Genome-Wide Distribution, Expression and Function Analysis of the U-Box Gene Family in Brassica oleracea L.
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatment
2.2. Retrieval and Identification of U-Box Family Genes in B. oleracea
2.3. Phylogenetic, Conservative Motif, and Structure Analysis
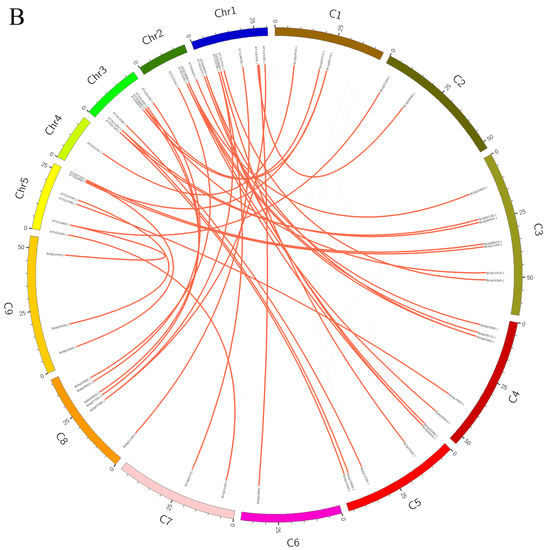
2.4. Chromosome Localization, Genome Collinearity, and Ka/Ks Ratio Analysis


2.5. Expression Analysis of U-Box Gene Family and RT-qPCR
2.6. Promoter Region Analysis of U-Box Differentially Expressed Genes
2.7. Interaction Network of B. oleracea U-Box Protein
2.8. Gene Ontology Annotation and KEGG Pathway Enrichment of B. oleracea U-Box Gene
3. Results and Discussion
3.1. Identification, Classification, and Phylogenetic Analysis of U-Box Gene Family
3.2. Collinear Analysis within the Genome of B. oleracea and between the Genomes of B. oleracea and A. thaliana
3.3. Phylogenetic, Conservative Motif, and Structural Analysis of the U-Box Gene Family in B. oleracea
3.4. Chromosome Localization of B. oleracea U-Box Gene
3.5. Interaction Network of U-Box Protein
3.6. Gene Ontology Annotation and KEGG Pathway Enrichment Analysis of U-Box genes in B. oleracea
3.7. Expression Analysis and RT-qPCR
3.8. Cis-Regulatory Element Analysis in the Promoter Regions of U-Box Differentially Expressed Genes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Haring, V.; Gray, J.E.; McClure, B.A.; Anderson, M.A.; Clarke, A.E. Self-incompatibility: A self-recognition system in plants. Science 1990, 250, 937–941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, T.; Mazzurco, M.; Sulaman, W.; Matias, D.D.; Goring, D.R. Binding of an arm repeat protein to the kinase domain of the S-locus receptor kinase. Proc. Natl. Acad. Sci. USA 1998, 95, 382–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deshaies, R.J.; Joazeiro, C.A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009, 78, 399–434. [Google Scholar] [CrossRef] [PubMed]
- Liquan, Z.; Yan, Z. Brassica oleracea L. self-incompatibility signal transduction element and conduction process. Crop J. 2015, 41, 1–14. [Google Scholar]
- Hatzfeld, M. The armadillo family of structure proteins. Int. Rev. Cytol. 1998, 186, 179–224. [Google Scholar]
- Trujillo, M. News from the PUB: Plant U-box type E3 ubiquitin ligases. J. Exp. Bot. 2018, 69, 371–384. [Google Scholar] [CrossRef] [Green Version]
- Wiborg, J.; O’Shea, C.; Skriver, K. Biochemical function of typical and variant Arabidopsis thaliana U-box E3 ubiquitin-protein ligases. Biochem. J. 2008, 413, 447–457. [Google Scholar] [CrossRef] [Green Version]
- Zeng, L.R.; Park, C.H.; Venu, R.C.; Gough, J.; Wang, G.L. Classification, expression pattern, and E3 ligase activity assay of rice U-box-containing proteins. Mol. Plant 2008, 1, 800–815. [Google Scholar] [CrossRef] [Green Version]
- Hu, H.; Dong, C.; Sun, D.; Hu, Y.; Xie, J. Genome-wide identification and analysis of U-Box E3 ubiquitin-protein ligase gene family in banana. Int. J. Mol. Sci. 2018, 19, 3874. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Duan, W.; Riquicho, A.R.; Jing, Z.; Liu, T.; Hou, X.; Li, Y. Genome-wide survey and expression analysis of the PUB family in Chinese cabbage (Brassica rapa ssp. pekinesis). Mol. Genet. Genom. 2015, 290, 2241–2260. [Google Scholar] [CrossRef]
