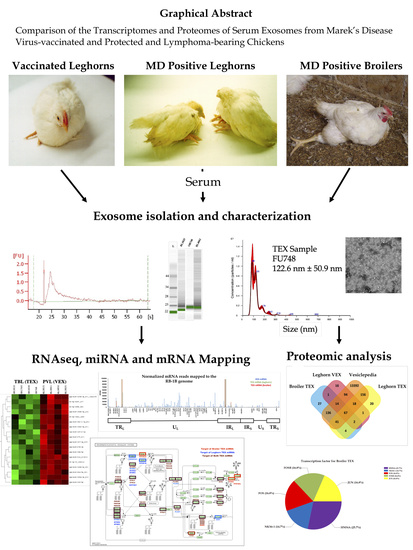
Comparison of the Transcriptomes and Proteomes of Serum Exosomes from Marek’s Disease Virus-Vaccinated and Protected and Lymphoma-Bearing Chickens
Abstract
:1. Introduction
2. Materials and Methods
2.1. Source of Serum Exosomes
2.2. Serum Exosome Purification
2.3. Nanoparticle Tracking Analysis
2.4. Transmission Electron Microscopy
2.5. Exosomal Small RNA Sequencing
2.5.1. Small RNA Library Preparation
2.5.2. Small RNA seq Data Analyses
2.5.3. MiRDeep2 Analyses of Precursor and Mature MDV-1 miRNAs
2.5.4. miRDB Prediction of miRNA Gene Targets
2.5.5. Gene Ontology and Pathway Enrichment Analysis
2.5.6. Geneious Mapping of Reads to the MDV Genome
2.5.7. Read Count Normalization and Comparisons
2.6. Proteomic Analysis of Exosomes
2.6.1. Sample Preparation
2.6.2. Protein Identification
3. Results
3.1. Characterization of Exosomes Isolated from Chicken Sera
3.2. Small RNAs Contained within Particles Conform to Exosomal Cargo
3.2.1. Cellular miRNAs Compose a Major Key Component of Exosomal Contents
3.2.2. MDV-Encoded miRNAs Differ in Abundance between VEX versus TEX
3.2.3. Target Pathways Predicted to Be Affected by VEX and TEX Cellular miRNAs
3.2.4. VEX and TEX Contain MDV mRNAs
3.3. Determining Protein Biomarkers of VEX and TEX
3.3.1. Enrichment Analysis of Exosomal Transcription Factors (TFs)
3.3.2. Putative Protein Biomarkers of VEX and TEX
4. Discussion
4.1. Abundant VEX-Borne miRNAs
4.1.1. The miR-146 Family
4.1.2. The miR-23~27~24 Cluster
4.1.3. miR-26a
4.1.4. VEX-Borne miRNA Targeted Cellular Pathways
4.2. TEX-Borne miRNAs
4.2.1. miR-92 and -363
4.2.2. Let-7i and -7g
4.2.3. miR-21
4.2.4. MDV-Encoded miRNAs
4.2.5. TEX-Borne miRNA Targeted Cellular Pathways
4.3. VEX/TEX-Borne MDV mRNA Expression
4.4. Proteomic Profiles of VEX and TEX
4.4.1. Potential VEX and TEX Biomarkers
4.4.2. TEX Contain MDV Tegument Proteins
4.4.3. VEX Contain MDV ICP4
4.4.4. Transcription Factor-Associated Pathways in VEX and TEX
5. Main Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Osterrieder, N.; Kamil, J.P.; Schumacher, D.; Tischer, B.K.; Trapp, S. Marek’s disease virus: from miasma to model. Nat. Rev. Microbiol. 2006, 4, 283–294. [Google Scholar] [CrossRef]
- Bertzbach, L.D.; Laparidou, M.; Härtle, S.; Etches, R.J.; Kaspers, B.; Schusser, B.; Kaufer, B.B. Unraveling the role of B cells in the pathogenesis of an oncogenic avian herpesvirus. Proc. Natl. Acad. Sci. USA 2018, 115, 11603–11607. [Google Scholar] [CrossRef]
- Jarosinski, K.W.; Osterrieder, N.; Nair, V.K.; Schat, K.A. Attenuation of Marek’s disease virus by deletion of open reading frame RLORF4 but not RLORF5a. J. Virol. 2005, 79, 11647–11659. [Google Scholar] [CrossRef] [PubMed]
- Trapp, S.; Parcells, M.S.; Kamil, J.P.; Schumacher, D.; Tischer, B.K.; Kumar, P.M.; Nair, V.K.; Osterrieder, N. A virus-encoded telomerase RNA promotes malignant T cell lymphomagenesis. J. Exp. Med. 2006, 203, 1307–1317. [Google Scholar] [CrossRef]
- Yao, Y.; Nair, V. Role of virus-encoded microRNAs in Avian viral diseases. Viruses 2014, 6, 1379–1394. [Google Scholar] [CrossRef] [PubMed]
- Jarosinski, K.; Kattenhorn, L.; Kaufer, B.; Ploegh, H.; Osterrieder, N. A herpesvirus ubiquitin-specific protease is critical for efficient T cell lymphoma formation. Proc. Natl. Acad. Sci. USA 2007, 104, 20025–20030. [Google Scholar] [CrossRef] [PubMed]
- Shack, L.A.; Buza, J.J.; Burgess, S.C. The neoplastically transformed (CD30hi) Marek’s disease lymphoma cell phenotype most closely resembles T-regulatory cells. Cancer Immunol. Immunother. 2008, 57, 1253–1262. [Google Scholar] [CrossRef]
