Internalization Characterization of Si Nanorod with Camouflaged Cell Membrane Proteins Reveals ATXN2 as a Negative Regulator
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fabrication of Nanorods
2.2. Characterization of SiNRs by SEM and TEM
2.3. Cell Culture
2.4. Cell Membrane Protein Extraction
2.5. SDS-PAGE and Silver Staining
2.6. Western Blot
2.7. LC-MS/MS
2.8. GO Enrichment Analysis
2.9. Membrane Protein Coating and Labeling
2.10. Quantitative Analysis on Nanorod Uptake
2.11. Immunostaining, EGF Binding Assay, Colocalization Assay, and Confocal Microscopy
2.12. Total RNA Isolation and qRT-PCR Analysis
2.13. Small Interfere RNA Transfection and Chemical Inhibitors Studies
2.14. Statistic Method
3. Results
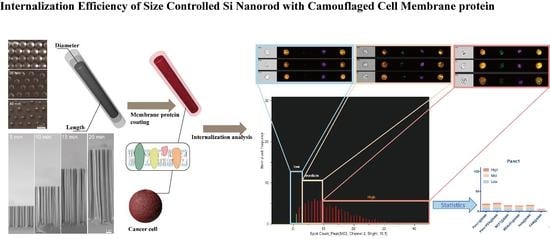
3.1. Fabrication of Size-Controllable SiNR Array
3.2. Nanofabrication of SiNR Coating with Membrane Protein
3.3. Proteomics Analysis of Coating Membrane Proteins Derived from Five Cancer Cell Lines
3.4. SiNR Internalization Efficiency Is also Determined by Target Cells
3.5. Gene Ontology on Cancer Cell Internalization
3.6. ATXN2 Is a Negative Regulator of Nanorod Internalization
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhao, Y. Nanomedicine in China. Adv. Healthc. Mater. 2018, 7, e1801051. [Google Scholar] [CrossRef] [Green Version]
- Pelaz, B.; Alexiou, C.; Alvarez-Puebla, R.A.; Alves, F.; Andrews, A.M.; Ashraf, S.; Balogh, L.P.; Ballerini, L.; Bestetti, A.; Brendel, C.; et al. Diverse Applications of Nanomedicine. ACS Nano 2017, 11, 2313–2381. [Google Scholar] [CrossRef] [Green Version]
- Albanese, A.; Tang, P.S.; Chan, W.C. The effect of nanoparticle size, shape, and surface chemistry on biological systems. Annu. Rev. Biomed. Eng. 2012, 14, 1–16. [Google Scholar] [CrossRef]
- Truong, N.P.; Whittaker, M.R.; Mak, C.W.; Davis, T.P. The importance of nanoparticle shape in cancer drug delivery. Expert. Opin. Drug Deliv. 2015, 12, 129–142. [Google Scholar] [CrossRef]
- Barua, S.; Yoo, J.W.; Kolhar, P.; Wakankar, A.; Gokarn, Y.R.; Mitragotri, S. Particle shape enhances specificity of antibody-displaying nanoparticles. Proc. Natl. Acad. Sci. USA 2013, 110, 3270–3275. [Google Scholar] [CrossRef] [Green Version]
- Fang, R.H.; Kroll, A.V.; Gao, W.; Zhang, L. Cell Membrane Coating Nanotechnology. Adv. Mater. 2018, 30, e1706759. [Google Scholar] [CrossRef] [PubMed]
- Kroll, A.V.; Fang, R.H.; Zhang, L. Biointerfacing and Applications of Cell Membrane-Coated Nanoparticles. Bioconjug. Chem. 2017, 28, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Zhang, P.; Hou, Y.; Ning, H.; Wang, Z.; Huang, J.; Gao, M. “Smart” Nanoprobes for Visualization of Tumor Microenvironments. Adv. Healthc. Mater. 2018, 7, e1800391. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Wang, C.; Zhao, F.; Gu, Z.; Zhao, Y. Application of Multifunctional Nanomaterials in Radioprotection of Healthy Tissues. Adv. Healthc. Mater. 2018, 7, e1800421. [Google Scholar] [CrossRef]
- Shi, J.; Kantoff, P.W.; Wooster, R.; Farokhzad, O.C. Cancer nanomedicine: Progress, challenges and opportunities. Nat. Rev. Cancer 2017, 17, 20–37. [Google Scholar] [CrossRef]