- Ma, X.; Mo, B.; Cao, X. New players in ABA signaling: Identification of PUB12/13 involved in degradation of ABA co-receptor ABI1. Sci. China Life Sci. 2015, 58, 1173–1174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, L.; Cheng, J.; Zhu, Y.; Ding, Y.; Meng, J.; Chen, Z.; Xie, Q.; Guo, Y.; Li, J.; Yang, S.; et al. Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat. Commun. 2015, 6, 8630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.W.; González-Lamothe, R.; Ewan, R.A.; Rowland, O.; Yoshioka, H.; Shenton, M.; Ye, H.; O’Donnell, E.; Jones, J.D.; Sadanandom, A. The E3 ubiquitin ligase activity of Arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell 2006, 18, 1084–1098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Qu, B.; Dou, S.; Li, L.; Yin, D.; Pang, Z.; Zhou, Z.; Tian, M.; Liu, G.; Xie, Q.; et al. The E3 ligase OsPUB15 interacts with the receptor-like kinase PID2 and regulates plant cell death and innate immunity. BMC Plant Biol. 2015, 15, 49. [Google Scholar] [CrossRef] [Green Version]
- Adler, G.; Mishra, A.K.; Maymon, T.; Raveh, D.; Bar-Zvi, D. Overexpression of Arabidopsis ubiquitin ligase AtPUB46 enhances tolerance to drought and oxidative stress. Plant Sci. 2018, 276, 220–228. [Google Scholar] [CrossRef]
- Peng, L.; Wan, X.; Huang, K.; Pei, L.; Xiong, J.; Li, X.; Wang, J. AtPUB48 E3 ligase plays a crucial role in the thermotolerance of Arabidopsis. Biochem. Biophys. Res. Commun. 2019, 509, 281–286. [Google Scholar] [CrossRef]
- Min, H.J.; Cui, L.H.; Oh, T.R.; Kim, J.H.; Kim, T.W.; Kim, W.T. OsBZR1 turnover mediated by OsSK22-regulated U-box E3 ligase OsPUB24 in rice BR response. Plant J. 2019, 99, 426–438. [Google Scholar] [CrossRef]
- Samuel, M.A.; Mudgil, Y.; Salt, J.N.; Delmas, F.; Ramachandran, S.; Chilelli, A.; Goring, D.R. Interactions between the S-domain receptor kinases and AtPUB-ARM E3 ubiquitin ligases suggest a conserved signaling pathway in Arabidopsis. Plant Physiol. 2008, 147, 2084–2095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yiping, W.; Yingying, W.; Boying, Y.; Zhao, Y.; Yiji, X. EXTRA-LARGE G PROTEINs interact with E3 ligases PUB4 and PUB2 and function in cytokinin and developmental processes. Plant Physiol. 2017, 173, 1235–1246. [Google Scholar]
- Wheeler, T.J.; Eddy, S.R. NHMMER: DNA homology search with profile HMMs. Bioinformatics 2013, 29, 2487–2489. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, 222–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Xia, R.; Chen, H. TBtools: A toolkit for biologists integrating various HTS-data handling tools with a user-friendly interface. BioRxiv 2018, 289660. [Google Scholar] [CrossRef]
- Gu, Z.; Cavalcanti, A.; Chen, F.C.; Bouman, P.; Li, W.H. Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol. Biol. Evol. 2002, 19, 256–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.P.; Wan, H.L.; Zhang, S.; Yu, J. Gamma-MYN: A new algorithm for estimating Ka and Ks with consideration of variable substitution rates. Biol. Direct 2009, 4, 20. [Google Scholar] [CrossRef] [Green Version]
- Hero, A.O.; Fleury, G.; Mears, A.J.; Swaroop, A. Multicriteria gene screening for analysis of differential expression with DNA microarrays. EURASIP J. Adv. Signal Process. 2004, 2004, 43–52. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, 106. [Google Scholar] [CrossRef] [Green Version]
- Qu, C.M.; Fu, F.Y. Differential accumulation of phenolic compounds and expression of related genes in black-and yellow-seeded Brassica napus. J. Exp. Bot. 2013, 64, 2885–2898. [Google Scholar] [CrossRef] [Green Version]
- Livaka, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Ai, C.; Kong, L. CGPS: A machine learning-based approach integrating multiple gene set analysis tools for better prioritization of biologically relevant pathways. J. Genet. Genom. 2018, 45, 489–504. [Google Scholar] [CrossRef]
- Zheng, N.; Shabek, N. Ubiquitin ligases: Structure, function, and regulation. Annu. Rev. Biochem. 2017, 86, 129–157. [Google Scholar] [CrossRef]
- Hertel, J.; Lindemeyer, M.; Missal, K.; Fried, C.; Tanzer, A.; Flamm, C.; Hofacker, I.L.; Stadler, P.F. The expansion of the metazoan microRNA repertoire. BMC Genom. 2006, 7, 25. [Google Scholar] [CrossRef] [Green Version]
- Freeling, M. Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 2009, 60, 433–453. [Google Scholar] [CrossRef]
