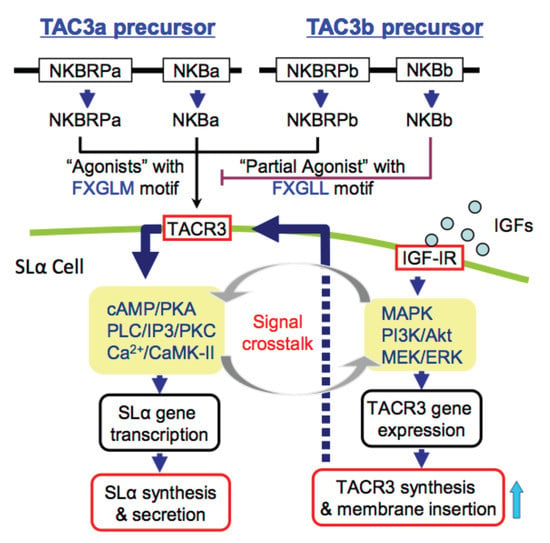
IGFs Potentiate TAC3-induced SLα Expression via Upregulation of TACR3 Expression in Grass Carp Pituitary Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Reagents
2.2. Molecular Cloning, Tissue Expression, and Structural Analysis of Grass Carp TACR3
2.3. Functional Expression of Grass Carp TACR3 in HEK-293 Cells
2.4. Receptor Binding Assay for Grass Carp TACR3
2.5. Fluorescence Imaging of TACR3 Binding
2.6. RT-PCR for TACR3 and IGF-IR Expression in Immune-Identified Somatolactin (SL) Cells
2.7. Measurement of SLα and TACR3 mRNA Expression
2.8. Western Blot for Signaling Kinases
2.9. Data Transformation and Statistics
3. Results
3.1. Molecular Cloning and Tissue Distribution of Grass Carp TACR3
3.2. Functional Expression of Grass Carp TACR3
3.3. IGF-Induced TACR3 mRNA Expression in Grass Carp Pituitary Cells
Synergistic Effects of IGF and TAC3 Gene Products on SLα mRNA Expression
3.4. Signal Transduction for the Synergistic Effect of IGFs and TACR3 Activation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AC | Adenylyl cyclase |
| Akt | protein kinase B |
| CaM | Calmodulin |
| CaMK-II | CaM-dependent protein kinase II |
| DAG | diacylglycerol |
| ERK | extracellular signal-regulated kinase |
| FAM | fluorescein amidite |
| GPCR | G protein coupled receptor |
| Gq | G-protein alpha subunit, group Q |
| IGF | insulin-like growth factor |
| IP3 | Inositol 1,4,5-triphosphate |
| LCM | laser capture microdissection |
| MAPK | mitogen-activated protein kinase |
| mTOR | mammalian target of rapamycin |
| TACR3 | tachykinin receptor 3 |
| NIL | neurointermediate lobe |
| NKB | neurokinin B |
| NKBRP | NKB-related peptide |
| PACAP | pituitary adenylate cyclase activating peptide |
| PI3K | phosphatidylinositol-3-kinase |
| PKC | Protein kinase C |
| PKA | Protein kinase A |
| PLC | phospholipase C |
| RLU | relative luminescence unit |
| SL | Somatolactin |
| SP | Substance P |
| TAC3 | tachykinin 3 |
| TACR3 | tachykinin receptor 3 |
| VSCC | voltage sensitive calcium channel |
References
- Satake, H.; Aoyama, M.; Sekiguchi, T.; Kawada, T. Insight into molecular and functional diversity of tachykinins and their receptors. Protein. Pept. Lett. 2013, 20, 615–627. [Google Scholar] [CrossRef] [PubMed]
- Guran, T.; Tolhurst, G.; Bereket, A.; Rocha, K.; Porter, K.; Turan, S.; Gribble, F.M.; Kotan, L.D.; Akcay, T.; Atay, Z.; et al. Hypogonadotropic hypogonadism due to a novel missense mutation in the first rxtracellular loop of the neurokinin B receptor. J. Clin. Endocr. Metab. 2009, 94, 3633–3639. [Google Scholar] [CrossRef] [PubMed]
- Topaloglu, A.K.; Reimann, F.; Guclu, M.; Yalin, A.S.; Kotan, L.D.; Porter, K.M.; Serin, A.; Mungan, N.O.; Cook, J.R.; Imamoglu, S.; et al. TAC3 and TACR3 mutations in familial hypogonadotropic hypogonadism reveal a key role for Neurokinin B in the central control of reproduction. Nat. Genet. 2009, 41, 354–358. [Google Scholar] [CrossRef] [PubMed]
- Young, J.; Bouligand, J.; Francou, B.; Raffin-Sanson, M.L.; Gaillez, S.; Jeanpierre, M.; Grynberg, M.; Kamenicky, P.; Chanson, P.; Brailly-Tabard, S.; et al. TAC3 and TACR3 defects cause hypothalamic congenital hypogonadotropic hypogonadism in humans. J. Clin. Endocrinol. Metab. 2010, 95, 2287–2295. [Google Scholar] [CrossRef] [PubMed]