- Wilhelm, S.; Tavares, A.J.; Dai, Q.; Ohta, S.; Audet, J.; Dvorak, H.F.; Chan, W.C.W. Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater. 2016, 1, 16014. [Google Scholar] [CrossRef]
- Kim, W.; Ng, J.K.; Kunitake, M.E.; Conklin, B.R.; Yang, P. Interfacing Silicon Nanowires with Mammalian Cells. J. Am. Chem. Soc. 2007, 129, 7228–7229. [Google Scholar] [CrossRef]
- Huang, Z.P.; Fang, H.; Zhu, J. Fabrication of silicon nanowire arrays with controlled diameter, length, and density. Adv. Mater. 2007, 19, 744–748. [Google Scholar] [CrossRef]
- Zimmerman, J.F.; Parameswaran, R.; Murray, G.; Wang, Y.; Burke, M.; Tian, B. Cellular uptake and dynamics of unlabeled freestanding silicon nanowires. Sci. Adv. 2016, 2, e1601039. [Google Scholar] [CrossRef]
- Ding, L.; Yao, C.; Yin, X.; Li, C.; Huang, Y.; Wu, M.; Wang, B.; Guo, X.; Wang, Y.; Wu, M. Size, Shape, and Protein Corona Determine Cellular Uptake and Removal Mechanisms of Gold Nanoparticles. Small 2018, e1801451. [Google Scholar] [CrossRef]
- Li, S.Y.; Cheng, H.; Xie, B.R.; Qiu, W.X.; Zeng, J.Y.; Li, C.X.; Wan, S.S.; Zhang, L.; Liu, W.L.; Zhang, X.Z. Cancer Cell Membrane Camouflaged Cascade Bioreactor for Cancer Targeted Starvation and Photodynamic Therapy. ACS Nano 2017, 11, 7006–7018. [Google Scholar] [CrossRef]
- Li, S.Y.; Cheng, H.; Qiu, W.X.; Zhang, L.; Wan, S.S.; Zeng, J.Y.; Zhang, X.Z. Cancer cell membrane-coated biomimetic platform for tumor targeted photodynamic therapy and hypoxia-amplified bioreductive therapy. Biomaterials 2017, 142, 149–161. [Google Scholar] [CrossRef]
- Gao, L.; Wang, H.; Nan, L.; Peng, T.; Sun, L.; Zhou, J.; Xiao, Y.; Wang, J.; Sun, J.; Lu, W.; et al. Erythrocyte Membrane-Wrapped pH Sensitive Polymeric Nanoparticles for Non-Small Cell Lung Cancer Therapy. Bioconjug. Chem. 2017, 28, 2591–2598. [Google Scholar] [CrossRef]
- Gao, M.; Liang, C.; Song, X.; Chen, Q.; Jin, Q.; Wang, C.; Liu, Z. Erythrocyte-Membrane-Enveloped Perfluorocarbon as Nanoscale Artificial Red Blood Cells to Relieve Tumor Hypoxia and Enhance Cancer Radiotherapy. Adv. Mater. 2017, 29. [Google Scholar] [CrossRef]
- Dehaini, D.; Wei, X.; Fang, R.H.; Masson, S.; Angsantikul, P.; Luk, B.T.; Zhang, Y.; Ying, M.; Jiang, Y.; Kroll, A.V.; et al. Erythrocyte-Platelet Hybrid Membrane Coating for Enhanced Nanoparticle Functionalization. Adv. Mater. 2017, 29. [Google Scholar] [CrossRef] [Green Version]
- Dai, Y.; Xu, C.; Sun, X.; Chen, X. Nanoparticle design strategies for enhanced anticancer therapy by exploiting the tumour microenvironment. Chem. Soc. Rev. 2017, 46, 3830–3852. [Google Scholar] [CrossRef]
- Zhang, N.; Li, M.; Sun, X.; Jia, H.; Liu, W. NIR-responsive cancer cytomembrane-cloaked carrier-free nanosystems for highly efficient and self-targeted tumor drug delivery. Biomaterials 2018, 159, 25–36. [Google Scholar] [CrossRef]
- Jiao, M.; Zhang, P.; Meng, J.; Li, Y.; Liu, C.; Luo, X.; Gao, M. Recent advancements in biocompatible inorganic nanoparticles towards biomedical applications. Biomater. Sci. 2018, 6, 726–745. [Google Scholar] [CrossRef]