- Wagner, A. Gene duplications, robustness and evolutionary innovations. BioEssays 2018, 30, 367–373. [Google Scholar] [CrossRef] [Green Version]
- Ren, R.; Wang, H.; Guo, C.; Zhang, N.; Zeng, L.; Chen, Y.; Ma, H.; Qi, J. Widespread whole genome duplications contribute to genome complexity and species diversity in angiosperms. Mol. Plant 2018, 11, 414–428. [Google Scholar] [CrossRef] [Green Version]
- Zhi, Z.; Jianghua, Y. Genome-wide comparison reveals divergence of cassava and rubber aquaporin family genes after the recent whole-genome duplication. BMC Genom. 2019, 20, 380. [Google Scholar]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Zhang, Z.; Vang, S.; Yu, J.; Wong, G.K.; Wang, J. Correlation between Ka/Ks and Ks is related to substitution model and evolutionary lineage. J. Mol. Evol. 2009, 68, 414–423. [Google Scholar] [CrossRef]
- De Bodt, S.; Maere, S.; Van de Peer, Y. Genome duplication and the origin of angiosperms. Trends Ecol. Evol. 2005, 20, 591–597. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.F.; Gaitatzes, C.; Saxena, K.; Neer, E.J. The WD repeat: A common architecture for diverse functions. Trends Biochem. Sci. 1999, 24, 181–185. [Google Scholar] [CrossRef]
- Li, D.; Roberts, R. WD-repeat proteins: Structure characteristics, biological function, and their involvement in human diseases. Cell Mol. Life Sci. 2001, 58, 2085–2097. [Google Scholar] [CrossRef] [PubMed]
- Aravind, L.; Koonin, E.V. The U box is a modified RING finger—A common domain in ubiquitination. Curr. Biol. 2000, 10, 132–134. [Google Scholar] [CrossRef] [Green Version]
- Andersen, P.; Kragelund, B.B.; Olsen, A.N.; Larsen, F.H.; Chua, N.H.; Poulsen, F.M.; Skriver, K. Structure and biochemical function of a prototypical Arabidopsis U-box domain. J. Biol. Chem. 2004, 279, 40053–40061. [Google Scholar] [CrossRef] [Green Version]
- Neer, E.J.; Schmidt, C.J.; Nambudripad, R.; Smith, T.F. The ancient regulatory-protein family of WD-repeat proteins. Nature 1994, 371, 297–300. [Google Scholar] [CrossRef]
- Stirnimann, C.U.; Petsalaki, E.; Russell, R.B.; Müller, C.W. WD40 proteins propel cellular networks. Trends Biochem. Sci. 2010, 35, 565–574. [Google Scholar] [CrossRef]
- Manning, G.; Plowman, G.D.; Hunter, T.; Sudarsanam, S. Evolution of protein kinase signaling from yeast to man. Trends Biochem. Sci. 2002, 27, 514–520. [Google Scholar] [CrossRef]
- Knighton, D.R.; Zheng, J.H.; Ten Eyck, L.F.; Ashford, V.A.; Xuong, N.H.; Taylor, S.S.; Sowadski, J.M. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 1991, 253, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Hanks, S.K.; Hunter, T. Protein kinases 6. The eukaryotic protein kinase superfamily: Kinase (catalytic) domain structure and classification. FASEB J. 1995, 9, 576–596. [Google Scholar] [CrossRef] [PubMed]
- Peifer, M.; Berg, S.; Reynolds, A.B. A repeating amino acid motif shared by proteins with diverse cellular roles. Cell 1994, 76, 789–791. [Google Scholar] [CrossRef]
- Groves, M.R.; Barford, D. Topological characteristics of helical repeat proteins. Curr. Opin. Struct. Biol. 1999, 9, 383–389. [Google Scholar] [CrossRef]
- Stone, S.L.; Anderson, E.M.; Mullen, R.T.; Goring, D.R. ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell 2003, 15, 885–898. [Google Scholar] [CrossRef] [Green Version]
- Mudgil, Y.; Shiu, S.H.; Stone, S.L.; Salt, J.N.; Goring, D.R. A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiol. 2004, 134, 59–66. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Minaker, S.; Roth, C.; Huang, S.; Hieter, P.; Lipka, V.; Wiermer, M.; Li, X. An E4 ligase facilitates polyubiquitination of plant immune receptor resistance proteins in Arabidopsis. Plant Cell 2014, 26, 485–496. [Google Scholar] [CrossRef] [Green Version]