- Parcells, M.S.; Burgess, S.C. Immunological aspects of Marek’s disease virus (MDV)-induced lymphoma progression. In Selected Aspects of Cancer Progression: Metastasis, Apoptosis and Immune Response; Kaiser, H.E., Nasir, A., Eds.; Springer: Dordrecht, The Netherlands, 2008; pp. 169–191. [Google Scholar]
- Omar, A.R.; Schat, K.A. Syngeneic Marek’s disease virus (MDV)-specific cell-mediated immune responses against immediate early, late, and unique MDV proteins. Virology 1996, 222, 87–99. [Google Scholar] [CrossRef]
- Omar, A.R.; Schat, K.A. Characterization of Marek’s disease herpesvirus-specific cytotoxic T lymphocytes in chickens inoculated with a non-oncogenic vaccine strain of MDV. Immunology 1997, 90, 579–585. [Google Scholar] [CrossRef]
- Lupiani, B.; Lee, L.F.; Cui, X.; Gimeno, I.; Anderson, A.; Morgan, R.W.; Silva, R.F.; Witter, R.L.; Kung, H.J.; Reddy, S.M. Marek’s disease virus-encoded Meq gene is involved in transformation of lymphocytes but is dispensable for replication. Proc. Natl. Acad. Sci. USA 2004, 101, 11815–11820. [Google Scholar] [CrossRef]
- Brown, A.C.; Nair, V.; Allday, M.J. Epigenetic regulation of the latency-associated region of Marek’s disease virus in tumor-derived T-cell lines and primary lymphoma. J. Virol. 2012, 86, 1683–1695. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Xu, H.; Yao, Y.; Smith, L.P.; Kgosana, L.; Green, J.; Petherbridge, L.; Baigent, S.J.; Nair, V. Critical role of the virus-encoded microRNA-155 ortholog in the induction of Marek’s disease lymphomas. PLoS Pathog. 2011, 7, e1001305. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yao, Y.; Xu, H.; Lambeth, L.; Smith, L.P.; Kgosana, L.; Wang, X.; Nair, V. A Functional MicroRNA-155 Ortholog Encoded by the Oncogenic Marek’s Disease Virus. J. Virol. 2009, 83, 489–492. [Google Scholar] [CrossRef] [PubMed]
- Lambeth, L.S.; Yao, Y.; Smith, L.P.; Zhao, Y.; Nair, V. MicroRNAs 221 and 222 target p27Kip1 in Marek’s disease virus-transformed tumour cell line MSB-1. J. Gen. Virol. 2009, 90, 1164–1171. [Google Scholar] [CrossRef] [PubMed]
- Neerukonda, S.N.; Egan, N.A.; Parcells, M.S. Exosomal Communication during Infection, Inflammation and Virus-Associated Pathology. J. Cancer Sci. 2017, 1, 1–13. [Google Scholar]
- Utsugi-Kobukai, S.; Fujimaki, H.; Hotta, C.; Nakazawa, M.; Minami, M. MHC class I-mediated exogenous antigen presentation by exosomes secreted from immature and mature bone marrow derived dendritic cells. Immunol. Lett. 2003, 89, 125–131. [Google Scholar] [CrossRef]
- Verweij, F.J.; van Eijndhoven, M.A.; Hopmans, E.S.; Vendrig, T.; Wurdinger, T.; Cahir-McFarland, E.; Kieff, E.; Geerts, D.; van der Kant, R.; Neefjes, J.; et al. LMP1 association with CD63 in endosomes and secretion via exosomes limits constitutive NF-κB activation. Embo J. 2011, 30, 2115–2129. [Google Scholar] [CrossRef]
- Walker, J.D.; Maier, C.L.; Pober, J.S. Cytomegalovirus-infected human endothelial cells can stimulate allogeneic CD4+ memory T cells by releasing antigenic exosomes. J. Immunol. 2009, 182, 1548–1559. [Google Scholar] [CrossRef]
- Whiteside, T.L. Immune modulation of T-cell and NK (natural killer) cell activities by TEXs (tumour-derived exosomes). Biochem. Soc. Trans. 2013, 41, 245–251.21. [Google Scholar] [CrossRef]
- Tavlarides-Hontz, P.; Kumar, P.M.; Amortegui, J.R.; Osterrieder, N.; Parcells, M.S. A deletion within glycoprotein L of Marek's disease virus (MDV) field isolates correlates with a decrease in bivalent MDV vaccine efficacy in contact-exposed chickens. Avian Dis 2009, 53, 287–296. [Google Scholar] [CrossRef]
