Physiological and Transcriptomic Analysis Reveals Distorted Ion Homeostasis and Responses in the Freshwater Plant Spirodela polyrhiza L. under Salt Stress
Abstract
:1. Introduction
2. Results
2.1. Physiological Changes of S. Polyrhiza under Salt Stress
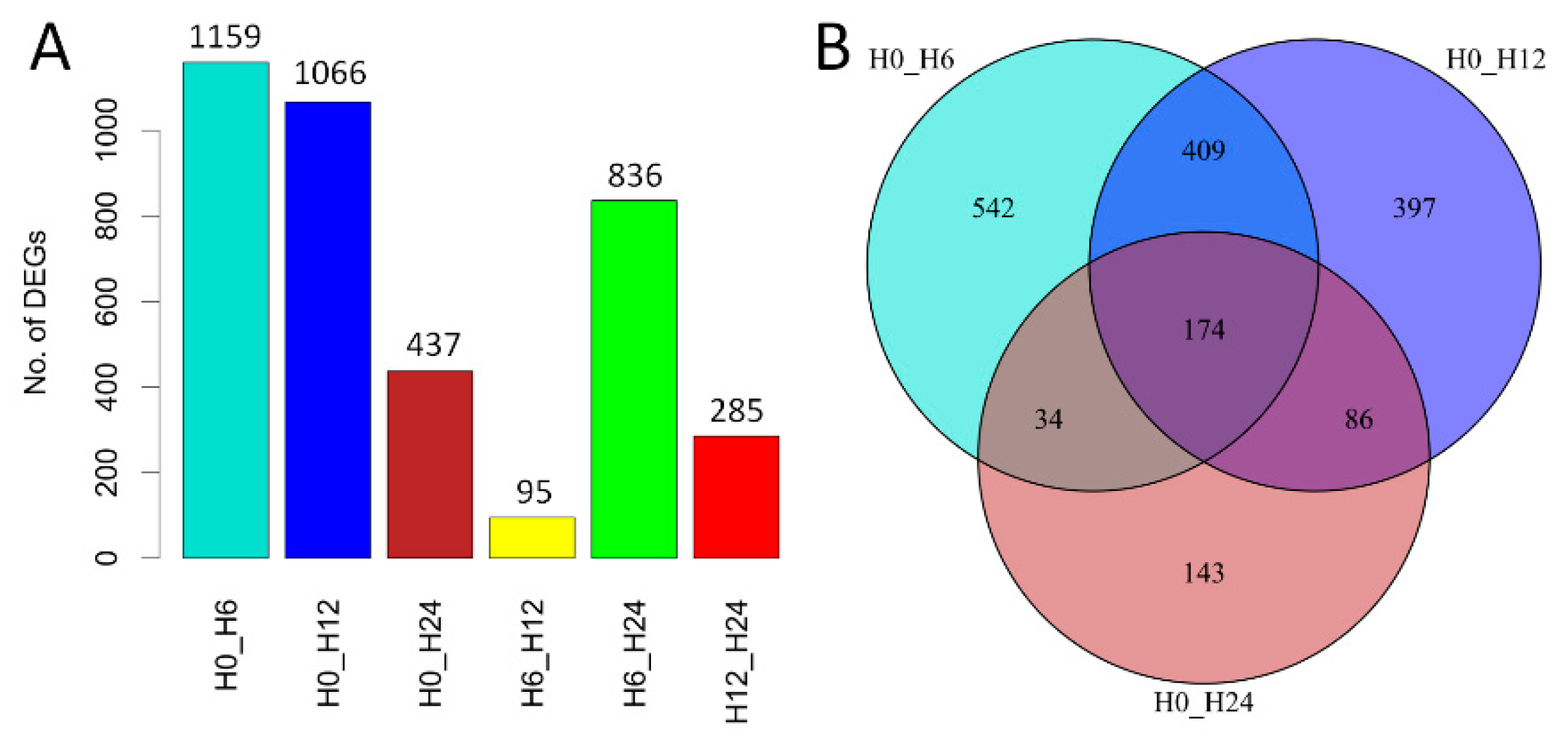
2.2. Transcriptome Changes in Response to Salt Stress
2.3. Expression Patterns of DE Genes and Functional Enrichment
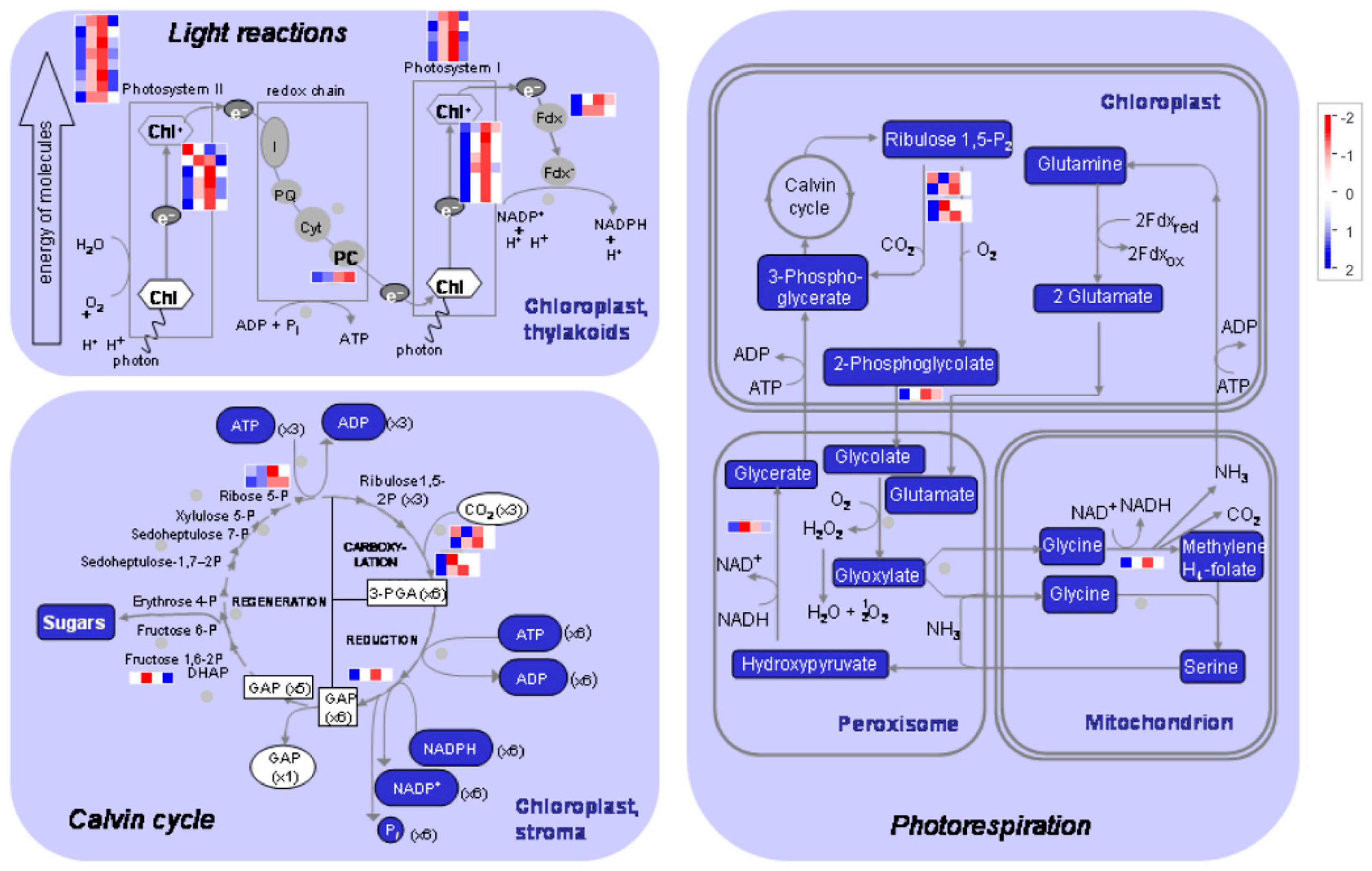
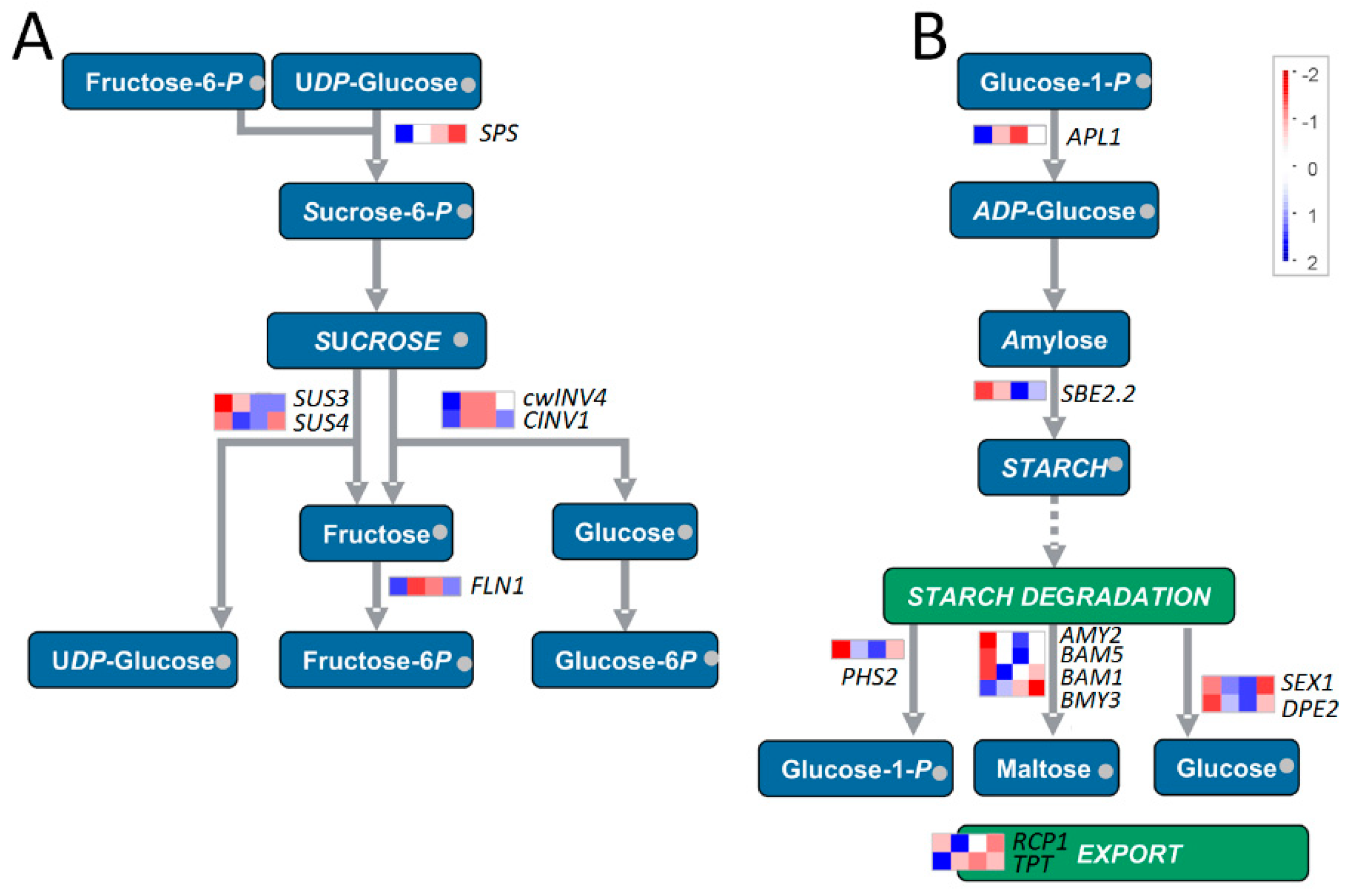
2.4. Photosynthesis, Tetrapyrrole Synthesis, and Carbohydrate Metabolism
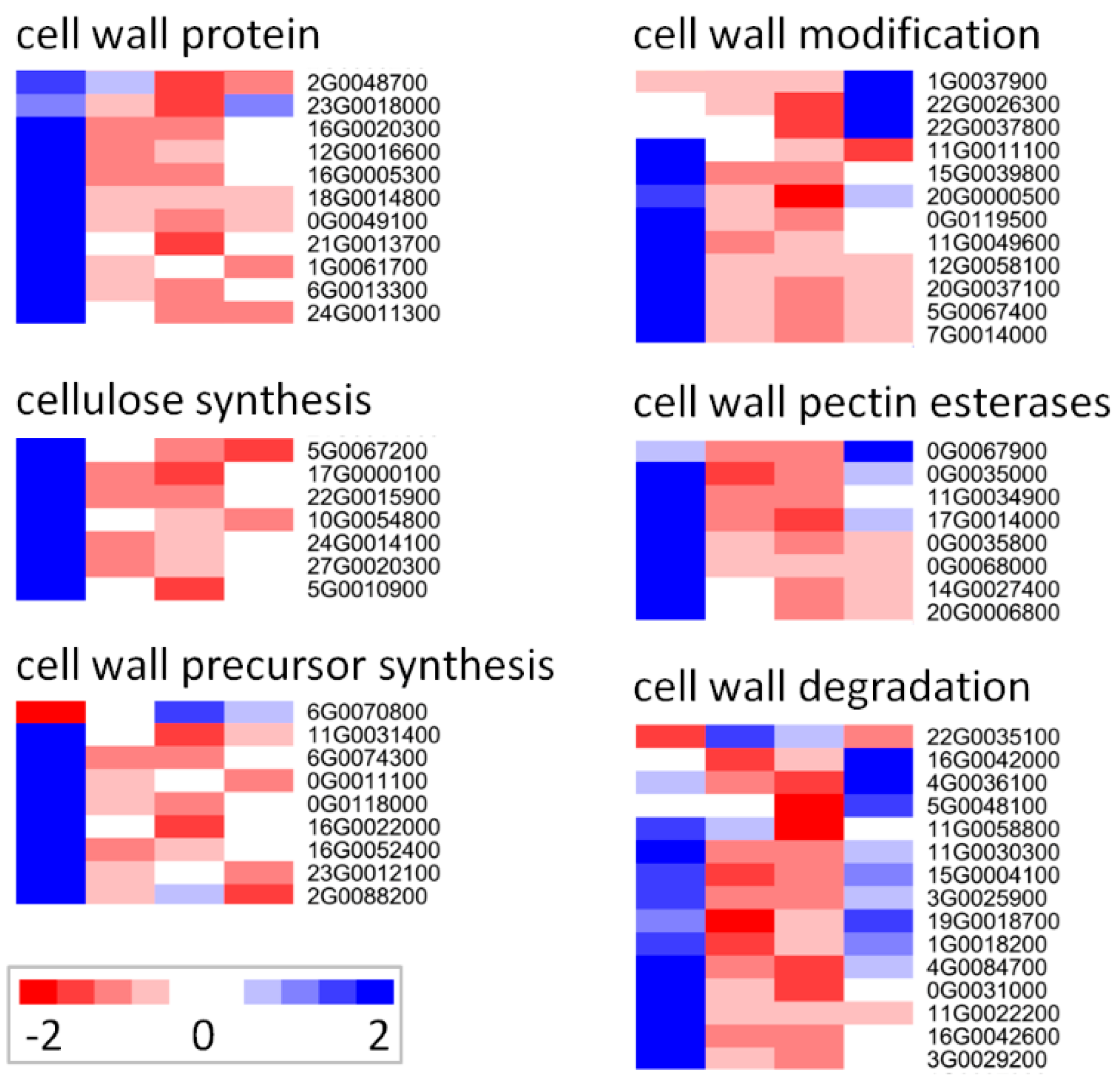
2.5. Response of Cell Wall-Related Genes
2.6. Response of Abiotic Stress-Related Genes
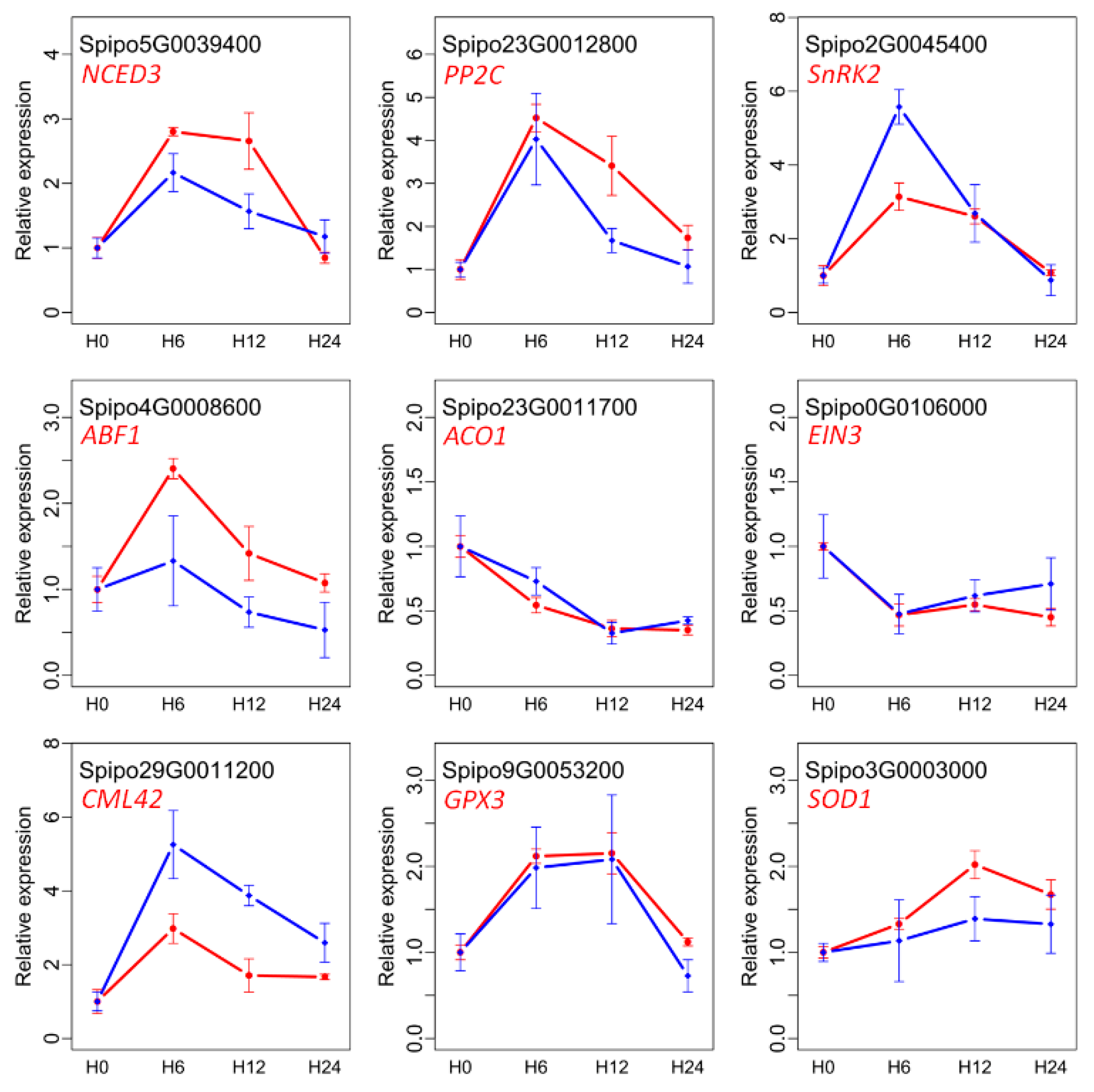
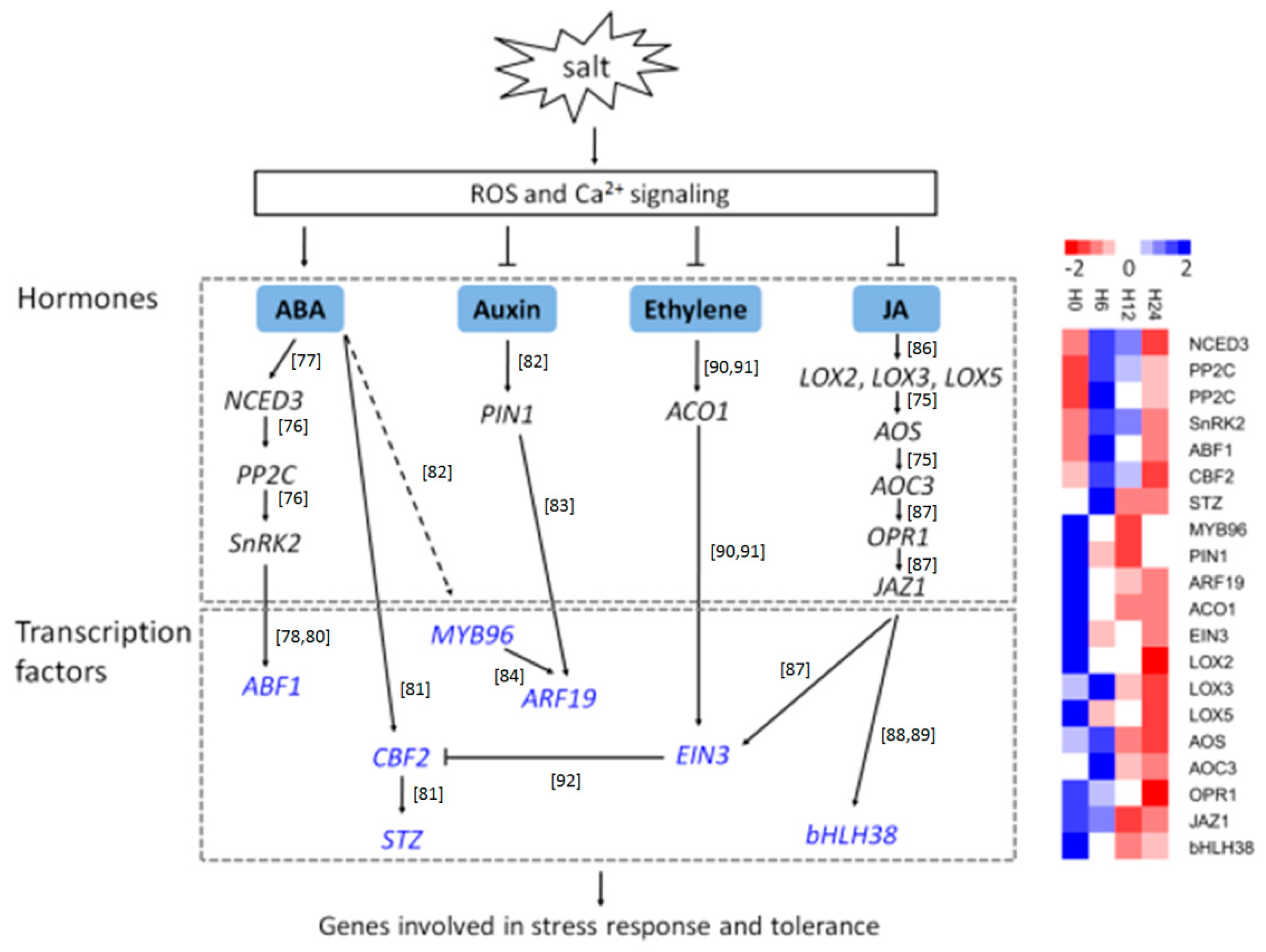
2.7. Roles of Genes Related to Hormone Biosynthesis
2.8. Roles of Transcription Factor (TF)
2.9. Responses of Cation Transporters That Maintain Ion Homeostasis during Salt Stress
2.10. Quantitative RT-PCR Validation of DE Genes
3. Discussion
3.1. Physiological Performance and Maintenance of Ion Homeostasis in S. Polyrhiza under Salt Stress
3.2. Expression of Genes Related to Cell Wall, Photosynthesis, and Carbohydrate Metabolism
3.3. Expression of Genes Related to Redox and Ca2+ Signaling
3.4. Regulatory Networks of Salt Stress in S. Polyrhiza
4. Methods
4.1. Plant Materials and Salt Treatment
4.2. RNA Extraction and Sequencing
4.3. RNA-Seq Data Analysis
4.4. Clustering and Functional Enrichment
4.5. Quantitative RT-PCR Analysis
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Appenroth, K.-J.; Borisjuk, N.; Lam, E. Telling duckweed apart: Genotyping technologies for the Lemnaceae. Chin. J. Appl. Environ. Biol. 2013, 19, 1–10. [Google Scholar]
- Cui, W.; Cheng, J.J. Growing duckweed for biofuel production: A review. Plant Biol. (Stuttg) 2015, 17, 16–23. [Google Scholar] [CrossRef] [PubMed]
- An, D.; Li, C.; Zhou, Y.; Wu, Y.; Wang, W. Genomes and Transcriptomes of Duckweeds. Front. Chem. 2018, 6, 230. [Google Scholar] [CrossRef] [PubMed]
- Landolt, E. Biosystematic Investigations in the Family of Duckweeds (Lemnaceae) Vol. 2: The Family of Lemnaceae: A Monographic Study—Morphology, Karyology, Ecology, Geographic Distribution, Nomenclature, Descriptions; Eidgenössische Technische Hochschule Zürich: Zürich, Switzerland, 1986. [Google Scholar]
- Cheng, T.S. NaCl-induced responses in giant duckweed. J. Aquat. Plant Manag. 2011, 49, 62–71. [Google Scholar]
- Chang, I.H.; Cheng, K.T.; Huang, P.C.; Lin, Y.Y.; Cheng, L.J.; Cheng, T.S. Oxidative stress in greater duckweed ( Spirodela polyrhiza ) caused by long-term NaCl exposure. Acta Physiol. Plant. 2012, 34, 1165–1176. [Google Scholar] [CrossRef]
- Liu, C.; Dai, Z.; Sun, H. Potential of duckweed (Lemna minor) for removal of nitrogen and phosphorus from water under salt stress. J. Environ. Manag. 2017, 187, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Oukarroum, A.; Bussotti, F.; Goltsev, V.; Kalaji, H.M. Correlation between reactive oxygen species production and photochemistry of photosystems I and II in Lemna gibba L. plants under salt stress. Environ. Exp. Bot. 2015, 109, 80–88. [Google Scholar] [CrossRef]