- Tu, D.; Li, W.; Ye, Y.; Brunger, A.T. Structure and function of the yeast U-box-containing ubiquitin ligase Ufd2p. Proc. Natl. Acad. Sci. USA 2007, 104, 15599–15606. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Windheim, M.; Roe, S.M.; Peggie, M.; Cohen, P.; Prodromou, C.; Pearl, L.H. Chaperoned ubiquitylation—crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol. Cell 2005, 20, 525–538. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Shen, G.; Yan, J.; He, C.; Zhang, H. AtCHIP functions as an E3 ubiquitin ligase of protein phosphatase 2A subunits and alters plant response to abscisic acid treatment. Plant J. 2006, 46, 649–657. [Google Scholar] [CrossRef]
- Yan, J.; Wang, J.; Li, Q.; Hwang, J.R.; Patterson, C.; Zhang, H. AtCHIP, a U-box-containing E3 ubiquitin ligase, plays a critical role in temperature stress tolerance in Arabidopsis. Plant Physiol. 2003, 132, 861–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, S.K.; Ryu, M.Y.; Song, C.; Kwak, J.M.; Kim, W.T. Arabidopsis PUB22 and PUB23 are homologous U-Box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant Cell 2008, 20, 1899–1914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, D.H.; Ahn, M.Y.; Park, K.Y.; Kim, E.Y.; Kim, W.T. The N-Terminal UND motif of the Arabidopsis U-Box E3 ligase PUB18 is critical for the negative regulation of ABA-mediated stomatal movement and determines its ubiquitination specificity for exocyst subunit Exo70B1. Plant Cell 2016, 28, 2952–2973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Florea, L.; Song, L.; Salzberg, S.L. Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Research 2013, 2, 188. [Google Scholar] [CrossRef]
- Osakabe, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S. ABA control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytol. 2014, 202, 35–49. [Google Scholar] [CrossRef]
- Simpson, S.D.; Nakashima, K.; Narusaka, Y.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Two different novel cis-acting elements of erd1, a clpA homologous Arabidopsis gene function in induction by dehydration stress and dark-induced senescence. Plant J. 2003, 33, 259–270. [Google Scholar] [CrossRef]
- Abe, H.; Yamaguchi-Shinozaki, K.; Urao, T.; Iwasaki, T.; Hosokawa, D.; Shinozaki, K. Role of MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 1997, 9, 1859–1868. [Google Scholar]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef] [Green Version]
- Bate, N.; Twell, D. Functional architecture of a late pollen promoter: Pollen-specific transcription is developmentally regulated by multiple stage-specific and co-dependent activator elements. Plant Mol. Biol. 1998, 37, 859–869. [Google Scholar] [CrossRef]
- Filichkin, S.A.; Leonard, J.M.; Monteros, A.; Liu, P.P.; Nonogaki, H. A novel endo-β-mannanase gene in tomato LeMAN5 is associated with anther and pollen development. Plant Physiol. 2004, 134, 1080–1087. [Google Scholar] [CrossRef] [Green Version]
- Dong, J.; Chen, C.; Chen, Z. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 2003, 51, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Chunhong, C.; Zhixiang, C. Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol. 2002, 129, 706–716. [Google Scholar]







| Group | B. oleracea Gene | BoPUB ID | Chr | Genomic Location | AtPUB ID | A. thaliana Gene | E-Value | Identity (%) | Transcriptome ID | E-Value | Identity (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| I | Bol019224 | BoPUB1 | C09 | 4,934,468–4,937,484 | AtPUB2 | AT5G67340.1 | 0 | 84.53% | Bo9g020280.1 | 0 | 98.56% |
| Bol014311 | BoPUB2 | Scaffold000175_P1 | 442,705–445,437 | AtPUB2 | AT5G67340.1 | 0 | 78.79% | Bo7g063700.1 | 0 | 99.19% | |
| Bol044176 | BoPUB3 | C04 | 24,020,183–24,022,584 | AtPUB3 | AT3G54790.1 | 0 | 90.02% | Bo4g118390.1 | 7.97 × 10−133 | 100.00% | |