- Nath Neerukonda, S.; Egan, N.A.; Patria, J.; Assakhi, I.; Tavlarides-Hontz, P.; Modla, S.; Munoz, E.R.; Hudson, M.B.; Parcells, M.S. Comparison of exosomes purified via ultracentrifugation (UC) and Total Exosome Isolation (TEI) reagent from the serum of Marek’s disease virus (MDV)-vaccinated and tumor-bearing chickens. J. Virol. Methods 2019, 263, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, R.; Bernhart, S.H.; Honer Zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Jarosinski, K.W.; Vautherot, J.-F. Differential expression of Marek’s disease virus (MDV) late proteins during in vitro and in situ replication: Role for pUL47 in regulation of the MDV UL46–UL49 gene locus. Virology 2015, 484, 213–226. [Google Scholar] [CrossRef] [PubMed]
- Friedlander, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Wong, N.; Wang, X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015, 43, D146–D152. [Google Scholar] [CrossRef] [PubMed]
- Dennis, G., Jr.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, P3. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Collins, J.R.; Alvord, W.G.; Roayaei, J.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007, 8, R183. [Google Scholar] [CrossRef] [PubMed]
- Roller, R.J.; Baines, J.D. Herpesvirus Nuclear Egress. In Cell Biology of Herpes Viruses; Osterrieder, K., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 143–169. [Google Scholar]
- McCarthy, F.M.; Burgess, S.C.; van den Berg, B.H.J.; Koter, M.D.; Pharr, G.T. Differential Detergent Fractionation for Non-electrophoretic Eukaryote Cell Proteomics. J. Proteome Res. 2005, 4, 316–324. [Google Scholar] [CrossRef]
- Pathan, M.; Keerthikumar, S.; Ang, C.-S.; Gangoda, L.; Quek, C.Y.J.; Williamson, N.A.; Mouradov, D.; Sieber, O.M.; Simpson, R.J.; Salim, A.; et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015, 15, 2597–2601. [Google Scholar] [CrossRef]
- Nahid, M.A.; Pauley, K.M.; Satoh, M.; Chan, E.K. miR-146a is critical for endotoxin-induced tolerance: Implication in Innate Immunity. J. Biol. Chem. 2009, 284, 34590–34599. [Google Scholar] [CrossRef]
- Taganov, K.D.; Boldin, M.P.; Chang, K.J.; Baltimore, D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef] [PubMed]
- Tulman, E.R.; Afonso, C.L.; Lu, Z.; Zsak, L.; Rock, D.L.; Kutish, G.F. The genome of a very virulent Marek’s disease virus. J. Virol. 2000, 74, 7980–7988. [Google Scholar] [CrossRef] [PubMed]
- Albright, B.S.; Kosinski, A.; Szczepaniak, R.; Cook, E.A.; Stow, N.D.; Conway, J.F.; Weller, S.K. The putative herpes simplex virus 1 chaperone protein UL32 modulates disulfide bond formation during infection. J. Virol. 2015, 89, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Heming, J.D.; Conway, J.F.; Homa, F.L. Herpesvirus Capsid Assembly and DNA Packaging. Adv. Anat. Embryol. Cell Biol. 2017, 223, 119–142. [Google Scholar] [PubMed]
- Pathan, M.; Fonseka, P.; Sanwlani, R.; Chitti, S.V.; Kang, T.; Mathivanan, S.; Hendrix, A.; Van Deun, J. Vesiclepedia 2019: a compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Res. 2018, 47, D516–D519. [Google Scholar] [CrossRef] [PubMed]
- Hurwitz, S.N.; Rider, M.A.; Bundy, J.L.; Liu, X.; Singh, R.K.; Meckes, D.G., Jr. Proteomic profiling of NCI-60 extracellular vesicles uncovers common protein cargo and cancer type-specific biomarkers. Oncotarget 2016, 7, 86999–87015. [Google Scholar] [CrossRef]
- Lu, L.-F.; Cho, S.; Wu, C.-J.; Yasuda, T.; Cruz, L.O.; Khan, A.A.; Lin, L.-L.; Nguyen, D.T.; Miller, M.; Lee, H.-M.; et al. MiR-23~27~24 clusters control effector T cell differentiation and function. J. Immunol. 2016, 196, 239–245. [Google Scholar]
- Zhu, S.; Pan, W.; Song, X.; Liu, Y.; Shao, X.; Tang, Y.; Liang, D.; He, D.; Wang, H.; Liu, W.; et al. The microRNA miR-23b suppresses IL-17-associated autoimmune inflammation by targeting TAB2, TAB3 and IKK-α. Nat. Med. 2012, 18, 1077. [Google Scholar] [CrossRef]
