Efficient Gene Disruption via Base Editing Induced Stop in Newt Pleurodeles waltl
Abstract
:1. Introduction
2. Materials and Methods
2.1. Synthesis of Base Editor mRNA and sgRNAs
2.2. Animal Care and Egg Injections
2.3. Animal Genotyping and Phenotyping
3. Results
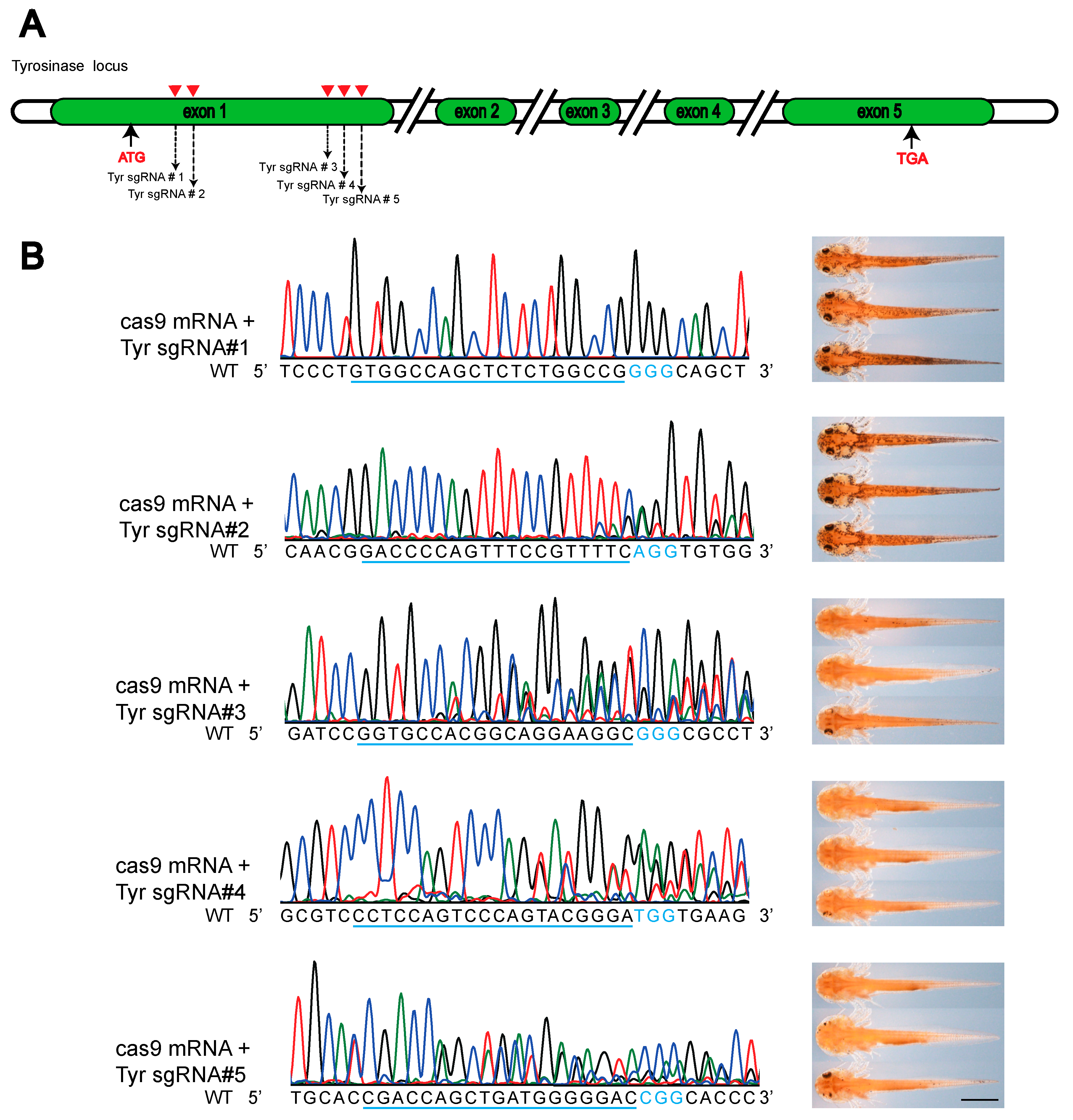
3.1. Strategy for Designing Premature Stop Codon for Target Gene Knockout in Newt
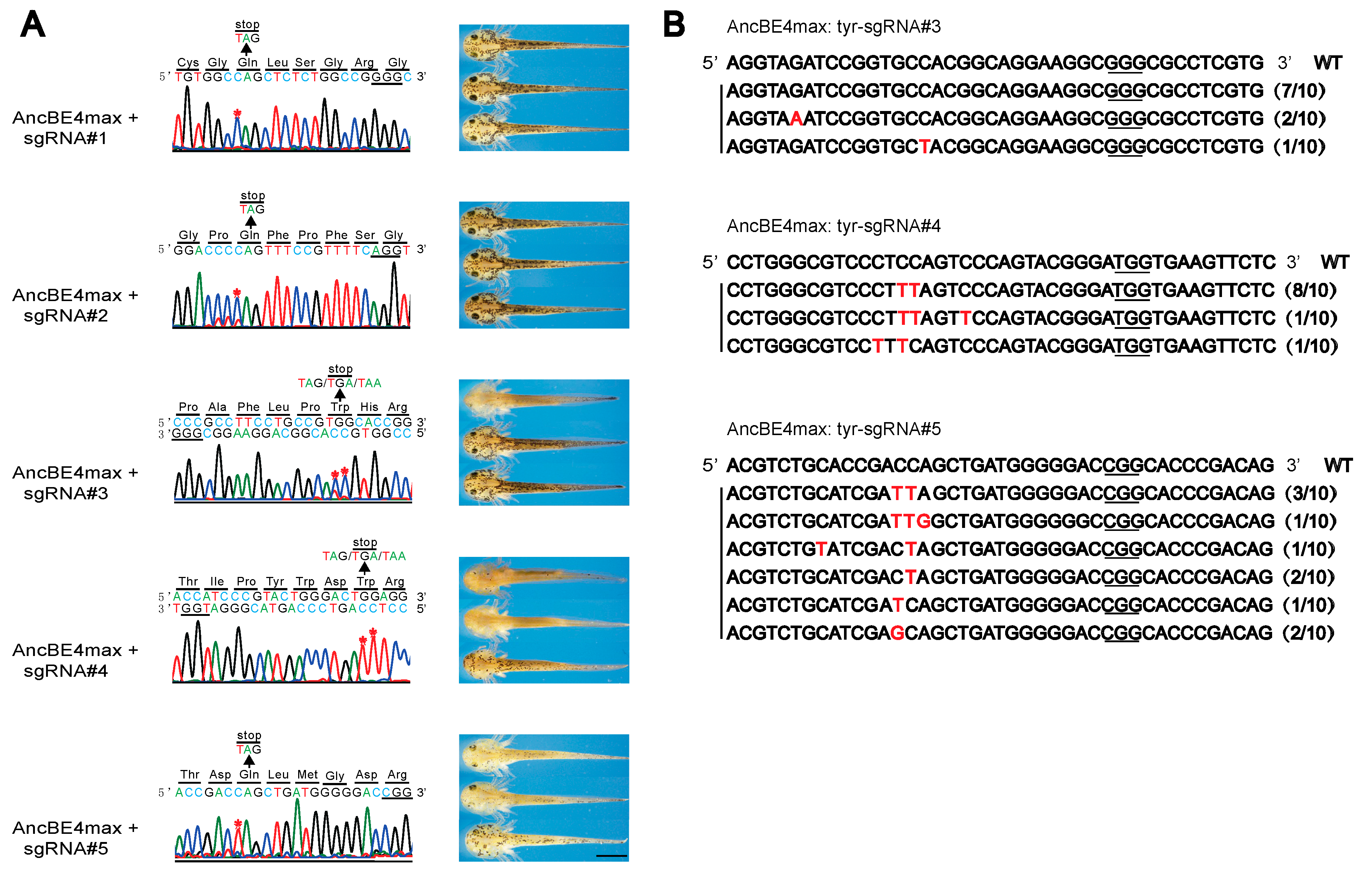
3.2. Generation of Albino Newt with Base Editing-induced Stop Codon in Tyrosinase Gene
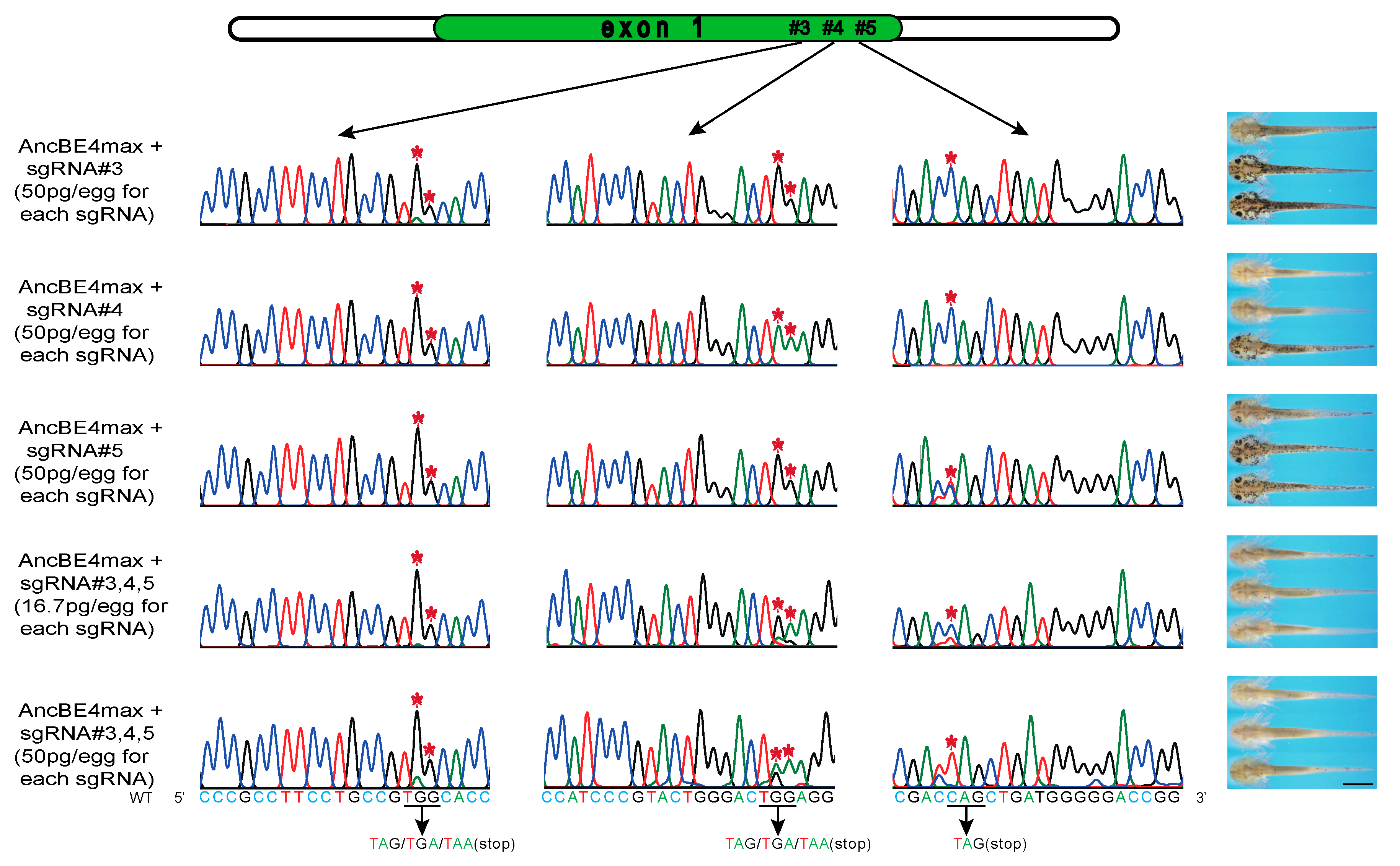
3.3. Multiplex sgRNAs Improves the Knockout Efficiency
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pasmans, F.; Martel, A. Amphibian Taxonomy, Anatomy, and Physiology. In Mader’s Reptile and Amphibian Medicine and Surgery; Elsevier: Amsterdam, The Netherlands, 2019; pp. 86–89. [Google Scholar]
- Brockes, J.P. Variation in Salamanders: An Essay on Genomes, Development, and Evolution. In Salamanders in Regeneration Research; Humana Press: New York, NY, USA, 2015; pp. 3–15. [Google Scholar]
- Nowoshilow, S.; Schloissnig, S.; Fei, J.F.; Dahl, A.; Pang, A.W.C.; Pippel, M.; Winkler, S.; Hastie, A.R.; Young, G.; Roscito, J.G.; et al. The axolotl genome and the evolution of key tissue formation regulators. Nature 2018, 554, 50–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elewa, A.; Wang, H.; Talavera-López, C.; Joven, A.; Brito, G.; Kumar, A.; Hameed, L.S.; Penrad-Mobayed, M.; Yao, Z.; Zamani, N.; et al. Reading and editing the Pleurodeles waltl genome reveals novel features of tetrapod regeneration. Nat. Commun. 2017, 8, 2286. [Google Scholar] [CrossRef] [PubMed]