- Sree, K.S.; Adelmann, K.; Garcia, C.; Lam, E.; Appenroth, K.J. Natural variance in salt tolerance and induction of starch accumulation in duckweeds. Planta 2015, 241, 1395–1404. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Zhang, Y.; Xiao, Y.; Liu, F.; Wang, M.; Zhu, X.; Liu, P.; Sun, Q.; Wang, W.; Peng, M.; et al. Transcriptome response of cassava leaves under natural shade. Sci. Rep. 2016, 6, 31673. [Google Scholar] [CrossRef] [Green Version]
- Fu, L.; Ding, Z.; Han, B.; Hu, W.; Li, Y.; Zhang, J. Physiological investigation and transcriptome analysis of polyethylene glycol (PEG)-induced dehydration stress in cassava. Int. J. Mol. Sci. 2016, 17, 283. [Google Scholar] [CrossRef]
- Wang, W.; Messing, J. Status of duckweed genomics and transcriptomics. Plant Biol. (Stuttg) 2015, 17, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Haberer, G.; Gundlach, H.; Glasser, C.; Nussbaumer, T.; Luo, M.C.; Lomsadze, A.; Borodovsky, M.; Kerstetter, R.A.; Shanklin, J.; et al. The Spirodela polyrhiza genome reveals insights into its neotenous reduction fast growth and aquatic lifestyle. Nat. Commun. 2014, 5, 3311. [Google Scholar] [CrossRef] [PubMed]
- An, D.; Zhou, Y.; Li, C.; Xiao, Q.; Wang, T.; Zhang, Y.; Wu, Y.; Li, Y.; Chao, D.Y.; Messing, J.; et al. Plant evolution and environmental adaptation unveiled by long-read whole-genome sequencing of Spirodela. Proc. Natl. Acad. Sci. USA 2019, 116, 18893–18899. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Fang, Y.; Xiao, Y.; Jin, Y.L.; Ma, X.R.; Zhao, Y.; He, K.Z.; Zhao, H.; Wang, H.Y. Comparative transcriptome analysis to investigate the high starch accumulation of duckweed (Landoltia punctata) under nutrient starvation. Biotechnol. Biofuels 2013, 6, 72. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Fang, Y.; Huang, M.; Jin, Y.; Sun, J.; Tao, X.; Zhang, G.; He, K.; Zhao, Y.; Zhao, H. Uniconazole-induced starch accumulation in the bioenergy crop duckweed (Landoltia punctata) I: Transcriptome analysis of the effects of uniconazole on chlorophyll and endogenous hormone biosynthesis. Biotechnol. Biofuels 2015, 8, 57. [Google Scholar] [CrossRef]
- Liu, Y.; Fang, Y.; Huang, M.; Jin, Y.; Sun, J.; Tao, X.; Zhang, G.; He, K.; Zhao, Y.; Zhao, H. Uniconazole-induced starch accumulation in the bioenergy crop duckweed (Landoltia punctata) II: Transcriptome alterations of pathways involved in carbohydrate metabolism and endogenous hormone crosstalk. Biotechnol. Biofuels 2015, 8, 64. [Google Scholar] [CrossRef]
- Xu, H.; Yu, C.; Xia, X.; Li, M.; Li, H.; Wang, Y.; Wang, S.; Wang, C.; Ma, Y.; Zhou, G. Comparative transcriptome analysis of duckweed (Landoltia punctata) in response to cadmium provides insights into molecular mechanisms underlying hyperaccumulation. Chemosphere 2018, 190, 154–165. [Google Scholar] [CrossRef]
- Wang, W.; Wu, Y.; Messing, J. RNA-Seq transcriptome analysis of Spirodela dormancy without reproduction. BMC Genom. 2014, 15, 60. [Google Scholar] [CrossRef]
