR3 | 36,428 | 16,731 | 1545 | 5658 |
| BdQJL1 | 37,831 | 16,365 | 1601 | 5455 |
| BdQJL2 | 36,892 | 16,614 | 1518 | 5625 |
| BdQJL3 | 37,677 | 16,124 | 1500 | 5391 |
| BdQOL1 | 37,624 | 16,125 | 1685 | 5291 |
| BdQOL2 | 35,766 | 16,028 | 1473 | 5339 |
| BdQOL3 | 36,421 | 16,189 | 1513 | 5383 |
| BsWR1 | 34,561 | 16,128 | 1357 | 5563 |
| BsWR2 | 35,245 | 16,801 | 1433 | 5777 |
| BsWR3 | 34,940 | 17,124 | 1417 | 5895 |
| BsWJL1 | 36,579 | 16,029 | 1513 | 5412 |
| BsWJL2 | 38,206 | 16,252 | 1470 | 5382 |
| BsWJL3 | 37,242 | 16,214 | 1529 | 5468 |
| BsWOL1 | 38,490 | 15,781 | 1812 | 5114 |
| BsWOL2 | 37,855 | 15,850 | 1767 | 5161 |
| BsWOL3 | 37,224 | 15,411 | 1713 | 5025 |
| BdWR1 | 35,174 | 16,653 | 1407 | 5738 |
| BdWR2 | 35,790 | 17,013 | 1483 | 5830 |
| BdWR3 | 36,108 | 16,838 | 1494 | 5766 |
| BdWJL1 | 36,001 | 15,875 | 1498 | 5328 |
| BdWJL2 | 37,751 | 16,557 | 1588 | 5538 |
| BdWJL3 | 36,854 | 15,976 | 1523 | 5360 |
| BdWOL1 | 36,841 | 16,041 | 1559 | 5325 |
| BdWOL2 | 36,942 | 15,973 | 1485 | 5335 |
| BdWOL3 | 35,714 | 15,746 | 1517 | 5284 |
Table 3.
Thirty-three and two differential alternative splicing genes detected in root, juvenile leaves and old leaves simultaneously in B. napus cultivars Qingyou10 and Westar10, respectively.
Table 3.
Thirty-three and two differential alternative splicing genes detected in root, juvenile leaves and old leaves simultaneously in B. napus cultivars Qingyou10 and Westar10, respectively.
| Cultivars | GeneID | Gene Description | DAS-Type |
|---|
| QY10 | BnaC09g39180D | alpha/beta-Hydrolases superfamily protein | A5SS |
| BnaA06g17710D | alpha-glucan phosphorylase 2 (PHS2) | RI |
| BnaC01g05800D | AME3 | RI |
| BnaC03g31650D | ATOZI1 | A3SS |
| BnaC09g20000D | calcineurin B-like protein 9 (CBL9) | ES |
| BnaC06g30700D | cAMP-regulated phosphoprotein 19-related protein | A3SS |
| BnaC05g02560D | casein kinase like 13 (CKL13) | A5SS |
| BnaC08g08360D | CONSTITUTIVE PHOTOMORPHOGENIC 9 (COP9) | RI |
| BnaA03g27430D | DEAD box RNA helicase family protein | RI |
| BnaA09g27350D | dormancy-associated protein-like 1 (DYL1) | RI |
| BnaA03g39560D | enhancer of ag-4 2 (hua2) | RI |
| BnaC03g57920D | FK506 binding protein 53 (fkbp53) | A3SS |
| BnaAnng37730D | glutathione S-transferase phi 8 (GSTF8) | ES |
| BnaA07g00230D | actin 7 (ACT7) | RI |
| BnaA04g12350D | glycine rich protein 7 (atgrp7) | A5SS |
| BnaC04g04470D | homeodomain GLABROUS 1 (HDG1) | RI |
| BnaA06g30540D | NADH-ubiquinone oxidoreductase B8 subunit, putative | ES |
| BnaA06g38980D | nitrilase 2 (NIT2) | ES |
| BnaC09g20910D | peptide transporter 2 (PTR2) | RI |
| BnaC05g18490D | Phosphoglucomutase/phosphomannomutase family protein | RI |
| BnaA07g16600D | PUR5 | A3SS |
| BnaC05g24350D | radical-induced cell death1 (rcd1) | RI |
| BnaA03g30650D | Ribosomal L29 family protein | ES |
| BnaC09g54460D | Ribosomal protein S13/S18 family | A3SS |
| BnaA07g25750D | RNA-binding (RRM/RBD/RNP motifs) family protein | RI |
| BnaC06g14780D | RSZ32 | A3SS |
| BnaA07g16660D | sedoheptulose-bisphosphatase (SBPASE) | ES |
| BnaC05g08610D | sugar transporter 1 (STP1) | A3SS |
| BnaC04g31660D | TLD-domain containing nucleolar protein | RI |
| BnaA03g07610D | Translation elongation factor EF1B | A3SS |
| BnaC04g56630D | unknown protein | RI |
| BnaCnng63660D | unknown protein | RI |
| BnaC03g03780D | VND-interacting 1 (VNI1) | A3SS |
| W10 | BnaCnng40950D | Cystatin/monellin superfamily protein | A3SS |
| BnaC07g51220D | nicotinate phosphoribosyltransferase 1 (NAPRT1) | A5SS |
Table 4.