- Cho, S.; Wu, C.-J.; Yasuda, T.; Cruz, L.O.; Khan, A.A.; Lin, L.-L.; Nguyen, D.T.; Miller, M.; Lee, H.-M.; Kuo, M.-L.; et al. miR-23∼27∼24 clusters control effector T cell differentiation and function. J. Exp. Med. 2016. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, X.F.; Lu, X.; Jia, H.L.; Liang, L.; Dong, Q.Z.; Ye, Q.H.; Qin, L.X. MicroRNA-26a suppresses angiogenesis in human hepatocellular carcinoma by targeting hepatocyte growth factor-cMet pathway. Hepatology 2014, 59, 1874–1885. [Google Scholar] [CrossRef]
- Wan, L.; Zhang, L.; Fan, K.; Wang, J. miR-27b targets LIMK1 to inhibit growth and invasion of NSCLC cells. Mol. Cell. Biochem. 2014, 390, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Wu, X.; Wu, D.; Wu, P.; Ni, C.; Zhang, Z.; Chen, Z.; Qiu, F.; Xu, J.; Huang, J. miRNA-27b Targets Vascular Endothelial Growth Factor C to Inhibit Tumor Progression and Angiogenesis in Colorectal Cancer. PLoS ONE 2013, 8, e60687. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Rho, J.G.; Kim, C.; Tak, H.; Lee, H.; Ji, E.; Ahn, S.; Shin, A.R.; Cho, H.-I.; Huh, Y.H.; et al. The miR-24-3p/p130Cas: A novel axis regulating the migration and invasion of cancer cells. Sci. Rep. 2017, 7, 44847. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Kluiver, J.; Koerts, J.; de Jong, D.; Rutgers, B.; Abdul Razak, F.R.; Terpstra, M.; Plaat, B.E.; Nolte, I.M.; Diepstra, A.; et al. miR-24-3p Is Overexpressed in Hodgkin Lymphoma and Protects Hodgkin and Reed-Sternberg Cells from Apoptosis. Am. J. Pathol. 2017, 187, 1343–1355. [Google Scholar] [CrossRef] [PubMed]
- Huse, J.T.; Brennan, C.; Hambardzumyan, D.; Wee, B.; Pena, J.; Rouhanifard, S.H.; Sohn-Lee, C.; le Sage, C.; Agami, R.; Tuschl, T.; et al. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009, 23, 1327–1337. [Google Scholar] [CrossRef] [PubMed]
- Sander, S.; Bullinger, L.; Klapproth, K.; Fiedler, K.; Kestler, H.A.; Barth, T.F.; Moller, P.; Stilgenbauer, S.; Pollack, J.R.; Wirth, T. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood 2008, 112, 4202–4212. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.F.; Tellam, R.L. MicroRNA-26a targets the histone methyltransferase Enhancer of Zeste homolog 2 during myogenesis. J. Biol. Chem. 2008, 283, 9836–9843. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Ju, W.Q.; Yuan, X.P.; Zhu, X.F.; Wang, D.P.; He, X.S. miR-26a regulates mouse hepatocyte proliferation via directly targeting the 3′ untranslated region of CCND2 and CCNE2. Hepatobiliary Pancreat. Dis. Int. 2016, 15, 65–72. [Google Scholar] [CrossRef]
- Zhuang, C.; Wang, P.; Huang, D.; Xu, L.; Wang, X.; Wang, L.; Hu, L. A double-negative feedback loop between EZH2 and miR-26a regulates tumor cell growth in hepatocellular carcinoma. Int. J. Oncol. 2016, 48, 1195–1204. [Google Scholar] [CrossRef]
- Lian, L.; Qu, L.; Chen, Y.; Lamont, S.J.; Yang, N. A systematic analysis of miRNA transcriptome in Marek’s disease virus-induced lymphoma reveals novel and differentially expressed miRNAs. PLoS ONE 2012, 7, e51003. [Google Scholar] [CrossRef]
- Li, X.; Lian, L.; Zhang, D.; Qu, L.; Yang, N. gga-miR-26a targets NEK6 and suppresses Marek’s disease lymphoma cell proliferation. Poult. Sci. 2014, 93, 1097–1105. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, A.; Ohno, S.; Wu, W.; Borjigin, N.; Fujita, K.; Aoki, T.; Ueda, S.; Takanashi, M.; Kuroda, M. miR-92 is a key oncogenic component of the miR-17–92 cluster in colon cancer. Cancer Sci. 2011, 102, 2264–2271. [Google Scholar] [CrossRef] [PubMed]