| Bol008980 | BoPUB4 | Scaffold000240 | 145,239–147,580 | AtPUB3 | AT3G54790.1 | 0 | 88.12% | Bo8g085910.1 | 0 | 100.00% | |
| Bol045811 | BoPUB5 | C08 | 34,875,756–34,878,494 | AtPUB4 | AT2G23140.1 | 0 | 86.94% | Bo8g100610.1 | 0 | 100.00% | |
| Bol031706 | BoPUB6 | Scaffold000053 | 637,493–643,779 | AtPUB4 | AT2G23140.1 | 0 | 87.92% | Bo4g151360.1 | 2.58 × 10−108 | 99.64% | |
| Bol028412 | BoPUB7 | C01 | 7,866,975–7,868,078 | AtPUB8 | AT4G21350.1 | 2 x 10−149 | 78.81% | Bo1g024100.1 | 0 | 98.11% | |
| Bol008152 | BoPUB8 | C01 | 38,471,230–38,472,813 | AtPUB9 | AT3G07360.1 | 0 | 84.52% | Bo1g147370.1 | 0 | 100.00% | |
| Bol007667 | BoPUB9 | C05 | 32,165,369–32,167,119 | AtPUB9 | AT3G07360.1 | 0 | 85.00% | Bo5g141600.1 | 0 | 100.00% | |
| Bol033996 | BoPUB10 | Scaffold000040 | 700,223–702,053 | AtPUB9 | AT3G07360.1 | 0 | 85.54% | Bo3g061180.1 | 0 | 100.00% | |
| Bol016165 | BoPUB11 | C07 | 4,511,321–4,514,149 | AtPUB10 | AT1G71020.1 | 0 | 80.22% | Bo6g112660.1 | 0 | 98.50% | |
| Bol012315 | BoPUB12 | C06 | 23,724,580–23,726,780 | AtPUB11 | AT1G23030.1 | 0 | 84.00% | Bo7g056480.1 | 0 | 97.94% | |
| Bol023511 | BoPUB13 | Scaffold000099_P1 | 524,243–526,409 | AtPUB11 | AT1G23030.1 | 0 | 86.88% | Bo5g039200.1 | 0 | 99.73% | |
| Bol033248 | BoPUB14 | C04 | 32,033,767–32,036,354 | AtPUB12 | AT2G28830.1 | 0 | 81.45% | Bo4g166850.1 | 0 | 100.00% | |
| Bol018774 | BoPUB15 | C01 | 15,040,202–15,042,653 | AtPUB13 | AT3G46510.1 | 0 | 88.01% | Bo1g070710.1 | 0 | 100.00% | |
| Bol041392 | BoPUB16 | C03 | 35,763,128–35,765,315 | AtPUB13 | AT3G46510.1 | 0 | 86.17% | Bo3g131520.1 | 0 | 100.00% | |
| Bol045384 | BoPUB17 | C08 | 29,129,169–29,134,639 | AtPUB14 | AT3G54850.1 | 0 | 85.61% | Bo8g087100.1 | 0 | 98.26% | |
| Bol035302 | BoPUB18 | Scaffold000035_P2 | 510,161–512,658 | AtPUB15 | AT5G42340.1 | 0 | 85.12% | Bo9g058200.1 | 0 | 99.39% | |
| Bol000236 | BoPUB19 | Scaffold000960 | 766–2784 | AtPUB16 | AT5G01830.1 | 0 | 81.20% | Bo7g016300.1 | 0 | 98.96% | |
| Bol008579 | BoPUB20 | C03 | 44,740,354–44,742,513 | AtPUB17 | AT1G29340.1 | 0 | 81.53% | Bo3g144000.1 | 0 | 99.31% | |
| Bol037877 | BoPUB21 | C04 | 35,662,808–35,664,799 | AtPUB17 | AT1G29340.1 | 0 | 75.45% | Bo4g182450.1 | 0 | 99.00% | |
| Bol041877 | BoPUB22 | C06 | 17,530,961–17,533,096 | AtPUB17 | AT1G29340.1 | 0 | 80.70% | Bo7g045530.1 | 0 | 99.63% | |
| Bol001099 | BoPUB23 | Scaffold000469 | 39,061–41,190 | AtPUB17 | AT1G29340.1 | 0 | 76.53% | Bo5g058680.1 | 0 | 99.62% | |
| Bol036718 | BoPUB24 | C05 | 25,576,732–25,578,822 | AtPUB18 | AT1G10560.1 | 0 | 81.99% | Bo5g012020.1 | 0 | 100.00% | |
| Bol036591 | BoPUB25 | C01 | 22,029,097–22,031,136 | AtPUB19 | AT1G60190.1 | 0 | 79.36% | Bo1g091760.1 | 0 | 98.77% | |
| Bol008036 | BoPUB26 | C02 | 41,702,893–41,704,551 | AtPUB38 | AT5G65200.1 | 0 | 78.67% | Bo00615s220.1 | 0 | 98.19% | |
| Bol024918 | BoPUB27 | C03 | 39,891,535–39,893,145 | AtPUB39 | AT3G47820.1 | 0 | 84.62% | Bo3g135910.1 | 0 | 100.00% | |
| Bol012875 | BoPUB28 | Scaffold000192 | 121,054–122,718 | AtPUB40 | AT5G40140.1 | 0 | 82.37% | Bo4g139680.1 | 0 | 97.12% | |
| Bol020697 | BoPUB29 | Scaffold000121_P2 | 233,911–235,575 | AtPUB41 | AT5G62560.1 | 0 | 80.55% | Bo2g164280.1 | 0 | 98.69% | |
| Bol019195 | BoPUB30 | Scaffold000133 | 1,178,856–1,180,623 | AtPUB41 | AT5G62560.1 | 0 | 81.64% | Bo9g015020.1 | 0 | 99.46% | |
| Bol034489 | BoPUB31 | C03 | 3,652,498–3,654,286 | AtPUB46 | AT5G18320.1 | 4 × 10−131 | 82.50% | Bo3g013070.1 | 0 | 98.23% | |
| Bol034491 | BoPUB32 | C03 | 3,671,228–3,673,202 | AtPUB46 | AT5G18320.1 | 9 x 10−131 | 78.61% | Bo3g013070.2 | 0 | 89.87% | |
| Bol019726 | BoPUB33 | C09 | 31,567,100–31,568,692 | AtPUB46 | AT5G18320.1 | 1 × 10−151 | 78.28 | Bo9g157270.1 | 0 | 100.00% | |
| II | Bol037390 | BoPUB34 | C08 | 24,303,890–24,311,884 | AtPUB32 | AT3G49060.1 | 0 | 90.48% | Bo8g077500.2 | 3.98 × 10−98 | 99.50% |