KEGG pathways of differential alternative splicing genes in root, juvenile leaves and old leaves of B. napus cultivars Qingyou10 and Westar10 under B deficient conditions.
Table 4.
KEGG pathways of differential alternative splicing genes in root, juvenile leaves and old leaves of B. napus cultivars Qingyou10 and Westar10 under B deficient conditions.
| Tissues | KEGG Pathways | Pathway ID | No. of DAS Gene | p-Value |
|---|
| QR | Spliceosome | ko03040 | 13 | 3.08E-06 |
| Carbon metabolism | ko01200 | 7 | 0.034913379 |
| Carbon fixation in photosynthetic organisms | ko00710 | 3 | 0.034913379 |
| Biosynthesis of amino acids | ko01230 | 6 | 0.034913379 |
| mRNA surveillance pathway | ko03015 | 4 | 0.034913379 |
| Glycolysis / Gluconeogenesis | ko00010 | 3 | 0.034913379 |
| Citrate cycle (TCA cycle) | ko00020 | 2 | 0.034913379 |
| Glyoxylate and dicarboxylate metabolism | ko00630 | 2 | 0.034913379 |
| Pyruvate metabolism | ko00620 | 2 | 0.034913379 |
| Sulfur metabolism | ko00920 | 1 | 0.034913379 |
| Nitrogen metabolism | ko00910 | 1 | 0.034913379 |
| Calcium signaling pathway | ko04020 | 1 | 0.034913379 |
| Galactose metabolism | ko00052 | 1 | 0.035225944 |
| MAPK signaling pathway | ko04010 | 1 | 0.035225944 |
| RNA transport | ko03013 | 2 | 0.037548325 |
| Arginine and proline metabolism | ko00330 | 1 | 0.038190214 |
| Starch and sucrose metabolism | ko00500 | 2 | 0.039194028 |
| Amino sugar and nucleotide sugar metabolism | ko00520 | 1 | 0.043604665 |
| Oxidative phosphorylation | ko00190 | 1 | 0.046236768 |
| Plant hormone signal transduction | ko04075 | 2 | 0.047198466 |
| QJL | Carbon fixation in photosynthetic organisms | ko00710 | 4 | 0.011345749 |
| Tryptophan metabolism | ko00380 | 3 | 0.011345749 |
| Pentose phosphate pathway | ko00030 | 3 | 0.011345749 |
| Glycolysis / Gluconeogenesis | ko00010 | 3 | 0.019619536 |
| Spliceosome | ko03040 | 4 | 0.019619536 |
| Glyoxylate and dicarboxylate metabolism | ko00630 | 2 | 0.019619536 |
| Pyruvate metabolism | ko00620 | 2 | 0.019619536 |
| ABC transporters | ko02010 | 1 | 0.019619536 |
| Carbon metabolism | ko01200 | 4 | 0.019619536 |
| Fructose and mannose metabolism | ko00051 | 1 | 0.019619536 |
| MAPK signaling pathway | ko04010 | 1 | 0.019619536 |
| Glycine, serine and threonine metabolism | ko00260 | 1 | 0.020680045 |
| Arginine and proline metabolism | ko00330 | 1 | 0.021716355 |
| Starch and sucrose metabolism | ko00500 | 2 | 0.021716355 |
| Peroxisome | ko04146 | 1 | 0.021716355 |
| Pentose and glucuronate interconversions | ko00040 | 1 | 0.021716355 |
| Glutathione metabolism | ko00480 | 1 | 0.021865611 |
| Amino sugar and nucleotide sugar metabolism | ko00520 | 1 | 0.024431115 |
| Plant hormone signal transduction | ko04075 | 2 | 0.026222312 |
| RNA transport | ko03013 | 1 | 0.02673548 |
| Biosynthesis of amino acids | ko01230 | 1 | 0.029409127 |
| QOL | Spliceosome | ko03040 | 7 | 0.030261395 |