- Wang, W.; Li, R.; Zhu, Q.; Tang, X.; Zhao, Q. Transcriptomic and physiological analysis of common duckweed Lemna minor responses to NH4(+) toxicity. BMC Plant Biol. 2016, 16, 92. [Google Scholar] [CrossRef]
- Van Hoeck, A.; Horemans, N.; Nauts, R.; Van Hees, M.; Vandenhove, H.; Blust, R. Lemna minor plants chronically exposed to ionising radiation: RNA-seq analysis indicates a dose rate dependent shift from acclimation to survival strategies. Plant Sci. 2017, 257, 84–95. [Google Scholar] [CrossRef]
- Yu, C.; Zhao, X.; Qi, G.; Bai, Z.; Wang, Y.; Wang, S.; Ma, Y.; Liu, Q.; Hu, R.; Zhou, G. Integrated analysis of transcriptome and metabolites reveals an essential role of metabolic flux in starch accumulation under nitrogen starvation in duckweed. Biotechnol. Biofuels 2017, 10, 167. [Google Scholar] [CrossRef] [PubMed]
- Nassoury, N.; Fritz, L.; Morse, D. Circadian changes in ribulose-1,5-bisphosphate carboxylase/oxygenase distribution inside individual chloroplasts can account for the rhythm in dinoflagellate carbon fixation. Plant Cell 2001, 13, 923–934. [Google Scholar] [CrossRef] [PubMed]
- Grabsztunowicz, M.; Gorski, Z.; Lucinski, R.; Jackowski, G. A reversible decrease in ribulose 1,5-bisphosphate carboxylase/oxygenase carboxylation activity caused by the aggregation of the enzyme’s large subunit is triggered in response to the exposure of moderate irradiance-grown plants to low irradiance. Physiol. Plant 2015, 154, 591–608. [Google Scholar] [CrossRef] [PubMed]
- Orzechowski, S. Starch metabolism in leaves. Acta Biochim. Pol. 2008, 55, 435–445. [Google Scholar] [PubMed]
- Wang, X.; Cnops, G.; Vanderhaeghen, R.; De Block, S.; Van Montagu, M.; Van Lijsebettens, M. AtCSLD3, a cellulose synthase-like gene important for root hair growth in arabidopsis. Plant Physiol. 2001, 126, 575–586. [Google Scholar] [CrossRef] [PubMed]
- Bernal, A.J.; Jensen, J.K.; Harholt, J.; Sorensen, S.; Moller, I.; Blaukopf, C.; Johansen, B.; de Lotto, R.; Pauly, M.; Scheller, H.V.; et al. Disruption of ATCSLD5 results in reduced growth, reduced xylan and homogalacturonan synthase activity and altered xylan occurrence in Arabidopsis. Plant J. 2007, 52, 791–802. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Elliott, J.E.; Williamson, R.E. Features of the primary wall CESA complex in wild type and cellulose-deficient mutants of Arabidopsis thaliana. J. Exp. Bot. 2008, 59, 2627–2637. [Google Scholar] [CrossRef]
- Chan, J.; Eder, M.; Crowell, E.F.; Hampson, J.; Calder, G.; Lloyd, C. Microtubules and CESA tracks at the inner epidermal wall align independently of those on the outer wall of light-grown Arabidopsis hypocotyls. J. Cell Sci. 2011, 124, 1088–1094. [Google Scholar] [CrossRef]
- Johnson, K.L.; Kibble, N.A.; Bacic, A.; Schultz, C.J. A fasciclin-like arabinogalactan-protein (FLA) mutant of Arabidopsis thaliana, fla1, shows defects in shoot regeneration. PLoS ONE 2011, 6, e25154. [Google Scholar] [CrossRef]
- Shi, H.; Kim, Y.; Guo, Y.; Stevenson, B.; Zhu, J.K. The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell 2003, 15, 19–32. [Google Scholar] [CrossRef]
- Li, G.L.; Li, B.; Liu, H.T.; Zhou, R.G. The responses of AtJ2 and AtJ3 gene expression to environmental stresses in Arabidopsis. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao 2005, 31, 47–52. [Google Scholar] [PubMed]