| Bol003480 | BoPUB35 | Scaffold000349 | 333,337–337,418 | AtPUB32 | AT3G49060.1 | 0 | 87.64% | Bo8g077500.1 | 0 | 89.62% | |
| Bol030222 | BoPUB36 | C04 | 3,124,409–3,127,799 | AtPUB33 | AT2G45910.1 | 0 | 86.31% | Bo4g025080.1 | 0 | 100.00% | |
| Bol021760 | BoPUB37 | C04 | 40,486,202–40,489,850 | AtPUB33 | AT2G45910.1 | 0 | 86.82% | Bo4g195910.1 | 0 | 98.80% | |
| Bol022718 | BoPUB38 | C06 | 7,979,725–7,989,744 | AtPUB34 | AT2G19410.1 | 0 | 85.09% | Bo7g005960.1 | 0 | 99.47% | |
| Bol003293 | BoPUB39 | Scaffold000357 | 194,515–201,007 | AtPUB34 | AT2G19410.1 | 4 × 10−130 | 79.34% | Bo7g062400.1 | 7.53 × 10−107 | 100.00% | |
| Bol039498 | BoPUB40 | C01 | 11,366,737–11,368,105 | AtPUB35 | AT4G25160.1 | 8 × 10−172 | 87.19% | Bo1g039750.1 | 0 | 100.00% | |
| Bol042214 | BoPUB41 | C06 | 43,741,380–43,743,553 | AtPUB35 | AT4G25160.1 | 5 × 10−92 | 97.06% | Bo7g110030.1 | 0 | 98.74% | |
| Bol045651 | BoPUB42 | C08 | 32,648,599–32,650,459 | AtPUB36 | AT3G61390.1 | 1 × 10−134 | 94.12% | Bo8g095910.1 | 3.16 × 10−35 | 100.00% | |
| Bol037509 | BoPUB43 | Scaffold000024 | 1,052,543–1,056,193 | AtPUB36 | AT3G61390.1 | 4 × 10−92 | 91.09% | Bo4g101400.1 | 0 | 99.88% | |
| Bol003429 | BoPUB44 | Scaffold000351 | 326,763–329,071 | AtPUB36 | AT3G61390.2 | 4.9 × 10−324 | 81.41% | Bo6g077670.1 | 0 | 99.87% | |
| Bol009241 | BoPUB45 | C03 | 42,826,034–42,829,000 | AtPUB50 | AT5G65500.2 | 0 | 85.15% | Bo3g099120.1 | 0 | 98.79% | |
| Bol019041 | BoPUB46 | Scaffold000133 | 89,160–92,092 | AtPUB50 | AT5G65500.1 | 0 | 85.44% | Bo9g018750.1 | 4.65 × 10−26 | 99.41% | |
| Bol017004 | BoPUB47 | C06 | 37,382,487–37,385,659 | AtPUB51 | AT5G61560.1 | 0 | 87.61% | Bo7g098800.1 | 0 | 99.75% | |
| Bol007790 | BoPUB48 | Scaffold000256 | 377,456–380,990 | AtPUB51 | AT5G61560.1 | 2 × 10−144 | 88.98% | Bo2g161490.1 | 0 | 99.35% | |
| Bol029355 | BoPUB49 | C09 | 17,336,879–17,341,604 | AtPUB53 | AT5G51270.2 | 1 × 10−93 | 80.92% | Bo2g050530.1 | 6.05 × 10−103 | 83.76% | |
| Bol045287 | BoPUB50 | Scaffold000001_P2 | 1,247,247–1,250,408 | AtPUB53 | AT5G51270.1 | 5 × 10−137 | 80.33% | Bo2g050530.2 | 0 | 100.00% | |
| Bol029573 | BoPUB51 | C03 | 13,545,754–13,549,222 | AtPUB54 | AT1G01680.1 | 3 × 10−38 | 81.06% | Bo3g039080.1 | 0 | 99.48% | |
| Bol040674 | BoPUB52 | C05 | 39,733–41,926 | AtPUB54 | AT1G01680.1 | 0 | 82.33% | Bo5g001070.1 | 5.90 × 10−116 | 98.72% | |
| Bol040676 | BoPUB53 | C05 | 45,765–48,401 | AtPUB55 | AT1G01660.1 | 1 × 10−131 | 89.67% | Bo5g001050.1 | 1.02 × 10−93 | 100.00% | |
| Bol013021 | BoPUB54 | C03 | 54,964,336–54,966,942 | AtPUB56 | AT1G01670.1 | 2 × 10−99 | 82.13% | Bo3g175980.2 | 0 | 99.06% | |
| Bol040675 | BoPUB55 | C05 | 43,012–45,429 | AtPUB56 | AT1G01670.1 | 0 | 79.74% | Bo5g001060.1 | 0 | 98.39% | |
| Bol000206 | BoPUB56 | Scaffold001134 | 1549–4155 | AtPUB56 | AT1G01670.1 | 2 × 10−99 | 76.52% | Bo3g175980.1 | 0 | 99.06% | |
| Bol013313 | BoPUB57 | C07 | 19,000,520–19,002,951 | AtPUB57 | AT1G56030.1 | 9 × 10−146 | 77.54% | Bo6g052570.1 | 0 | 99.54% | |
| Bol037990 | BoPUB58 | C05 | 4,401,778–4,406,478 | - | AT5G57035.1 | 4 × 10−167 | 89.16% | Bo9g130630.1 | 0 | 99.79% | |
| III | Bol021972 | BoPUB59 | C01 | 18,150,442–18,151,785 | AtPUB3 | AT3G49810.1 | 0 | 87.31% | Bo1g081270.1 | 0 | 100.00% |
| Bol009126 | BoPUB60 | C04 | 8,207,638–8,208,912 | AtPUB22 | AT3G52450.1 | 0 | 89.63% | Bo4g125390.1 | 0 | 100.00% | |
| Bol005035 | BoPUB61 | C06 | 38,049,040–38,050,311 | AtPUB22 | AT3G52450.1 | 0 | 87.78% | Bo7g101510.1 | 0 | 100.00% | |
| Bol025043 | BoPUB62 | C08 | 27,739,954–27,741,261 | AtPUB22 | AT3G52450.1 | 0 | 89.14% | Bo8g080650.1 | 0 | 99.92% | |
| Bol039769 | BoPUB63 | C03 | 8,905,416–8,906,642 | AtPUB23 | AT2G35930.1 | 0 | 88.13% | Bo3g029820.1 | 0 | 99.02% | |