- Joven, A.; Elewa, A.; Simon, A. Model systems for regeneration: salamanders. Development 2019, 146, dev167700. [Google Scholar] [CrossRef] [PubMed]
- Flowers, G.P.; Timberlake, A.T.; McLean, K.C.; Monaghan, J.R.; Crews, C.M. Highly efficient targeted mutagenesis in axolotl using Cas9 RNA-guided nuclease. Development 2014, 141, 2165–2171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fei, J.F.; Schuez, M.; Tazaki, A.; Taniguchi, Y.; Roensch, K.; Tanaka, E.M. CRISPR-mediated genomic deletion of Sox2 in the axolotl shows a requirement in spinal cord neural stem cell amplification during tail regeneration. Stem. Cell Rep. 2014, 3, 444–459. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Hayashi, T.; Inoue, T.; Agata, K.; Hirayama, M.; Suzuki, M.; Shigenobu, S.; Takeuchi, T.; Yamamoto, T.; Suzuki, K.I.T. Cas9 ribonucleoprotein complex allows direct and rapid analysis of coding and noncoding regions of target genes in Pleurodeles waltl development and regeneration. Dev. Biol. 2018, 443, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Eid, A.; Alshareef, S.; Mahfouz, M.M. CRISPR base editors: Genome editing without double-stranded breaks. Biochem. J. 2018, 475, 1955–1964. [Google Scholar] [CrossRef] [PubMed]