- Wu, J.R.; Wang, T.Y.; Weng, C.P.; Duong, N.K.T.; Wu, S.J. AtJ3, a specific HSP40 protein, mediates protein farnesylation-dependent response to heat stress in Arabidopsis. Planta 2019. [Google Scholar] [CrossRef] [PubMed]
- Xin, Z.; Mandaokar, A.; Chen, J.; Last, R.L.; Browse, J. Arabidopsis ESK1 encodes a novel regulator of freezing tolerance. Plant J. 2007, 49, 786–799. [Google Scholar] [CrossRef] [PubMed]
- Goh, C.H.; Nam, H.G.; Park, Y.S. Stress memory in plants: A negative regulation of stomatal response and transient induction of rd22 gene to light in abscisic acid-entrained Arabidopsis plants. Plant J. 2003, 36, 240–255. [Google Scholar] [CrossRef] [PubMed]
- Mitchum, M.G.; Yamaguchi, S.; Hanada, A.; Kuwahara, A.; Yoshioka, Y.; Kato, T.; Tabata, S.; Kamiya, Y.; Sun, T.P. Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J. 2006, 45, 804–818. [Google Scholar] [CrossRef]
- Gigli-Bisceglia, N.; Engelsdorf, T.; Strnad, M.; Vaahtera, L.; Khan, G.A.; Yamoune, A.; Alipanah, L.; Novak, O.; Persson, S.; Hejatko, J.; et al. Cell wall integrity modulates Arabidopsis thaliana cell cycle gene expression in a cytokinin- and nitrate reductase-dependent manner. Development 2018, 145. [Google Scholar] [CrossRef] [PubMed]
- Serrano, R.; Rodriguez-Navarro, A. Ion homeostasis during salt stress in plants. Curr. Opin. Cell Biol. 2001, 13, 399–404. [Google Scholar] [CrossRef]
- Olias, R.; Eljakaoui, Z.; Li, J.; De Morales, P.A.; Marin-Manzano, M.C.; Pardo, J.M.; Belver, A. The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs. Plant Cell Environ. 2009, 32, 904–916. [Google Scholar] [CrossRef] [PubMed]
- Yokoi, S.; Quintero, F.J.; Cubero, B.; Ruiz, M.T.; Bressan, R.A.; Hasegawa, P.M.; Pardo, J.M. Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. Plant J. 2002, 30, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Yang, H.; Xue, Y.; Kong, D.; Ye, R.; Li, C.; Zhang, J.; Theprungsirikul, L.; Shrift, T.; Krichilsky, B.; et al. OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 2014, 514, 367–371. [Google Scholar] [CrossRef]
- Wang, P.; Li, Z.; Wei, J.; Zhao, Z.; Sun, D.; Cui, S. A Na+/Ca2+ exchanger-like protein (AtNCL) involved in salt stress in Arabidopsis. J. Biol. Chem. 2012, 287, 44062–44070. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Ding, Y.; Xu, Y.; Li, Z.; Jin, Y.; He, K.; Fang, Y.; Zhao, H. Responses of Landoltia punctata to cobalt and nickel: Removal, growth, photosynthesis, antioxidant system and starch metabolism. Aquat. Toxicol. 2017, 190, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Shi, H.J.; Wang, M.L.; Cui, L.; Zhao, H.; Zhao, Y. Effect of nitrogen and phosphorus deficiency on transcriptional regulation of genes encoding key enzymes of starch metabolism in duckweed (Landoltia punctata). Plant Physiol. Biochem. 2015, 86, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Gu, W.; Dai, Z.; Li, J.; Jiang, H.; Zhang, Q. Boron accumulation by Lemna minor L. under salt stress. Sci. Rep. 2018, 8, 8954. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, J.; Sun, L. Structure of the hyperosmolality-gated calcium-permeable channel OSCA1.2. Nat. Commun. 2018, 9, 5060. [Google Scholar] [CrossRef]