- Biran, J.; Palevitch, O.; Ben-Dor, S.; Levavi-Sivan, B. Neurokinin Bs and neurokinin B receptors in zebrafish-potential role in controlling fish reproduction. Proc. Natl. Acad. Sci. USA 2012, 109, 10269–10274. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Li, S.; Liu, Y.; Qi, X.; Chen, H.; Cheng, C.H.; Liu, X.; Zhang, Y.; Lin, H. The evolution of tachykinin/tachykinin receptor (TAC/TACR) in vertebrates and molecular identification of the TAC3/TACR3 system in zebrafish (Danio rerio). Mol. Cell. Endocrinol. 2012, 361, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Salem, M.; Zhou, W.; Sato-Shimizu, M.; Ye, G.; Smitz, J.; Peng, C. Neurokinin B exerts direct effects on the ovary to stimulate estradiol production. Endocrinology 2016, 157, 3355–3365. [Google Scholar] [CrossRef]
- Hu, G.F.; He, M.L.; Ko, W.K.; Lin, C.Y.; Wong, A.O.L. Novel pituitary actions of TAC3 gene products in fish model: -Receptor specificity and signal transduction for prolactin and somatolactin alpha regulation by neurokinin B (NKB) and NKB-related peptide in carp pituitary cells. Endocrinology 2014, 155, 3582–3596. [Google Scholar] [CrossRef]
- Meyer, A.; Van de Peer, Y. From 2R to 3R: Evidence for a fish-specific genome duplication (FSGD). Bioessays 2005, 27, 937–945. [Google Scholar] [CrossRef]
- Frago, L.M.; Chowen, J.A. Basic physiology of the growth hormone/insulin-like growth factor axis. Adv. Exp. Med. Biol. 2005, 567, 1–25. [Google Scholar]
- Le Roith, D. The insulin-like growth factor system. Exp. Diabesity Res. 2003, 4, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Daughaday, W.H. Growth hormone axis overview: Somatomedin hypothesis. Pediatr. Nephrol. 2000, 14, 537–540. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, S.; Ayson, F.G.; Kawauchi, H. Growth regulation by insulin-like growth factor-I in fish. Biosci. Biotechnol. Biochem. 2000, 64, 1553–1562. [Google Scholar] [CrossRef] [PubMed]
- Rousseau, K.; Huang, Y.S.; Le Belle, N.; Vidal, B.; Marchelido, J.; Epelbaum, J.; Dufour, S. Long-term inhibitory effects of somatostatin and insulin-like growth factor 1 on growth hormone release by serum free primary culture of pituitary cells from European eel (Anguilla anguilla). Neuroendocrinology 1998, 67, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Huo, L.; Fu, G.; Wang, X.; Ko, W.K.; Wong, A.O. Modulation of calmodulin gene expression as a novel mechanism for growth hormone feedback control by insulin-like growth factor in grass carp pituitary cells. Endocrinology 2005, 146, 3821–3835. [Google Scholar] [CrossRef] [PubMed]
- Ono, M.; Takayama, Y.; Rand-Weaver, M.; Sakata, S.; Yasunaga, T.; Noso, T.; Kawauchi, H. cDNA cloning of somatolactin, a pituitary protein related to growth hormone and prolactin. Proc. Natl. Acad. Sci. USA 1990, 87, 4330–4334. [Google Scholar] [CrossRef]
- Zhu, Y.; Stiller, J.W.; Shaner, M.P.; Baldini, A.; Scemama, J.L.; Capehart, A.A. Cloning of somatolactin alpha and beta cDNAs in zebrafish and phylogenetic analysis of two distinct somatolactin subtypes in fish. J. Endocrinol. 2004, 182, 509–518. [Google Scholar] [CrossRef]
- Kawauchi, H.; Sower, S.A.; Moriyama, S. The neuroendocrine regulation of prolactin and somatolactin secretion in fish. Fish Physiol. 2009, 28, 197–234. [Google Scholar]
- Jiang, Q.; Ko, W.K.; Wong, A.O. Insulin-like growth factor as a novel stimulator for somatolactin secretion and synthesis in carp pituitary cells via activation of MAPK cascades. Am. J. Physiol. Endocrinol. Metab. 2011, 301, E1208–E1219. [Google Scholar] [CrossRef] [Green Version]