- Rees, H.A.; Liu, D.R. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018, 19, 770–788. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.-S. An Updated Review of Tyrosinase Inhibitors. Int. J. Mol. Sci. 2009, 10, 2440–2475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joven, A.; Kirkham, M.; Simon, A. Husbandry of Spanish Ribbed Newts (Pleurodeles waltl). In Salamanders in Regeneration Research; Humana Press: New York, NY, USA, 2015; pp. 47–70. [Google Scholar]
- Shi, D.L.; Boucaut, J.C. The chronological development of the urodele amphibian Pleurodeles waltl (Michah). Int. J. Dev. Biol. 1995, 39, 427–441. [Google Scholar] [PubMed]
- Shrivastav, M.; De Haro, L.P.; Nickoloff, J.A. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008, 18, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Shepard, D.B.; Chong, R.A.; Arriaza, J.L.; Hall, K.; Castoe, T.A.; Feschotte, C.; Pollock, D.D.; Mueller, R.L. LTR retrotransposons contribute to genomic gigantism in plethodontid salamanders. Genome Biol. Evol. 2012, 4, 168–183. [Google Scholar] [CrossRef] [PubMed]
- Zuo, E.; Sun, Y.; Wei, W.; Yuan, T.; Ying, W.; Sun, H.; Yuan, L.; Steinmetz, L.M.; Li, Y.; Yang, H. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 2019, 364, 289–292. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Zong, Y.; Gao, Q.; Zhu, Z.; Wang, Y.; Qin, P.; Liang, C.; Wang, D.; Qiu, J.L.; Zhang, F.; et al. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science 2019, 364, 292–295. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Sun, Y.; Yan, R.; Liu, Y.; Zuo, E.; Gu, C.; Han, L.; Wei, Y.; Hu, X.; Zeng, R.; et al. Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature 2019, 571, 275–278. [Google Scholar] [CrossRef] [PubMed]