- Fahmideh, L.; Fooladvand, Z. Isolation and semi quantitative PCR of Na(+)/H(+) antiporter (SOS1 and NHX) genes under salinity stress in Kochia scoparia. Biol. Proced. Online 2018, 20, 11. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Sun, J.; Cao, P.; Ren, L.; Liu, C.; Chen, S.; Chen, F.; Jiang, J. Variation in tissue Na(+) content and the activity of SOS1 genes among two species and two related genera of Chrysanthemum. BMC Plant Biol. 2016, 16, 98. [Google Scholar] [CrossRef]
- Pittman, J.K.; Hirschi, K.D. CAX-ing a wide net: Cation/H(+) transporters in metal remediation and abiotic stress signalling. Plant Biol. (Stuttg) 2016, 18, 741–749. [Google Scholar] [CrossRef]
- Kim, C.K.; Han, J.S.; Lee, H.S.; Oh, J.Y.; Shigaki, T.; Park, S.H.; Hirschi, K. Expression of an Arabidopsis CAX2 variant in potato tubers increases calcium levels with no accumulation of manganese. Plant Cell Rep. 2006, 25, 1226–1232. [Google Scholar] [CrossRef]
- Do, T.D.; Chen, H.; Hien, V.T.; Hamwieh, A.; Yamada, T.; Sato, T.; Yan, Y.; Cong, H.; Shono, M.; Suenaga, K.; et al. Ncl synchronously regulates Na+, K+, and Cl- in soybean and greatly increases the grain yield in saline field conditions. Sci. Rep. 2016, 6, 19147. [Google Scholar] [CrossRef]
- Pilot, G.; Pratelli, R.; Gaymard, F.; Meyer, Y.; Sentenac, H. Five-group distribution of the Shaker-like K+ channel family in higher plants. J. Mol. Evol. 2003, 56, 418–434. [Google Scholar] [CrossRef] [PubMed]
- Obata, T.; Kitamoto, H.K.; Nakamura, A.; Fukuda, A.; Tanaka, Y. Rice shaker potassium channel OsKAT1 confers tolerance to salinity stress on yeast and rice cells. Plant Physiol. 2007, 144, 1978–1985. [Google Scholar] [CrossRef] [PubMed]
- Pilot, G.; Gaymard, F.; Mouline, K.; Cherel, I.; Sentenac, H. Regulated expression of Arabidopsis shaker K+ channel genes involved in K+ uptake and distribution in the plant. Plant Mol. Biol. 2003, 51, 773–787. [Google Scholar] [CrossRef] [PubMed]
- Sze, H.; Padmanaban, S.; Cellier, F.; Honys, D.; Cheng, N.H.; Bock, K.W.; Conejero, G.; Li, X.; Twell, D.; Ward, J.M.; et al. Expression patterns of a novel AtCHX gene family highlight potential roles in osmotic adjustment and K+ homeostasis in pollen development. Plant Physiol. 2004, 136, 2532–2547. [Google Scholar] [CrossRef] [PubMed]
- Cellier, F.; Conejero, G.; Ricaud, L.; Luu, D.T.; Lepetit, M.; Gosti, F.; Casse, F. Characterization of AtCHX17, a member of the cation/H+ exchangers, CHX family, from Arabidopsis thaliana suggests a role in K+ homeostasis. Plant J. 2004, 39, 834–846. [Google Scholar] [CrossRef] [PubMed]
- da Silva, H.A.; de Oliveira, D.F.A.; Avelino, A.P.; de Macedo, C.E.C.; Barros-Galvao, T.; Voigt, E.L. Salt stress differentially regulates mobilisation of carbon and nitrogen reserves during seedling establishment of Pityrocarpa moniliformis. Plant Biol. (Stuttg) 2019. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.K.; Bhargava, P.; Thapar, R.; Rai, L.C. Salinity-induced physiological and proteomic changes in Anabaena doliolum. Environ. Exp. Bot. 2008, 64, 49–57. [Google Scholar] [CrossRef]