- Rozengurt, E.; Sinnett-Smith, J.; Kisfalvi, K. Crosstalk between insulin/insulin-like growth factor-1 receptors and G protein-coupled receptor signaling systems: A novel target for the antidiabetic drug metformin in pancreatic cancer. Clin. Cancer Res. 2010, 16, 2505–2511. [Google Scholar] [CrossRef]
- Kisfalvi, K.; Rey, O.; Young, S.H.; Sinnett-Smith, J.; Rozengurt, E. Insulin potentiates Ca2+ signaling and phosphatidylinositol 4,5-bisphosphate hydrolysis induced by Gq protein-coupled agonists through an mTOR-dependent pathway. Endocrinology 2007, 148, 3246–3257. [Google Scholar] [CrossRef]
- Kisfalvi, K.; Eibl, G.; Sinnett-Smith, J.; Rozengurt, E. Metformin disrupts crosstalk between G protein-coupled receptor and isulin receptor signaling systems and inhibits pancreatic cancer growth. Cancer Res. 2009, 69, 6539–6545. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Ofuji, K.; Chikama, T.; Nishida, T. Combined effects of substance P and insulin-like growth factor-1 on corneal epithelial wound closure of rabbit in vivo. Curr. Eye Res. 1997, 16, 275–278. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Chikama, T.; Nishida, T. Up-regulation of integrin α5 expression by combination of substance P and insulin-like growth factor-1 in rabbit corneal epithelial cells. Biochem. Bioph. Res. Co. 1998, 246, 777–782. [Google Scholar] [CrossRef]
- Yamada, N.; Yanai, R.; Inui, M.; Nishida, T. Sensitizing effect of substance P on corneal epithelial migration induced by IGF-I, fibronectin, or interleukin-6. Invest. Ophthalmol. Vis. Sci. 2005, 46, 833–839. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.F.; He, M.L.; Wong, A.O.L. Novel functional role of NK3R expression in the potentiating effects on somatolactin α autoregulation in grass carp pituitary cells. Sci. Rep. 2016, 6, 36102. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: An automated protein homology- modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef]
- Cheng, Z.J.; Garvin, D.; Paguio, A.; Stecha, P.; Wood, K.; Fan, F. Luciferase reporter assay system for deciphering GPCR pathways. Curr. Chem. Genomics 2010, 4, 84–91. [Google Scholar] [CrossRef]
- Lu, H.L.; Kersch, C.N.; Taneja-Bageshwar, S.; Pietrantonio, P.V. A calcium bioluminescence assay for functional analysis of mosquito (Aedes aegypti) and Tick (Rhipicephalus microplus) G protein-coupled receptors. J. Vis. Exp. 2011, 50, e2732. [Google Scholar] [CrossRef]
- Wong, A.O.; Ng, S.; Lee, E.K.; Leung, R.C.; Ho, W.K. Somatostatin inhibits (D-Arg6, Pro9-NEt) salmon gonadotropin-releasing hormone- and dopamine D1- stimulated growth hormone release from perifused pituitary cells of Chinese grass carp, Ctenopharyngodon idellus. Gen. Comp. Endocrinol. 1998, 110, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Ko, W.K.; Ho, W.K.; Stojilkovic, S.S.; Wong, A.O. Novel aspects of growth hormone (GH) autoregulation: GH-induced GH gene expression in grass carp pituitary cells through autocrine/paracrine mechanisms. Endocrinology 2004, 145, 4615–4628. [Google Scholar] [CrossRef] [PubMed]
- Grachev, P.; Li, X.F.; Kinsey-Jones, J.S.; di Domenico, A.L.; Millar, R.P.; Lightman, S.L.; O’Byrne, K.T. Suppression of the GnRH pulse generator by neurokinin B involves a kappa-opioid receptor-dependent mechanism. Endocrinology 2012, 153, 4894–4904. [Google Scholar] [CrossRef] [PubMed]