- Landais, S.; Landry, S.; Legault, P.; Rassart, E. Oncogenic Potential of the miR-106-363 Cluster and Its Implication in Human T-Cell Leukemia. Cancer Res. 2007, 67, 5699–5707. [Google Scholar] [CrossRef] [PubMed]
- Mogilyansky, E.; Rigoutsos, I. The miR-17/92 cluster: A comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013, 20, 1603–1614. [Google Scholar] [CrossRef] [PubMed]
- Mu, P.; Han, Y.-C.; Betel, D.; Yao, E.; Squatrito, M.; Ogrodowski, P.; de Stanchina, E.; D’Andrea, A.; Sander, C.; Ventura, A. Genetic dissection of the miR-17∼92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009, 23, 2806–2811. [Google Scholar] [CrossRef] [PubMed]
- Sampson, V.B.; Rong, N.H.; Han, J.; Yang, Q.; Aris, V.; Soteropoulos, P.; Petrelli, N.J.; Dunn, S.P.; Krueger, L.J. MicroRNA Let-7a Down-regulates MYC and Reverts MYC-Induced Growth in Burkitt Lymphoma Cells. Cancer Res. 2007, 67, 9762–9770. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Kumar, M.; Choudhury, S.N.; Becker Buscaglia, L.E.; Barker, J.R.; Kanakamedala, K.; Liu, M.-F.; Li, Y. Loss of the miR-21 allele elevates the expression of its target genes and reduces tumorigenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 10144–10149. [Google Scholar] [CrossRef]
- Stik, G.; Dambrine, G.; Pfeffer, S.; Rasschaert, D. The Oncogenic MicroRNA OncomiR-21 Overexpressed during Marek’s Disease Lymphomagenesis Is Transactivated by the Viral Oncoprotein Meq. J. Virol. 2013, 87, 80–93. [Google Scholar] [CrossRef]
- Melnik, B.C. MiR-21: An environmental driver of malignant melanoma? J. Transl. Med. 2015, 13, 202. [Google Scholar] [CrossRef]
- Parnas, O.; Corcoran, D.L.; Cullen, B.R. Analysis of the mRNA targetome of microRNAs expressed by Marek’s disease virus. MBio 2014, 5, e01060-13. [Google Scholar] [CrossRef]
- Hicks, J.A.; Trakooljul, N.; Liu, H.-C. Alterations in cellular and viral microRNA and cellular gene expression in Marek’s disease virus-transformed T-cell lines treated with sodium butyrate. Poult. Sci. 2018. [Google Scholar] [CrossRef] [PubMed]
- Wymann, M. PI3Ks—Drug Targets in Inflammation and Cancer. In Phosphoinositides I: Enzymes of Synthesis and Degradation; Balla, T., Wymann, M., York, J.D., Eds.; Springer: Dordrecht, The Netherlands, 2012; pp. 111–181. [Google Scholar]
- Czernek, L.; Chworos, A.; Duechler, M. The Uptake of Extracellular Vesicles is Affected by the Differentiation Status of Myeloid Cells. Scand. J. Immunol. 2015, 82, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Naquet, P.; Pitari, G.; Dupre, S.; Galland, F. Role of the Vnn1 pantetheinase in tissue tolerance to stress. Biochem. Soc. Trans. 2014, 42, 1094–1100. [Google Scholar] [CrossRef]
- Kaskow, B.J.; Proffitt, J.M.; Blangero, J.; Moses, E.K.; Abraham, L.J. Diverse biological activities of the vascular non-inflammatory molecules—The Vanin pantetheinases. Biochem. Biophys. Res. Commun. 2012, 417, 653–658. [Google Scholar] [CrossRef]
- Veiga, I.B.; Jarosinski, K.W.; Kaufer, B.B.; Osterrieder, N. Marek’s disease virus (MDV) ubiquitin-specific protease (USP) performs critical functions beyond its enzymatic activity during virus replication. Virology 2013, 437, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Guan, J.; Zheng, C.; Zheng, X. The herpes simplex virus 1 UL36USP deubiquitinase suppresses DNA repair in host cells via deubiquitination of proliferating cell nuclear antigen. J. Biol. Chem. 2017, 292, 8472–8483. [Google Scholar] [CrossRef] [PubMed]
- Ye, R.; Su, C.; Xu, H.; Zheng, C. Herpes Simplex Virus 1 Ubiquitin-Specific Protease UL36 Abrogates NF-κB Activation in DNA Sensing Signal Pathway. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Ratliff, M.L.; Mishra, M.; Frank, M.B.; Guthridge, J.M.; Webb, C.F. The Transcription Factor ARID3a Is Important for In Vitro Differentiation of Human Hematopoietic Progenitors. J. Immunol. 2016, 196, 614–623. [Google Scholar] [CrossRef]