- Qiu, W.X.; Zhang, M.K.; Liu, L.H.; Gao, F.; Zhang, L.; Li, S.Y.; Xie, B.R.; Zhang, C.; Feng, J.; Zhang, X.Z. A self-delivery membrane system for enhanced anti-tumor therapy. Biomaterials 2018, 161, 81–94. [Google Scholar] [CrossRef]
- Hu, C.M.; Fang, R.H.; Wang, K.C.; Luk, B.T.; Thamphiwatana, S.; Dehaini, D.; Nguyen, P.; Angsantikul, P.; Wen, C.H.; Kroll, A.V.; et al. Nanoparticle biointerfacing by platelet membrane cloaking. Nature 2015, 526, 118–121. [Google Scholar] [CrossRef]
- Hu, Q.; Sun, W.; Qian, C.; Wang, C.; Bomba, H.N.; Gu, Z. Anticancer Platelet-Mimicking Nanovehicles. Adv. Mater. 2015, 27, 7043–7050. [Google Scholar] [CrossRef]
- Thanh Nguyen, T.D.; Pitchaimani, A.; Koirala, M.B.; Muhammad, F.; Aryal, S. Engineered biomimetic nanoabsorbent for cellular detoxification of chemotherapeutics. RSC Adv. 2016, 6, 33003–33008. [Google Scholar] [CrossRef]
- Hu, C.M.; Zhang, L.; Aryal, S.; Cheung, C.; Fang, R.H.; Zhang, L. Erythrocyte membrane-camouflaged polymeric nanoparticles as a biomimetic delivery platform. Proc. Natl. Acad. Sci. USA 2011, 108, 10980–10985. [Google Scholar] [CrossRef] [Green Version]
- Parodi, A.; Quattrocchi, N.; van de Ven, A.L.; Chiappini, C.; Evangelopoulos, M.; Martinez, J.O.; Brown, B.S.; Khaled, S.Z.; Yazdi, I.K.; Enzo, M.V.; et al. Synthetic nanoparticles functionalized with biomimetic leukocyte membranes possess cell-like functions. Nat. Nanotechnol. 2013, 8, 61–68. [Google Scholar] [CrossRef]
- Cao, H.; Dan, Z.; He, X.; Zhang, Z.; Yu, H.; Yin, Q.; Li, Y. Liposomes Coated with Isolated Macrophage Membrane Can Target Lung Metastasis of Breast Cancer. ACS Nano 2016, 10, 7738–7748. [Google Scholar] [CrossRef]
- Kang, T.; Zhu, Q.; Wei, D.; Feng, J.; Yao, J.; Jiang, T.; Song, Q.; Wei, X.; Chen, H.; Gao, X.; et al. Nanoparticles Coated with Neutrophil Membranes Can Effectively Treat Cancer Metastasis. ACS Nano 2017, 11, 1397–1411. [Google Scholar] [CrossRef]
- Tang, J.; Shen, D.; Caranasos, T.G.; Wang, Z.; Vandergriff, A.C.; Allen, T.A.; Hensley, M.T.; Dinh, P.U.; Cores, J.; Li, T.S.; et al. Therapeutic microparticles functionalized with biomimetic cardiac stem cell membranes and secretome. Nat. Commun. 2017, 8, 13724. [Google Scholar] [CrossRef]
- Gao, C.; Lin, Z.; Jurado-Sanchez, B.; Lin, X.; Wu, Z.; He, Q. Stem Cell Membrane-Coated Nanogels for Highly Efficient In Vivo Tumor Targeted Drug Delivery. Small 2016, 12, 4056–4062. [Google Scholar] [CrossRef]
- Rao, L.; Bu, L.L.; Cai, B.; Xu, J.H.; Li, A.; Zhang, W.F.; Sun, Z.J.; Guo, S.S.; Liu, W.; Wang, T.H.; et al. Cancer Cell Membrane-Coated Upconversion Nanoprobes for Highly Specific Tumor Imaging. Adv. Mater. 2016, 28, 3460–3466. [Google Scholar] [CrossRef]
- Chen, Z.; Zhao, P.; Luo, Z.; Zheng, M.; Tian, H.; Gong, P.; Gao, G.; Pan, H.; Liu, L.; Ma, A.; et al. Cancer Cell Membrane-Biomimetic Nanoparticles for Homologous-Targeting Dual-Modal Imaging and Photothermal Therapy. ACS Nano 2016, 10, 10049–10057. [Google Scholar] [CrossRef]