| Bol011695 | BoPUB64 | C04 | 3,917,811–3,919,031 | AtPUB23 | AT2G35930.1 | 0 | 88.69% | Bo4g036170.1 | 0 | 99.10% | |
| Bol037760 | BoPUB65 | C04 | 36,502,645–36,503,883 | AtPUB23 | AT2G35930.1 | 0 | 89.55% | Bo4g185810.1 | 0 | 99.76% | |
| Bol007270 | BoPUB66 | C01 | 36,294,678–36,296,310 | AtPUB24 | AT3G11840.1 | 0 | 82.21% | Bo1g139780.1 | 0 | 97.57% | |
| Bol010350 | BoPUB67 | C05 | 30,543,476–30,545,255 | AtPUB24 | AT3G11840.1 | 0 | 83.26% | Bo5g135090.1 | 0 | 98.08% | |
| Bol003032 | BoPUB68 | Scaffold000366 | 135,192–137,357 | AtPUB24 | AT3G11840.1 | 0 | 78.76% | Bo3g064620.1 | 0 | 98.21% | |
| Bol031005 | BoPUB69 | C01 | 30,733,021–30,734,274 | AtPUB25 | AT3G19380.1 | 0 | 82.42% | Bo1g114760.1 | 0 | 100.00% | |
| Bol018091 | BoPUB70 | C05 | 23,579,947–23,581,212 | AtPUB25 | AT3G19380.1 | 0 | 82.11% | Bo5g115230.1 | 0 | 98.18% | |
| Bol023451 | BoPUB71 | C08 | 927,641–928,912 | AtPUB26 | AT1G49780.1 | 0 | 85.99% | Bo8g011300.1 | 0 | 99.61% | |
| Bol015885 | BoPUB72 | C03 | 30,604,113–30,605,372 | AtPUB27 | AT5G64660.1 | 0 | 86.47% | Bo3g101050.1 | 0 | 99.68% | |
| Bol019081 | - | Scaffold000133 | 386,596–386,904 | AtPUB27 | AT5G64660.1 | 6 × 10−25 | 81.32% | Bo9g018240.1 | 1.44 × 10−122 | 99.58% | |
| Bol030961 | BoPUB73 | C01 | 31,316,399–31,317,652 | AtPUB29 | AT3G18710.1 | 7 × 10−144 | 79,67% | Bo1g116350.1 | 0 | 100.00% | |
| Bol022877 | BoPUB74 | C03 | 23,114,355–23,115,629 | AtPUB29 | AT3G18710.1 | 6 × 10−151 | 82.50% | Bo3g071710.1 | 0 | 99.29% | |
| Bol002996 | BoPUB75 | Scaffold000367 | 136,856–138,127 | AtPUB29 | AT3G18710.1 | 3 × 10−121 | 81.29% | Bo5g117170.1 | 0 | 99.29% | |
| Bol005722 | BoPUB76 | C08 | 82,316–83,647 | AtPUB31 | AT5G65920.1 | 0 | 80.37% | Bo9g018990.1 | 0 | 99.92% | |
| IV | Bol028991 | BoPUB77 | C01 | 1,265,612–1,268,155 | AtPUB5 | AT4G36550.1 | 0 | 84.71% | Bo1g005580.1 | 6.09 × 10−15 | 100.00% |
| Bol018658 | - | C06 | 47,377,417–47,379,131 | AtPUB5 | AT4G36550.1 | 8 × 10−75 | 82.40% | Bo7g118020.1 | 0 | 98.65% | |
| Bol016339 | BoPUB78 | Scaffold000156_P2 | 219,216–221,685 | AtPUB5 | AT4G36550.1 | 0 | 82.34% | Bo3g154110.1 | 0 | 98.77% | |
| Bol016347 | BoPUB79 | Scaffold000156_P2 | 281,312–283,639 | AtPUB5 | AT4G36550.1 | 0 | 78.78% | Bo3g154230.1 | 0 | 99.86% | |
| Bol005289 | BoPUB80 | C03 | 43,163,200–43,166,241 | AtPUB6 | AT1G24330.1 | 0 | 85.35% | Bo3g141280.1 | 0 | 100.00% | |
| Bol041756 | BoPUB81 | C06 | 16,043,256–16,046,166 | AtPUB6 | AT1G24330.1 | 0 | 85.86% | Bo7g050100.1 | 7.04 × 10−54 | 99.77% | |
| Bol010460 | BoPUB82 | C02 | 11,448,388–11,451,770 | AtPUB7 | AT1G67530.1 | 0 | 87.04% | Bo2g055940.1 | 0 | 99.32% | |
| Bol008441 | BoPUB83 | C03 | 43,797,474–43,800,768 | AtPUB45 | AT1G27910.1 | 0 | 86.75% | Bo3g142960.1 | 0 | 99.48% | |
| Bol002093 | BoPUB84 | Scaffold000404 | 92,644–95,688 | AtPUB45 | AT1G27910.1 | 0 | 89.30% | Bo5g052370.1 | 0 | 100.00% | |
| V | Bol023972 | BoPUB85 | C07 | 7,790,036–7,793,449 | AtPUB42 | AT1G68940.1 | 0 | 88.81% | Bo6g108490.1 | 6.60 × 10−26 | 99.47% |
| Bol027639 | BoPUB86 | C07 | 1,710,724–1,713,541 | AtPUB43 | AT1G76390.1 | 0 | 83.79% | Bo8g070380.1 | 0 | 98.13% | |
| Bol007008 | BoPUB87 | C08 | 21,666,213–21,669,713 | AtPUB44 | AT1G20780.1 | 0 | 76.65% | Bo8g070380.2 | 0 | 98.13% | |
| Bol025462 | BoPUB88 | Scaffold000087_P1 | 203,738–207,119 | AtPUB44 | AT1G20780.1 | 0 | 84.14% | Bo7g061370.1 | 0 | 99.09% | |
| Bol011584 | - | C08 | 2,691,591–2,692,573 | - | AT1G04555.1 | 4 × 10−43 | 83.00% | Bo8g003030.1 | 0 | 99.33% | |
| VI | Bol007668 | BoPUB89 | C05 | 32,161,132–32,163,836 | AtCHIP | AT3G07370.1 | 1 × 10−140 | 88.48% | Bo5g141580.1 | 0 | 99.30% |
| Bol033995 | BoPUB90 | Scaffold000040 | 696,720–698,064 | AtCHIP | AT3G07370.1 | 0 | 86.73% | Bo3g061190.1 | 7.27 × 10−124 | 100.00% | |
| Bol040899 | BoPUB91 | C05 | 1,223,432–1,227,503 | MAC3A/59 | AT1G04510.1 | 0 | 91.68% | Bo5g004640.1 | 0 | 99.81% | |