- Goodman, R.L.; Hileman, S.M.; Nestor, C.C.; Porter, K.L.; Connors, J.M.; Hardy, S.L.; Milar, R.P.; Cernea, M.; Coolen, L.M.; Lehman, M.N. Kisspeptin, neurokinin B, and dynorphin act in the arcuate nucleus to control activity of the GnRH pulse generator in ewes. Endocrinology 2013, 154, 4259–4269. [Google Scholar] [CrossRef] [PubMed]
- Ruka, K.A.; Burger, L.L.; Moenter, S.M. Regulation of arcuate neurons coexpressing kisspeptin, neurokinin B, and dynorphin by modulators of neurokinin 3 and κ-opioid receptors in adult male mice. Endocrinology 2013, 154, 2761–2771. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, Y.; Matsuyama, H.; Shiina, T.; Takewaki, T.; Furness, J.B. Tachykinins and their functions in the gastrointestinal tract. Cell. Mol. Life Sci. 2008, 65, 295–311. [Google Scholar] [CrossRef] [PubMed]
- Thompson, G.W.; Hoover, D.B.; Ardell, J.L.; Armour, J.A. Canine intrinsic cardiac neurons involved in cardiac regulation possess NK1, NK2, and NK3 receptors. Am. J. Physiol. 1998, 275, R1683–R1689. [Google Scholar] [CrossRef] [PubMed]
- Lessard, A.; Campos, M.M.; Neugebauer, W.; Couture, R. Implication of nigral tachykinin NK3 receptors in the maintenance of hypertension in spontaneously hypertensive rats: A pharmacologic and autoradiographic study. Br. J. Pharmacol. 2003, 138, 554–563. [Google Scholar] [CrossRef]
- Mastrangelo, D.; Mathison, R.; Huggel, H.J.; Dion, S.; D’Orleans-Juste, P.; Rhaleb, N.E.; Drapeau, G.; Rovero, P.; Regoli, D. The rat isolated portal vein: A preparation sensitive to neurokinins, particularly neurokinin B. Eur. J. Pharmacol. 1987, 134, 321–326. [Google Scholar] [CrossRef]
- Cejudo Roman, A.; Pinto, F.M.; Dorta, I.; Almeida, T.A.; Hernandez, M.; Illanes, N.; Tena-Sempere, M.; Candenas, L. Analysis of the expression of neurokinin B, kisspeptin, and their cognate receptors NK3R and KISS1R in the human female genital tract. Fertil. Steril. 2012, 97, 1213–1219. [Google Scholar] [CrossRef]
- Pinto, F.M.; Ravina, C.G.; Subiran, N.; Cejudo-Roman, A.; Fernandez-Sanchez, M.; Irazusta, J.; Garrido, N.; Candenas, L. Autocrine regulation of human sperm motility by tachykinins. Reprod. Biol. Endocrinol. 2010, 8, 104. [Google Scholar] [CrossRef] [PubMed]
- Ravina, C.G.; Seda, M.; Pinto, F.M.; Orea, A.; Fernandez-Sanchez, M.; Pintado, C.O.; Candenas, M.L. A role for tachykinins in the regulation of human sperm motility. Hum. Reprod. 2007, 22, 1617–1625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almeida, T.A.; Rojo, J.; Nieto, P.M.; Pinto, F.M.; Hernandez, M.; Martin, J.D.; Canenas, M.L. Tachykinins and tachykinin receptors: Structure and activity relationships. Curr. Med. Chem. 2004, 11, 2045–2081. [Google Scholar] [CrossRef] [PubMed]
- Shigemoto, R.; Yokota, Y.; Tsuchida, K.; Nakanishi, S. Cloning and expression of a rat neurokinin-K receptor cDNA. J. Biol. Chem. 1990, 265, 623–628. [Google Scholar] [PubMed]
- Lelbach, A.; Muzes, G.; Feher, J. The insulin-like growth factor system: IGFs, IGF binding proteins and IGFBP-proteases. Acta Physiol Hung. 2005, 92, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Fruchtman, S.; Jackson, L.; Borski, R. Insulin-like growth factor I disparately regulates prolactin and growth hormone synthesis and secretion: Studies using the teleost pituitary model. Endocrinology 2000, 141, 2886–2894. [Google Scholar] [CrossRef] [PubMed]