- Kohn, D.B.; Porteus, M.H.; Scharenberg, A.M. Ethical and regulatory aspects of genome editing. Blood 2016, 127, 2553–2560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pickar-Oliver, A.; Gersbach, C.A. The next generation of CRISPR-Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 2019, 20, 490–507. [Google Scholar] [CrossRef] [PubMed]
| sgRNAs 1 | Injected Eggs | Survived Embryos | Phenotype 2 | ||
|---|---|---|---|---|---|
| Strong | Weak | None | |||
| sgRNA#3 (50 pg) | 115 | 82 | 7 (8.5%) | 1 (1.2%) | 74 (90.2%) |
| sgRNA#4 (50 pg) | 143 | 62 | 31 (50%) | 10 (16.1%) | 21 (33.9%) |
| sgRNA#5 (50 pg) | 128 | 32 | 2 (6.3%) | 2 (6.3%) | 28 (87.5%) |
| sgRNA#3 #4 #5 (16.7 pg each) | 106 | 40 | 28 (70%) | 8 (20%) | 4 (10%) |
| sgRNA#3 #4 #5 (50 pg each) | 128 | 81 | 53 (65.4%) | 15 (18.5%) | 13 (16.1%) |
| Control (ancBE4max only) | 108 | 92 | 0 | 0 | 92 (100%) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, H.; Peng, Z.; Ren, R.; Wang, H. Efficient Gene Disruption via Base Editing Induced Stop in Newt Pleurodeles waltl. Genes 2019, 10, 837. https://doi.org/10.3390/genes10110837
Cai H, Peng Z, Ren R, Wang H. Efficient Gene Disruption via Base Editing Induced Stop in Newt Pleurodeles waltl. Genes. 2019; 10(11):837. https://doi.org/10.3390/genes10110837
Chicago/Turabian StyleCai, Hao, Zhelun Peng, Ruimin Ren, and Heng Wang. 2019. "Efficient Gene Disruption via Base Editing Induced Stop in Newt Pleurodeles waltl" Genes 10, no. 11: 837. https://doi.org/10.3390/genes10110837