- Gong, D.H.; Wang, G.Z.; Si, W.T.; Zhou, Y.; Liu, Z.; Jia, J. Effects of salt stress on photosynthetic pigments and activity of ribulose-1,5-bisphosphate carboxylase/oxygenase in Kalidium foliatum. Russ. J. Plant Physiol. 2018, 65, 98–103. [Google Scholar] [CrossRef]
- Rinne, P.; Tuominen, H.; Junttila, O. Seasonal changes in bud dormancy in relation to bud morphology, water and starch content, and abscisic acid concentration in adult trees of Betula pubescens. Tree Physiol. 1994, 14, 549–561. [Google Scholar] [CrossRef]
- Howard, T.P.; Fahy, B.; Craggs, A.; Mumford, R.; Leigh, F.; Howell, P.; Greenland, A.; Smith, A.M. Barley mutants with low rates of endosperm starch synthesis have low grain dormancy and high susceptibility to preharvest sprouting. New Phytol. 2012, 194, 158–167. [Google Scholar] [CrossRef]
- Xie, R.; Pan, X.; Zhang, J.; Ma, Y.; He, S.; Zheng, Y.; Ma, Y. Effect of salt-stress on gene expression in citrus roots revealed by RNA-seq. Funct. Integr. Genom. 2018, 18, 155–173. [Google Scholar] [CrossRef] [PubMed]
- Kaashyap, M.; Ford, R.; Kudapa, H.; Jain, M.; Edwards, D.; Varshney, R.; Mantri, N. Differential regulation of genes involved in root morphogenesis and cell wall modification is associated with salinity tolerance in chickpea. Sci. Rep. 2018, 8, 4855. [Google Scholar] [CrossRef] [PubMed]
- Yaish, M.W.; Patankar, H.V.; Assaha, D.V.M.; Zheng, Y.; Al-Yahyai, R.; Sunkar, R. Genome-wide expression profiling in leaves and roots of date palm (Phoenix dactylifera L.) exposed to salinity. BMC Genom. 2017, 18, 246. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Hu, Y.; Huo, P.; Zhang, Q.; Chen, X.; Zhang, Z. Transcriptome analysis of hexaploid hulless oat in response to salinity stress. PLoS ONE 2017, 12, e0171451. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Schumaker, K.S.; Zhu, J.K. Cell signaling during cold, drought, and salt stress. Plant Cell 2002, 14, S165–S183. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Lv, D.; Wang, P.; Wang, X.C.; Chen, J.; Miao, C.; Song, C.P. An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell 2006, 18, 2749–2766. [Google Scholar] [CrossRef]
- Wang, Y.; Wisniewski, M.; Meilan, R.; Uratsu, S.L.; Cui, M.G.; Dandekar, A.; Fuchigami, L. Ectopic expression of Mn-SOD in Lycopersicon esculentum leads to enhanced tolerance to salt and oxidative stress. J. Appl. Hortic. 2007, 9, 3–8. [Google Scholar]
- Ranty, B.; Aldon, D.; Cotelle, V.; Galaud, J.P.; Thuleau, P.; Mazars, C. Calcium sensors as key hubs in plant responses to biotic and abiotic stresses. Front. Plant Sci. 2016, 7, 327. [Google Scholar] [CrossRef]
- Mehlmer, N.; Wurzinger, B.; Stael, S.; Hofmann-Rodrigues, D.; Csaszar, E.; Pfister, B.; Bayer, R.; Teige, M. The Ca(2+) -dependent protein kinase CPK3 is required for MAPK-independent salt-stress acclimation in Arabidopsis. Plant J. 2010, 63, 484–498. [Google Scholar] [CrossRef]