- Lau, H.H.; Ng, N.H.J.; Loo, L.S.W.; Jasmen, J.B.; Teo, A.K.K. The molecular functions of hepatocyte nuclear factors – In and beyond the liver. J. Hepatol. 2018, 68, 1033–1048. [Google Scholar] [CrossRef]
- Rahrmann, E.P.; Wolf, N.K.; Otto, G.M.; Heltemes-Harris, L.; Ramsey, L.B.; Shu, J.; LaRue, R.S.; Linden, M.A.; Rathe, S.K.; Starr, T.K.; et al. Sleeping Beauty Screen Identifies RREB1 and Other Genetic Drivers in Human B-cell Lymphoma. Mol. Cancer Res. 2018. [Google Scholar] [CrossRef]
- Kiss, M.; Czimmerer, Z.; Nagy, G.; Bieniasz-Krzywiec, P.; Ehling, M.; Pap, A.; Poliska, S.; Boto, P.; Tzerpos, P.; Horvath, A.; et al. Retinoid X receptor suppresses a metastasis-promoting transcriptional program in myeloid cells via a ligand-insensitive mechanism. Proc. Natl. Acad. Sci. USA 2017, 114, 10725–10730. [Google Scholar] [CrossRef] [PubMed]
- Núñez, V.; Alameda, D.; Rico, D.; Mota, R.; Gonzalo, P.; Cedenilla, M.; Fischer, T.; Boscá, L.; Glass, C.K.; Arroyo, A.G.; et al. Retinoid X receptor α controls innate inflammatory responses through the up-regulation of chemokine expression. Proc. Natl. Acad. Sci. USA 2010, 107, 10626–10631. [Google Scholar] [CrossRef] [PubMed]
- Khachigian, L.M. The Yin and Yang of YY1 in tumor growth and suppression. Int. J. Cancer 2018, 143, 460–465. [Google Scholar] [CrossRef] [PubMed]
- Gruffat, H.; Manet, E.; Sergeant, A. MEF2-mediated recruitment of class II HDAC at the EBV immediate early gene BZLF1 links latency and chromatin remodeling. Embo Rep. 2002, 3, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liu, P.; Borras, A.; Chatila, T.; Speck, S.H. Cyclosporin A-sensitive induction of the Epstein-Barr virus lytic switch is mediated via a novel pathway involving a MEF2 family member. Embo J. 1997, 16, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Pon, J.R.; Marra, M.A. MEF2 transcription factors: Developmental regulators and emerging cancer genes. Oncotarget 2016, 7, 2297–2312. [Google Scholar] [CrossRef]
- Jones, D.; Lee, L.; Liu, J.L.; Kung, H.J.; Tillotson, J.K. Marek disease virus encodes a basic-leucine zipper gene resembling the fos/jun oncogenes that is highly expressed in lymphoblastoid tumors. Proc. Natl. Acad. Sci. USA 1992, 89, 4042–4046. [Google Scholar] [CrossRef]
- Levy, A.M.; Gilad, O.; Xia, L.; Izumiya, Y.; Choi, J.; Tsalenko, A.; Yakhini, Z.; Witter, R.; Lee, L.; Cardona, C.J.; et al. Marek’s disease virus Meq transforms chicken cells via the v-Jun transcriptional cascade: A converging transforming pathway for avian oncoviruses. Proc. Natl. Acad. Sci. USA 2005, 102, 14831–14836. [Google Scholar] [CrossRef]







| Serum Exosome Source | Bird Tag# | Raw Reads | Mapped to Gallus gallus | % | miRNAs Mapped to MDV | % | mRNAs Mapped to MDV | % |
|---|---|---|---|---|---|---|---|---|
| leghorn VEX | BL3815 | 27,168,090 | 5,539,665 | 20.4 | 478 | 0.009 | 11,995 | 0.22 |
| BL3841 | 16,544,388 | 12,264,873 | 74.1 | 224 | 0.002 | 29,586 | 0.24 | |
| BL3850 | 11,567,549 | 73,94,774 | 63.9 | 6895 | 0.09 | 36,084 | 0.49 | |
| BL3825 | 8,516,052 | 4,885,568 | 57.4 | 1864 | 0.04 | 8335 | 0.17 | |
| Means | 15,949,020 | 7,563,369 | 47.4 | 2365 | 0.03 | 21,500 | 0.28 | |
| leghorn TEX | OR1810 | 18,024,380 | 9,038,453 | 50.15 | 47,893 | 0.53 | 64,636 | 0.72 |
| OR1760 | 13,826,014 | 8,725,701 | 63.11 | 4126 | 0.05 | 314 | 0.003 | |
| OR1838 | 26,009,675 | 8,559,904 | 32.91 | 4399 | 0.05 | 534 | 0.006 | |
| OR748 | 16,422,100 | 8,321,176 | 50.67 | 9594 | 0.12 | 56,629 | 0.68 | |
| Means | 18,570,542 | 8,661,309 | 46.64 | 16,503 | 0.19 | 30,528 | 0.35 | |