| Bol027262 | BoPUB92 | C04 | 20,160,334–20,165,996 | MAC3B/60 | AT2G33340.1 | 0 | 90.63% | Bo4g043800.1 | 0 | 99.51% | |
| Bol037940 | BoPUB93 | C04 | 35,183,234–35,187,431 | MAC3B/60 | AT2G33340.1 | 0 | 91.66% | Bo4g181820.1 | 0 | 100.00% | |
| VII | Bol005348 | BoPUB94 | Scaffold000301 | 209,407–210,723 | AtPUB21 | AT5G37490.1 | 0 | 83.91% | Bo4g143870.1 | 0 | 98.98% |
| Bol004317 | BoPUB95 | Scaffold000326 | 51,300–53,990 | AtPUB49 | AT5G67530.1 | 0 | 90.74% | Bo3g096470.1 | 0 | 99.05% | |
| Bol044051 | BoPUB96 | C09 | 39,181,374–39,183,068 | AtPUB62 | AT5G05230.1 | 0 | 85.04% | Bo9g177290.1 | 0 | 98.44% | |
| Bol034488 | BoPUB97 | C03 | 3,645,792–3,651,503 | ATAPY2 | AT5G18280.1 | 0 | 92.86% | Bo3g013060.1 | 4.81 × 10−37 | 99.56% | |
| Bol045810 | - | C08 | 34,872,922–34,874,905 | ATNRAMP3 | AT2G23150.1 | 0 | 83.64% | Bo8g100600.1 | 0 | 100.00% | |
| Bol021231 | BoPUB98 | C02 | 3,528,313–3,533,241 | MUSE3/1 | AT5G15400.1 | 0 | 88.72% | Bo2g012540.1 | 0 | 99.17% | |
| Bol030454 | BoPUB99 | C09 | 33,424,858–33,429,958 | MUSE3/1 | AT5G15400.1 | 0 | 88.06% | Bo9g164140.1 | 0 | 99.17% |
| Motif | E-Value | Sites | Width | Best Possible Match |
|---|---|---|---|---|
| 1 | 4.6 × 10−1084 | 82 | 24 | ELMKDPVIVASGQTYERASIZKWL |
| 2 | 3.5 × 10−631 | 83 | 21 | LSHTTLTPNHTLRSLIQEWCE |
| 3 | 2.0 × 10−463 | 34 | 40 | IPPLVDLLENGTPRGKKDAATALFNLSIYQENKGRIVRAG |
| 4 | 2.4 × 10−413 | 91 | 11 | IPEEFRCPISL |
| 5 | 1.9 × 10−367 | 54 | 29 | MEARENAAATLFSLSVLEENKHKIGSSGA |
| 6 | 1.3 × 10−353 | 59 | 29 | GVVPSLVKJSQNGTERARRKAASLLKLLR |
| 7 | 3.2 × 10−248 | 64 | 11 | GHNTCPKTKQV |
| 8 | 1.1 × 10−223 | 58 | 21 | LLRSGSPRGKENAVAVLLQLC |
| 9 | 3.3 × 10−365 | 17 | 50 | LAKRNTDNRIVIAEAGAIPLLVNLLKSEDSATQENAVTALLNLSIYENNK |
| 10 | 1.1 × 10−178 | 12 | 32 | DLKSRKAIYVSREAPDCCHIWFTCKGYLIHTR |
| 11 | 6.9 × 10−203 | 28 | 31 | BSVEEAILELIYESGIKKLVMGAAADRHYSR |
| 12 | 4.2 × 10−195 | 11 | 50 | LQEMLQLGVVAKLCLVLQVSCGNKTKEKAKELLKLHARVWKESPCIPRNL |
| 13 | 2.1 × 10−182 | 14 | 41 | DEKIYVAVNKDVEESKSTLVWALRNLGGKKJCJLHVHQPIS |
| 14 | 2.4 × 10−176 | 9 | 50 | QCAEGRAEFLNHGAAIAVVTKKILRVSQIASDRAVRVLLSVGRFCATPAL |
| 15 | 1.4 × 10−168 | 49 | 21 | VEEALAVLANLASHPEGKEAI |
| NCBI-Gene ID: | Pathway ID | Description | Gene Name | Bo U-Box Transcriptome ID |
|---|---|---|---|---|
| hsa:27339 | hsa04120 | Ubiquitin mediated proteolysis | PRPF19, NMP200, PRP19, PSO4, SNEV, UBOX4, hPSO4 | Bo5g004640.1 Bo4g181820.1 Bo4g043800.1 |
| hsa03040 | Spliceosome | |||
| hsa:10277 | hsa04120 | Ubiquitin mediated proteolysis | UBE4B, E4, HDNB1, UBOX3, UFD2, UFD2A | Bo9g164140.1 Bo2g012540.1 |
| hsa04141 | Protein processing in endoplasmic reticulum | |||
| hsa:10273 | hsa04120 | Ubiquitin mediated proteolysis | STUB1, CHIP, HSPABP2, SCAR16, SDCCAG7, UBOX1 | Bo5g141580.1 Bo5g135090.1 Bo3g061190.1 |
| hsa04141 | Protein processing in endoplasmic reticulum | |||
| hsa:23759 | hsa04120 | Ubiquitin mediated proteolysis | PPIL2, CYC4, CYP60, Cyp-60, UBOX7, hCyP-60 | Bo3g096470.1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, D.; Xie, Q.; Liu, Q.; Zuo, T.; Zhang, H.; Zhang, Y.; Lian, X.; Zhu, L. Genome-Wide Distribution, Expression and Function Analysis of the U-Box Gene Family in Brassica oleracea L. Genes 2019, 10, 1000. https://doi.org/10.3390/genes10121000
Hu D, Xie Q, Liu Q, Zuo T, Zhang H, Zhang Y, Lian X, Zhu L. Genome-Wide Distribution, Expression and Function Analysis of the U-Box Gene Family in Brassica oleracea L. Genes. 2019; 10(12):1000. https://doi.org/10.3390/genes10121000
Chicago/Turabian StyleHu, Dengke, Qinqin Xie, Qianying Liu, Tonghong Zuo, Hecui Zhang, Yizhong Zhang, Xiaoping Lian, and Liquan Zhu. 2019. "Genome-Wide Distribution, Expression and Function Analysis of the U-Box Gene Family in Brassica oleracea L." Genes 10, no. 12: 1000. https://doi.org/10.3390/genes10121000