- Xu, J.; Tian, Y.S.; Peng, R.H.; Xiong, A.S.; Zhu, B.; Jin, X.F.; Gao, F.; Fu, X.Y.; Hou, X.L.; Yao, Q.H. AtCPK6, a functionally redundant and positive regulator involved in salt/drought stress tolerance in Arabidopsis. Planta 2010, 231, 1251–1260. [Google Scholar] [CrossRef]
- Magnan, F.; Ranty, B.; Charpenteau, M.; Sotta, B.; Galaud, J.P.; Aldon, D. Mutations in AtCML9, a calmodulin-like protein from Arabidopsis thaliana, alter plant responses to abiotic stress and abscisic acid. Plant J. 2008, 56, 575–589. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.S.; Hung, M.J.; Cheng, Y.I.; Cheng, L.J. Calcium-induced proline accumulation contributes to amelioration of NaCl injury and expression of glutamine synthetase in greater duckweed (Spirodela polyrhiza L.). Aquat. Toxicol. 2013, 144–145, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.T.; Ma, S.L.; Bai, L.P.; Zhang, L.; Ma, H.; Jia, P.; Liu, J.; Zhong, M.; Guo, Z.F. Signal transduction during cold, salt, and drought stresses in plants. Mol. Biol. Rep. 2012, 39, 969–987. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015, 20, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 2016, 7, 571. [Google Scholar] [CrossRef] [PubMed]
- Roychoudhury, A.; Paul, S.; Basu, S. Cross-talk between abscisic acid-dependent and abscisic acid-independent pathways during abiotic stress. Plant Cell Rep. 2013, 32, 985–1006. [Google Scholar] [CrossRef] [PubMed]
- Uno, Y.; Furihata, T.; Abe, H.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 2000, 97, 11632–11637. [Google Scholar] [CrossRef] [PubMed]
- Furihata, T.; Maruyama, K.; Fujita, Y.; Umezawa, T.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. USA 2006, 103, 1988–1993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujii, H.; Verslues, P.E.; Zhu, J.K. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 2007, 19, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Fowler, S.; Thomashow, M.F. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 2002, 14, 1675–1690. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Liu, H.; Xiong, L. Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front. Plant Sci. 2013, 4, 397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Li, R.J.; Han, T.T.; Cai, W.; Fu, Z.W.; Lu, Y.T. Salt stress reduces root meristem size by nitric oxide-mediated modulation of auxin accumulation and signaling in Arabidopsis. Plant Physiol. 2015, 168, 343–356. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Xiang, F.; Qiao, M.; Park, J.Y.; Lee, Y.N.; Kim, S.G.; Lee, Y.H.; Park, W.J.; Park, C.M. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 2009, 151, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Guo, J.; Zhu, A.; Zhang, L.; Zhang, M. Exogenous jasmonic acid can enhance tolerance of wheat seedlings to salt stress. Ecotoxicol. Environ. Saf. 2014, 104, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Dong, W.; Zhang, N.; Ai, X.; Wang, M.; Huang, Z.; Xiao, L.; Xia, G. A wheat allene oxide cyclase gene enhances salinity tolerance via jasmonate signaling. Plant Physiol. 2014, 164, 1068–1076. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Zhang, C.; Gu, M.; Bai, Z.; Zhang, W.; Qi, T.; Cheng, Z.; Peng, W.; Luo, H.; Nan, F.; et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 2009, 21, 2220–2236. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; An, F.; Feng, Y.; Li, P.; Xue, L.; A, M.; Jiang, Z.; Kim, J.M.; To, T.K.; Li, W.; et al. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 12539–12544. [Google Scholar] [CrossRef] [PubMed]