| broiler TEX | BL4178 | 15,878,894 | 9,747,686 | 61.39 | 22,533 | 0.23 | 57,285 | 0.58 |
| BL4047 | 16,472,545 | 12,121,502 | 73.59 | 89,040 | 0.73 | 66,270 | 0.54 | |
| BL4062 | 14,407,601 | 9,424,620 | 65.41 | 148,011 | 1.57 | 55,003 | 0.58 | |
| BL4183 | 26,533,369 | 16,334,950 | 61.56 | 72,162 | 0.44 | 87,058 | 0.53 | |
| Means | 18,323,102 | 11,907,190 | 65.0 | 82,937 | 0.70 | 66,404 | 0.55 | |
| Totals | 211,370,657 | 112,358,872 | 53.16 | 409,687 | 0.36 | 473,729 | 0.42 |
| G. gallus miRNA | p-Value | Log2(VEX/TEX) | Leghorn TEX | Leghorn VEX |
|---|---|---|---|---|
| gga-miR-146b-5p | 7.21 × 10−5 | 5.06 | 2046 | 68,424 |
| PC-3p-53 200112 | 1.35 × 10−3 | 5.01 | 1134 | 36,566 |
| gga-miR-10b-5p | 2.03 × 10−3 | 4.38 | 21,594 | 449,498 |
| gga-miR-2188-5p | 3.24 × 10−2 | 3.65 | 16,457 | 207,103 |
| gga-miR-27b-3p | 3.24 × 10−4 | 3.63 | 8156 | 100,917 |
| gga-miR-99a-5p | 4.96 × 10−2 | 2.46 | 200,908 | 1,105,652 |
| gga-miR-26a-5p | 4.67 × 10−3 | 2.17 | 10,858 | 48,877 |
| gga-miR-146c-5p | 1.49 × 10−3 | 1.89 | 142,564 | 526,725 |
| gga-miR-24-3p | 7.26 × 10−2 | 1.43 | 3313 | 8949 |
| gga-miR-146a-5p | 3.91 × 10−2 | 1.16 | 18,000 | 40,291 |
| gga-miR-23b-3p | 6.65 × 10−2 | 1.16 | 3365 | 7535 |
| gga-miR-30e-5p | 4.99 × 10−2 | 0.88 | 4247 | 7789 |
| gga-miR-199-3p | 4.71 × 10−2 | 0.66 | 12,166 | 19,208 |
| gga-miR-142-5p | 3.59 × 10−2 | −1.16 | 11,024 | 4947 |
| gga-miR-125b-5p | 4.55 × 10−2 | −1.32 | 116,830 | 46,809 |
| gga-miR-181a-5p | 5.70 × 10−2 | −1.44 | 31,255 | 11,509 |
| gga-let-7 | 1.28 × 10−2 | −1.83 | 20,148 | 5681 |
| gga-miR-363-3p | 2.27 × 10−2 | −1.89 | 37,382 | 10,120 |
| gga-miR-92-3p | 4.55 × 10−3 | −2.17 | 60,566 | 13,491 |
| gga-miR-101-3p | 3.40 × 10−3 | −2.23 | 9426 | 2013 |
| gga-let-7g-5p | 6.86 × 10−4 | −2.24 | 11,384 | 2417 |
| gga-miR-451 | 2.47 × 10−3 | −2.33 | 588,775 | 117,350 |
| gga-miR-148a-3p | 6.94 × 10−3 | −2.44 | 113,250 | 20,858 |
| gga-miR-21-5p | 4.20 × 10−3 | −2.88 | 120,353 | 16,302 |
| gga-miR-7 | 1.86 × 10−3 | −3.21 | 47,490 | 5122 |
| gga-miR-15c-5p | 1.50 × 10−2 | −3.39 | 5628 | 537 |
| gga-miR-16-5p | 4.77 × 10−4 | −3.45 | 10,178 | 934 |
| gga-miR-16c-5p | 7.74 × 10−5 | −4.03 | 20,103 | 1231 |
| G.gallus miRNA | p-Value | Log2(VEX/TEX) | Broiler TEX | Leghorn VEX |
|---|---|---|---|---|
| gga-miR-99a-5p | 1.08 × 10−2 | 3.58 | 92,713 | 1,105,652 |
| gga-miR-2188-5p | 3.56 × 10−2 | 3.52 | 18,115 | 207,103 |
| gga-miR-10b-5p | 1.73 × 10−2 | 2.89 | 60,577 | 449,498 |
| tgu-miR-1388-5p | 1.20 × 10−2 | 1.81 | 14,403 | 50,359 |
| gga-let-7f-5p | 8.60 × 10−3 | 1.70 | 4454 | 14,519 |
| gga-miR-146b-5p | 1.58 × 10−2 | 1.70 | 21,014 | 68,424 |
| gga-miR-26a-5p | 2.81 × 10−2 | 1.53 | 16,930 | 48,877 |
| gga-miR-27b-3p | 1.99 × 10−2 | 1.49 | 35,868 | 100,917 |
| gga-miR-30e-5p | 3.23 × 10−2 | 1.41 | 13,857 | 36,839 |
| gga-miR-146c-5p | 8.41 × 10−3 | 1.34 | 208,239 | 526,725 |
| gga-miR-23b-3p | 6.49 × 10−2 | 1.06 | 3625 | 7535 |
| gga-miR-30e-5p | 8.75 × 10−2 | 0.82 | 4423 | 7789 |
| gga-miR-24-3p | 4.70 × 10−2 | 0.72 | 5445 | 8949 |
| gga-miR-199-3p | 8.75 × 10−2 | −0.53 | 27,729 | 19,208 |
| tgu-miR-425-5p | 3.19 × 10−2 | −0.78 | 5509 | 3198 |
| gga-miR-92-3p | 9.08 × 10−2 | −0.85 | 24,402 | 13,491 |
| gga-let-7i | 2.78 × 10−2 | −0.86 | 10,306 | 5,681 |
| gga-miR-181a-5p | 3.78 × 10−2 | −0.88 | 21,235 | 11,509 |
| gga-miR-126-3p | 7.58 × 10−2 | −1.05 | 10,493 | 5052 |
| gga-miR-148a-3p | 4.52 × 10−2 | −1.15 | 46,178 | 20,858 |
| tgu-miR-363-3p | 6.53 × 10−3 | −1.31 | 25,070 | 10,120 |
| gga-miR-21-5p | 4.23 × 10−3 | −1.35 | 41,470 | 16,302 |
| gga-miR-122-5p | 2.98 × 10−2 | −1.56 | 13,321 | 4527 |
| gga-miR-142-5p | 4.89 × 10−3 | −2.08 | 20,957 | 4947 |