| AMPK signaling pathway | ko04152 | 5 | 0.030261395 |
| Pyruvate metabolism | ko00620 | 3 | 0.047340426 |
| Tryptophan metabolism | ko00380 | 2 | 0.049084834 |
| WR | Ribosome | ko03010 | 2 | 0.000561484 |
| WJL | Peroxisome | ko04146 | 2 | 0.027237847 |
| AMPK signaling pathway | ko04152 | 2 | 0.042503393 |
| Glyoxylate and dicarboxylate metabolism | ko00630 | 1 | 0.190757264 |
| Glycolysis / Gluconeogenesis | ko00010 | 1 | 0.278858201 |
| WOL | Starch and sucrose metabolism | ko00500 | 3 | 0.034551184 |
| Amino sugar and nucleotide sugar metabolism | ko00520 | 2 | 0.076377304 |
| Pentose phosphate pathway | ko00030 | 1 | 0.165990643 |
| Galactose metabolism | ko00052 | 1 | 0.173014536 |
Table 5.
The number of differential alternative splicing genes and differential expressed genes in the root, juvenile leaf and old leaf of B. napus cultivars Qingyou10 and Westar10 under boron deficient conditions.
Table 5.
The number of differential alternative splicing genes and differential expressed genes in the root, juvenile leaf and old leaf of B. napus cultivars Qingyou10 and Westar10 under boron deficient conditions.
| Sample Names | DAS Genes | DE Genes | Overlap |
|---|
| BsQR vs. BdQR | 179 | 3404 | 32 |
| BsQJL vs. BdQJL | 223 | 1482 | 30 |
| BsQOL vs. BdQOL | 178 | 1364 | 8 |
| BsWR vs. BdWR | 32 | 2053 | 7 |
| BsWJL vs. BdWJL | 47 | 1054 | 1 |
| BsWOL vs. BdWOL | 85 | 1181 | 13 |
| BsQR vs. BsWR | 174 | 3253 | 10 |
| BsQJL vs. BsWJL | 217 | 744 | 4 |
| BsQOL vs. BsWOL | 589 | 2769 | 31 |
| BdQR vs. BdWR | 37 | 102 | 3 |
| BdQJL vs. BdWJL | 49 | 51 | 0 |
| BdQOL vs. BdWOL | 62 | 196 | 0 |
Table 6.
Different types of target genes and gene interaction in the splicing factor gene network of B. napus cultivars Qingyou10 and Westar10 under boron deficient conditions.
Table 6.
Different types of target genes and gene interaction in the splicing factor gene network of B. napus cultivars Qingyou10 and Westar10 under boron deficient conditions.
| Gene Name | Target Genes | Z-Score | Gene Description | DAS Type-Regulation |
|---|
| Boron Deficient/Boron Sufficient |
|---|
| QR | QJL | QOL | WR | WJL | WOL |
|---|
| BnaC06g14780D | BnaC04g52770D | 0.92 | Basic-leucine zipper (bZIP) transcription factor | RI | - | - | - | - | - |
| | BnaC01g37580D | 0.91 | Protein kinase domain | ES | - | ES | ES | - | - |
| | BnaA09g52970D | 0.88 | Expansin | RI | - | - | - | - | - |
| | BnaA01g14590D | 0.87 | Zinc finger, RING-type | - | - | RI | - | - | - |
| | BnaC04g12670D | 0.86 | Folate-biopterin transporter | - | A3SS | - | - | - | - |
| | BnaA05g30860D | 0.83 | Glycosyl hydrolase family 100 | - | - | ES | - | - | - |
| | BnaC02g31500D | 0.80 | Pectinacetylesterase | - | - | - | - | ES | - |
| | BnaA07g32180D | 0.80 | VPS35 homolog B | RI | - | RI | - | - | - |
| | BnaA01g30320D | 0.80 | Phosphoglycerate kinase | A3SS | - | - | - | - | - |