- Toda, Y.; Tanaka, M.; Ogawa, D.; Kurata, K.; Kurotani, K.; Habu, Y.; Ando, T.; Sugimoto, K.; Mitsuda, N.; Katoh, E.; et al. RICE SALT SENSITIVE3 forms a ternary complex with JAZ and class-C bHLH factors and regulates jasmonate-induced gene expression and root cell elongation. Plant Cell 2013, 25, 1709–1725. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Wang, Z.; Song, X.; Xu, J.; Jiang, C.; Zhao, Y.; Ma, C.; Zhang, H. Transcriptomic profiling revealed an important role of cell wall remodeling and ethylene signaling pathway during salt acclimation in Arabidopsis. Plant Mol. Biol. 2014, 86, 303–317. [Google Scholar] [CrossRef]
- Xu, J.; Li, Y.; Wang, Y.; Liu, H.; Lei, L.; Yang, H.; Liu, G.; Ren, D. Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J. Biol. Chem. 2008, 283, 26996–27006. [Google Scholar] [CrossRef]
- Chen, D.; Ma, X.; Li, C.; Zhang, W.; Xia, G.; Wang, M. A wheat aminocyclopropane-1-carboxylate oxidase gene, TaACO1, negatively regulates salinity stress in Arabidopsis thaliana. Plant Cell Rep. 2014, 33, 1815–1827. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Tian, S.; Hou, L.; Huang, X.; Zhang, X.; Guo, H.; Yang, S. Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 2012, 24, 2578–2595. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Huang, M.; Han, B.; Sun, X.; Sree, K.S.; Appenroth, K.J.; Zhang, J. Flower induction, microscope-aided cross-pollination, and seed production in the duckweed Lemna gibba with discovery of a male-sterile clone. Sci. Rep. 2017, 7, 3047. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Thimm, O.; Blasing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Kruger, P.; Selbig, J.; Muller, L.A.; Rhee, S.Y.; Stitt, M. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef]
- Saeed, A.I.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thiagarajan, M.; et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques 2003, 34, 374–378. [Google Scholar] [CrossRef]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, L.; Ding, Z.; Sun, X.; Zhang, J. Physiological and Transcriptomic Analysis Reveals Distorted Ion Homeostasis and Responses in the Freshwater Plant Spirodela polyrhiza L. under Salt Stress. Genes 2019, 10, 743. https://doi.org/10.3390/genes10100743
Fu L, Ding Z, Sun X, Zhang J. Physiological and Transcriptomic Analysis Reveals Distorted Ion Homeostasis and Responses in the Freshwater Plant Spirodela polyrhiza L. under Salt Stress. Genes. 2019; 10(10):743. https://doi.org/10.3390/genes10100743
Chicago/Turabian StyleFu, Lili, Zehong Ding, Xuepiao Sun, and Jiaming Zhang. 2019. "Physiological and Transcriptomic Analysis Reveals Distorted Ion Homeostasis and Responses in the Freshwater Plant Spirodela polyrhiza L. under Salt Stress" Genes 10, no. 10: 743. https://doi.org/10.3390/genes10100743