| gga-miR-199-5p | 1.00 × 10−2 | −2.18 | 77,490 | 17,139 |
| gga-miR-140-3p | 1.07 × 10−4 | −2.19 | 10,748 | 2359 |
| Leghorn TEX/Leghorn VEX | Broiler TEX/Leghorn VEX | ||||
|---|---|---|---|---|---|
| miRNA Origin Cluster | MDV-1-miR- | Fold Change | p-Value | Fold Change | p-Value |
| meq cluster 1 | -M9 | 9.41 | 0.22 | 39.43 | 0.07 |
| -M5 | 1.47 | 0.60 | 15.49 | 0.02 | |
| -M12 | 3.44 | 0.37 | 49.00 | 0.08 | |
| -M3 | 13.04 | 0.26 | 43.70 | 0.07 | |
| -M2 | 2.19 | 0.39 | 40.60 | 0.03 | |
| -M4 | 10.36 | 0.33 | 28.99 | 0.03 | |
| meq cluster 2 | -M11 | 4.27 | 0.16 | 69.20 | 0.01 |
| -M31 | 0.00 | 0.19 | - | - | |
| -M1 | 1.20 | 0.89 | 19.10 | 0.04 | |
| LAT cluster | -M8 | 3.85 | 0.02 | 13.83 | 0.08 |
| -M13 | 4.33 | 0.32 | 10.78 | 0.05 | |
| -M6 | 7.63 | 0.28 | 41.15 | 0.04 | |
| -M7 | 22.45 | 0.16 | 37.38 | 0.01 | |
| -M10 | 2.18 | 0.36 | 27.65 | 0.06 | |
| Protein Name (Accession#) | VEX (NI) 1 | TEX (NI) 1 | RatioVEX/TEX | p-Value |
|---|---|---|---|---|
| Protocadherin-17 (XP_015132312.1) | 2.31 | 0 | 11,109:1 | 0.015 |
| Collagen -1 (XXII) chain (XP_015138660.1) | 1.13 | 0 | 10,825:1 | 0.048 |
| IgGFc-binding protein-like protein (XP_428412.5) | 0.6 | 0 | 2851:1 | 0.035 |
| Insulin-like growth factor-binding protein complex acid labile subunit isoform (XP_425222.3) | 0.24 | 0 | 1159:1 | 0.014 |
| Tenascin isoforms (XP_015134827.1) | 0.15 | 0 | 998:1 | 0.014 |
| Insulin-like growth factor-binding protein complex acid labile subunit isoform (XP_425222.3) | 0.24 | 0 | 851:1 | 0.043 |
| Muellerian-inhibiting factor precursor (NP_990361.1) | 0.21 | 0 | 733:1 | 0.043 |
| Collagen -1 (XXII) chain (XP_015138660.1) | 1.13 | 0.04 | 27:1 | 0.039 |
| Transferrin Receptor (XP_015146868.1) | 0.1 | 0.01 | 12:1 | 0.005 |
| C-reactive protein, pentraxin-related precursor (NP_001300649.1) | 2.13 | 0.43 | 4.9:1 | 0.014 |
| Protein Name (Accession#) | VEX (NI) 1 | TEX (NI) 1 | Ratio TEX/VEX | p-Value |
|---|---|---|---|---|
| Histone H4 (XP_425458.2) | 0 | 9.94 | 44,448:1 | 0.015 |
| -2-antiplasmin isoform X2 (XP_015151528.1) | 0 | 4.36 | 19,487:1 | 0.014 |
| HSP90-α (NP_001034377.1) | 0 | 3.12 | 13,971:1 | 0.029 |
| Pantetheinase isoform X1 (XP_015139593.1) | 0 | 2.77 | 12,388:1 | 0.005 |
| Peroxiredoxin-1 (NP_001258861.1) | 0 | 2.46 | 11,102:1 | 0.043 |
| Deleted in malignant brain tumors 1 protein-like isoform X2 (DMBTL) (XP_015156078.1) | 0 | 11.8 | 52,741:1 | 0.048 |
| Ceruloplasmin (XP_015147339.1) | 0 | 1.85 | 8265:1 | 0.014 |
| Pantetheinase isoform X3 (XP_015139595.1) | 0 | 1.51 | 6734:1 | 0.013 |
| Clusterin precursor (NP_990231.1) | 0 | 1.3 | 5815:1 | 0.013 |
| Mannose-binding lectin precursor (NP_989680.2) | 0 | 1.23 | 5515:1 | 0.01 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neerukonda, S.N.; Tavlarides-Hontz, P.; McCarthy, F.; Pendarvis, K.; Parcells, M.S. Comparison of the Transcriptomes and Proteomes of Serum Exosomes from Marek’s Disease Virus-Vaccinated and Protected and Lymphoma-Bearing Chickens. Genes 2019, 10, 116. https://doi.org/10.3390/genes10020116
Neerukonda SN, Tavlarides-Hontz P, McCarthy F, Pendarvis K, Parcells MS. Comparison of the Transcriptomes and Proteomes of Serum Exosomes from Marek’s Disease Virus-Vaccinated and Protected and Lymphoma-Bearing Chickens. Genes. 2019; 10(2):116. https://doi.org/10.3390/genes10020116
Chicago/Turabian StyleNeerukonda, Sabari Nath, Phaedra Tavlarides-Hontz, Fiona McCarthy, Kenneth Pendarvis, and Mark S. Parcells. 2019. "Comparison of the Transcriptomes and Proteomes of Serum Exosomes from Marek’s Disease Virus-Vaccinated and Protected and Lymphoma-Bearing Chickens" Genes 10, no. 2: 116. https://doi.org/10.3390/genes10020116