- Fang, R.H.; Hu, C.M.; Luk, B.T.; Gao, W.; Copp, J.A.; Tai, Y.; O’Connor, D.E.; Zhang, L. Cancer cell membrane-coated nanoparticles for anticancer vaccination and drug delivery. Nano Lett. 2014, 14, 2181–2188. [Google Scholar] [CrossRef]
- Sun, H.; Su, J.; Meng, Q.; Yin, Q.; Chen, L.; Gu, W.; Zhang, P.; Zhang, Z.; Yu, H.; Wang, S.; et al. Cancer-Cell-Biomimetic Nanoparticles for Targeted Therapy of Homotypic Tumors. Adv. Mater. 2016, 28, 9581–9588. [Google Scholar] [CrossRef]
- Feng, L.; Yang, X.; Shi, X.; Tan, X.; Peng, R.; Wang, J.; Liu, Z. Polyethylene glycol and polyethylenimine dual-functionalized nano-graphene oxide for photothermally enhanced gene delivery. Small 2013, 9, 1989–1997. [Google Scholar] [CrossRef]
- Huang, Z.P.; Geyer, N.; Werner, P.; de Boor, J.; Gosele, U. Metal-Assisted Chemical Etching of Silicon: A Review. Adv. Mater. 2011, 23, 285–308. [Google Scholar] [CrossRef]
- Sun, T.; Tsang, W.M. 4 - Nanowires for biomedical applications. In Nanobiomaterials; Narayan, R., Ed.; Woodhead Publishing: Sawston, UK, 2018. [Google Scholar]
- Wagner, R.S.; Ellis, W.C. Vapor-liquid-solid mechanism of single crystal growth. Appl. Phys. Lett. 1964, 4, 89–90. [Google Scholar] [CrossRef]
- Mai, J.C.; Shen, H.; Watkins, S.C.; Cheng, T.; Robbins, P.D. Efficiency of protein transduction is cell type-dependent and is enhanced by dextran sulfate. J. Biol. Chem. 2002, 277, 30208–30218. [Google Scholar] [CrossRef]
- He, B.; Yang, D.; Qin, M.; Zhang, Y.; He, B.; Dai, W.; Wang, X.; Zhang, Q.; Zhang, H.; Yin, C. Increased cellular uptake of peptide-modified PEGylated gold nanoparticles. Biochem. Biophys. Res. Commun. 2017, 494, 339–345. [Google Scholar] [CrossRef]
- Dash, B.C.; Rethore, G.; Monaghan, M.; Fitzgerald, K.; Gallagher, W.; Pandit, A. The influence of size and charge of chitosan/polyglutamic acid hollow spheres on cellular internalization, viability and blood compatibility. Biomaterials 2010, 31, 8188–8197. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, R.; Singh, V.; Jurney, P.; Shi, L.; Sreenivasan, S.V.; Roy, K. Mammalian cells preferentially internalize hydrogel nanodiscs over nanorods and use shape-specific uptake mechanisms. Proc. Natl. Acad. Sci. USA 2013, 110, 17247–17252. [Google Scholar] [CrossRef] [Green Version]
- Lesniak, A.; Salvati, A.; Santos-Martinez, M.J.; Radomski, M.W.; Dawson, K.A.; Aberg, C. Nanoparticle adhesion to the cell membrane and its effect on nanoparticle uptake efficiency. J. Am. Chem. Soc. 2013, 135, 1438–1444. [Google Scholar] [CrossRef]
- Hinde, E.; Thammasiraphop, K.; Duong, H.T.; Yeow, J.; Karagoz, B.; Boyer, C.; Gooding, J.J.; Gaus, K. Pair correlation microscopy reveals the role of nanoparticle shape in intracellular transport and site of drug release. Nat. Nanotechnol. 2017, 12, 81–89. [Google Scholar] [CrossRef]
- Yu, M.; Wang, J.; Yang, Y.; Zhu, C.; Su, Q.; Guo, S.; Sun, J.; Gan, Y.; Shi, X.; Gao, H. Rotation-Facilitated Rapid Transport of Nanorods in Mucosal Tissues. Nano Lett. 2016, 16, 7176–7182. [Google Scholar] [CrossRef]
- Decuzzi, P.; Godin, B.; Tanaka, T.; Lee, S.Y.; Chiappini, C.; Liu, X.; Ferrari, M. Size and shape effects in the biodistribution of intravascularly injected particles. J. Control. Release 2010, 141, 320–327. [Google Scholar] [CrossRef]
- Amin, M.L.; Joo, J.Y.; Yi, D.K.; An, S.S. Surface modification and local orientations of surface molecules in nanotherapeutics. J. Control. Release 2015, 207, 131–142. [Google Scholar] [CrossRef]
- Todorova, N.; Chiappini, C.; Mager, M.; Simona, B.; Patel, I.I.; Stevens, M.M.; Yarovsky, I. Surface presentation of functional peptides in solution determines cell internalization efficiency of TAT conjugated nanoparticles. Nano Lett. 2014, 14, 5229–5237. [Google Scholar] [CrossRef]
- Hundal, H.S.; Maxwell, D.L.; Ahmed, A.; Darakhshan, F.; Mitsumoto, Y.; Klip, A. Subcellular distribution and immunocytochemical localization of Na,K-ATPase subunit isoforms in human skeletal muscle. Mol. Membr. Biol. 1994, 11, 255–262. [Google Scholar] [CrossRef]
- Lampugnani, M.G. Endothelial cell-to-cell junctions: Adhesion and signaling in physiology and pathology. Cold Spring Harb. Perspect. Med. 2012, 2. [Google Scholar] [CrossRef]
- Bausch, D.; Thomas, S.; Mino-Kenudson, M.; Fernandez-del, C.C.; Bauer, T.W.; Williams, M.; Warshaw, A.L.; Thayer, S.P.; Kelly, K.A. Plectin-1 as a novel biomarker for pancreatic cancer. Clin. Cancer Res. 2011, 17, 302–309. [Google Scholar] [CrossRef]
- Wiche, G.; Osmanagic-Myers, S.; Castanon, M.J. Networking and anchoring through plectin: A key to IF functionality and mechanotransduction. Curr. Opin. Cell Biol. 2015, 32, 21–29. [Google Scholar] [CrossRef]
- Karantza, V. Keratins in health and cancer: More than mere epithelial cell markers. Oncogene 2011, 30, 127–138. [Google Scholar] [CrossRef]
- Stengel, K.; Zheng, Y. Cdc42 in oncogenic transformation, invasion, and tumorigenesis. Cell Signal. 2011, 23, 1415–1423. [Google Scholar] [CrossRef] [Green Version]
- Lastres-Becker, I.; Brodesser, S.; Lutjohann, D.; Azizov, M.; Buchmann, J.; Hintermann, E.; Sandhoff, K.; Schurmann, A.; Nowock, J.; Auburger, G. Insulin receptor and lipid metabolism pathology in ataxin-2 knock-out mice. Hum. Mol. Genet. 2008, 17, 1465–1481. [Google Scholar] [CrossRef] [Green Version]
- Nonis, D.; Schmidt, M.H.; van de Loo, S.; Eich, F.; Dikic, I.; Nowock, J.; Auburger, G. Ataxin-2 associates with the endocytosis complex and affects EGF receptor trafficking. Cell Signal. 2008, 20, 1725–1739. [Google Scholar] [CrossRef]
- Bar, D.Z.; Charar, C.; Dorfman, J.; Yadid, T.; Tafforeau, L.; Lafontaine, D.L.; Gruenbaum, Y. Cell size and fat content of dietary-restricted Caenorhabditis elegans are regulated by ATX-2, an mTOR repressor. Proc. Natl. Acad. Sci. USA 2016, 113, E4620–E4629. [Google Scholar] [CrossRef]
- Bertrand, Y.; Demeule, M.; Michaud-Levesque, J.; Beliveau, R. Melanotransferrin induces human melanoma SK-Mel-28 cell invasion in vivo. Biochem. Biophys. Res. Commun. 2007, 353, 418–423. [Google Scholar] [CrossRef]
- Suryo Rahmanto, Y.; Dunn, L.L.; Richardson, D.R. The melanoma tumor antigen, melanotransferrin (p97): A 25-year hallmark--from iron metabolism to tumorigenesis. Oncogene 2007, 26, 6113–6124. [Google Scholar] [CrossRef]
- Demeule, M.; Bertrand, Y.; Michaud-Levesque, J.; Jodoin, J.; Rolland, Y.; Gabathuler, R.; Beliveau, R. Regulation of plasminogen activation: A role for melanotransferrin (p97) in cell migration. Blood 2003, 102, 1723–1731. [Google Scholar] [CrossRef]
- Alvarez, R.H.; Kantarjian, H.M.; Cortes, J.E. The role of Src in solid and hematologic malignancies: Development of new-generation Src inhibitors. Cancer 2006, 107, 1918–1929. [Google Scholar] [CrossRef]
- Martin, G.S. The hunting of the Src. Nat. Rev. Mol. Cell Biol. 2001, 2, 467–475. [Google Scholar] [CrossRef]
- Ivanov, A.I. Pharmacological inhibition of endocytic pathways: Is it specific enough to be useful? Methods Mol. Biol. 2008, 440, 15–33. [Google Scholar] [CrossRef]
- Ji, X.; Wang, H.; Song, B.; Chu, B.; He, Y. Silicon Nanomaterials for Biosensing and Bioimaging Analysis. Front. Chem. 2018, 6, 38. [Google Scholar] [CrossRef] [Green Version]
- De Rose, R.; Zelikin, A.N.; Johnston, A.P.R.; Sexton, A.; Chong, S.-F.; Cortez, C.; Mulholland, W.; Caruso, F.; Kent, S.J. Binding, Internalization, and Antigen Presentation of Vaccine-Loaded Nanoengineered Capsules in Blood. Adv. Mater. 2008, 20, 4698–4703. [Google Scholar] [CrossRef]
- Park, D.; Tosello-Trampont, A.C.; Elliott, M.R.; Lu, M.; Haney, L.B.; Ma, Z.; Klibanov, A.L.; Mandell, J.W.; Ravichandran, K.S. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature 2007, 450, 430–434. [Google Scholar] [CrossRef] [Green Version]
- Phanse, Y.; Ramer-Tait, A.E.; Friend, S.L.; Carrillo-Conde, B.; Lueth, P.; Oster, C.J.; Phillips, G.J.; Narasimhan, B.; Wannemuehler, M.J.; Bellaire, B.H. Analyzing cellular internalization of nanoparticles and bacteria by multi-spectral imaging flow cytometry. J. Vis. Exp. 2012. [Google Scholar] [CrossRef]
- Doherty, G.J.; McMahon, H.T. Mechanisms of Endocytosis. Annu. Rev. Biochem. 2009, 78, 857–902. [Google Scholar] [CrossRef] [Green Version]
- Canton, I.; Battaglia, G. Endocytosis at the nanoscale. Chem. Soc. Rev. 2012, 41, 2718–2739. [Google Scholar] [CrossRef]
- Sarkar, K.; Madras, G.; Chatterjee, K. Dendron conjugation to graphene oxide using click chemistry for efficient gene delivery. RSC Adv. 2015, 5, 50196–50211. [Google Scholar] [CrossRef] [Green Version]
- Piperno, A.; Scala, A.; Mazzaglia, A.; Neri, G.; Pennisi, R.; Sciortino, M.T.; Grassi, G. Cellular Signaling Pathways Activated by Functional Graphene Nanomaterials. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef]








| SiNR | Diameter (μm) | Length (μm) |
|---|---|---|
| L-size | 0.412 ± 0.027 | 3.495 ± 0.181 |
| M-size | 0.409 ± 0.010 | 2.229 ± 0.049 |
| S-size | 0.105 ± 0.011 | 0.656 ± 0.126 |
| Cell line | Genes |
|---|---|
| Hela | GNAI1 CAV2 SYAP1 LYN JAK1 |
| MDA231 | SAA1 LOXL2 VTN AHI1 HIP1 THBS1 TGFB2 ITGA5 FAM129B EIF3E |
| PDX | MELTF SRC |
| MCF7 | BCL2 GSK3B BAX RHOT2 |
| Panc-1 | ATXN2 VASP ARPC5L ARPC5 DCTN1 PXN LIMA1 MYH10 |
| Cell Type | GO ID | GO Term | Adjust p-Value |
|---|---|---|---|
| Panc-1 | 0001725 | stress fiber | 6.87 × 10−4 |
| 0003779 | actin binding | 1.06 × 10−4 | |
| 0005916 | fascia adherens | 7.08 × 10−4 | |
| 0007015 | actin filament organization | 7.46 × 10−4 | |
| 0008064 | regulation of actin polymerization or depolymerization | 1.36 × 10−3 | |
| 0008154 | actin polymerization or depolymerization | 1.78 × 10−3 | |
| 0030041 | actin filament polymerization | 4.13 × 10−3 | |
| 0030833 | regulation of actin filament polymerization | 2.83 × 10−3 | |
| 0030838 | positive regulation of actin filament polymerization | 2.86 × 10−3 | |
| 0045806 | negative regulation of endocytosis | 5.65 × 10−3 | |
| 0051495 | positive regulation of cytoskeleton organization | 2.49 × 10−3 | |
| 0097517 | contractile actin filament bundle | 6.87 × 10−4 | |
| 1902903 | regulation of supramolecular fiber organization | 4.18 × 10−3 | |
| 1902905 | positive regulation of supramolecular fiber organization | 4.21 × 10−4 | |
| Panc-PDX | 0031579 | membrane raft organization | 5.72 × 10−3 |
| 0034446 | substrate adhesion-dependent cell spreading | 1.90 × 10−3 | |
| 0035635 | entry of bacterium into host cell | 7.88 × 10−3 | |
| 1900024 | regulation of substrate adhesion-dependent cell spreading | 8.61 × 10−4 | |
| MCF-7 | 1905710 | positive regulation of membrane permeability | 5.32 × 10−5 |
| MDA231 | 0019898 | extrinsic component of membrane | 6.14 × 10−3 |
| 0051015 | actin filament binding | 4.10 × 10−3 | |
| 0005178 | integrin binding | 3.51 × 10−3 | |
| 0005884 | actin filament | 1.13 × 10−3 | |
| 0006898 | receptor-mediated endocytosis | 1.93 × 10−3 | |
| 0050840 | extracellular matrix binding | 8.92 × 10−3 | |
| 0033627 | cell adhesion mediated by integrin | 7.46 × 10−3 | |
| 0048259 | regulation of receptor-mediated endocytosis | 2.20 × 10−3 | |
| 0048260 | positive regulation of receptor-mediated endocytosis | 1.96 × 10−3 | |
| Hela | 0031234 | extrinsic component of cytoplasmic side of plasma membrane | 4.50 × 10−3 |
| 0044406 | adhesion of symbiont to host | 5.66 × 10−3 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Y.; Dai, J.; Kong, N.; Liu, J.; Gong, J.; Yao, Y. Internalization Characterization of Si Nanorod with Camouflaged Cell Membrane Proteins Reveals ATXN2 as a Negative Regulator. Cells 2019, 8, 931. https://doi.org/10.3390/cells8080931
Lu Y, Dai J, Kong N, Liu J, Gong J, Yao Y. Internalization Characterization of Si Nanorod with Camouflaged Cell Membrane Proteins Reveals ATXN2 as a Negative Regulator. Cells. 2019; 8(8):931. https://doi.org/10.3390/cells8080931
Chicago/Turabian StyleLu, Yi, Jing Dai, Na Kong, Jianghuai Liu, Jinkang Gong, and Yuan Yao. 2019. "Internalization Characterization of Si Nanorod with Camouflaged Cell Membrane Proteins Reveals ATXN2 as a Negative Regulator" Cells 8, no. 8: 931. https://doi.org/10.3390/cells8080931
APA StyleLu, Y., Dai, J., Kong, N., Liu, J., Gong, J., & Yao, Y. (2019). Internalization Characterization of Si Nanorod with Camouflaged Cell Membrane Proteins Reveals ATXN2 as a Negative Regulator. Cells, 8(8), 931. https://doi.org/10.3390/cells8080931