- Baker, D.M.; Davies, B.; Dickhoff, W.W.; Swanson, P. Insulin-like growth factor I increases follicle-stimulating hormone (FSH) content and gonadotropin-releasing hormone-stimulated FSH release from coho salmon pituitary cells in vitro. Biol Reprod. 2000, 63, 865–871. [Google Scholar] [CrossRef]
- Huang, Y.S.; Rousseau, K.; Le Belle, N.; Vidal, B.; Burzawa-Gerard, E.; Marchelidon, J.; Dufour, S. Insulin-like growth factor-I stimulates gonadotropin production from eel pituitary cells: A possible metabolic signal for induction of puberty. J. Endocrinol. 1998, 159, 43–52. [Google Scholar] [CrossRef]
- Jiang, Q.; He, M.L.; Wang, X.Y.; Wong, A.O.L. Grass carp somatolactin: II. Pharmacological study on postreceptor signaling mechanisms for PACAP-induced somatolactin α and β gene expression. Am. J. Physiol. Endocrinol. Metab. 2008, 295, E477–E490. [Google Scholar] [CrossRef]
- Matsuda, K.; Nejigaki, Y.; Satoh, M.; Shimaura, C.; Tanaka, M.; Kawamoto, K.; Uchiyama, M.; Kawauchi, H.; Shioda, S.; Takahashi, A. Effect of pituitary adenylate cyclase-activating polypeptide (PACAP) on prolactin and somatolactin release from the goldfish pituitary in vitro. Regul. Pept. 2008, 145, 72–79. [Google Scholar] [CrossRef]
- Jiang, Q.; Wong, A.O. Somatostatin-28 inhibitory action on somatolactin α and β gene expression in goldfish. Am. J. Physiol. Endocrinol. Metab. 2014, 307, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Wong, A.O.L. Signal transduction mechanisms for autocrine/paracrine regulation of somatolactin-α secretion and synthesis in carp pituitary cells by somatolactin-α and -β. Am. J. Physiol. Endocrinol. Metab. 2013, 304, E176–E186. [Google Scholar] [CrossRef] [PubMed]
- Chitnis, M.M.; Yuen, J.S.; Protheroe, A.S.; Pollak, M.; Macaulay, V.M. The type 1 insulin-like growth factor receptor pathway. Clin. Cancer Res. 2008, 14, 6364–6370. [Google Scholar] [CrossRef] [PubMed]
- Siddle, K. Signaling by insulin and IGF receptors: Supporting acts and new players. J. Mol. Endocrinol. 2011, 47, R1–R10. [Google Scholar] [CrossRef] [PubMed]







| Tachykinins | TAC1 | TAC3a | TAC3b | |||
|---|---|---|---|---|---|---|
| Luciferase Reporters | SP | NKA | NKBa | NKBRPa | NKBb | NKBRPb |
| ED50 (nM) for NK3R-mediated luciferase activity | ||||||
| CRE-Luc | 701.8 ± 50.1 | 1512.3 ± 25.1 | 26.9 ± 3.2 | 17.9 ± 2.5 | >10 µM | 15.9 ± 2.9 |
| AP1-Luc | 954.1 ± 33.3 | 719.2 ± 39.3 | 36.5 ± 7.8 | 27.5 ± 5.3 | >10 µM | 27.1 ± 2.3 |
| NFAT-Luc | 1071.3 ±65.4 | 1154.4 ± 30.8 | 37.1 ± 8.1 | 30.1 ± 2.2 | >10 µM | 23.9 ± 6.2 |
| SRE-Luc | 1060.2 ± 34.8 | 752.8 ± 20.5 | 15.5 ± 1.5 | 22.3 ± 5.8 | >10 µM | 12.7 ± 3.8 |
| ED50 (nM) for Ca2+ responses under Ca2+-containing/Ca2+-free condition | ||||||
| Ca2+-free | 492.4 ± 48.2 | 480.8 ± 26.5 | 31.8 ± 3.6 | 38.6 ± 2.8 | >10 µM | 40.4 ± 7.8 |
| Ca2+-containing | 487.1 ± 37.7 | 392.3 ± 23.4 | 50.3 ± 8.4 | 31.7 ± 4.9 | >10 µM | 33.2 ± 3.8 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, G.; He, M.; Ko, W.K.W.; Ye, C.; Hu, Q.; Wong, A.O.L. IGFs Potentiate TAC3-induced SLα Expression via Upregulation of TACR3 Expression in Grass Carp Pituitary Cells. Cells 2019, 8, 887. https://doi.org/10.3390/cells8080887
Hu G, He M, Ko WKW, Ye C, Hu Q, Wong AOL. IGFs Potentiate TAC3-induced SLα Expression via Upregulation of TACR3 Expression in Grass Carp Pituitary Cells. Cells. 2019; 8(8):887. https://doi.org/10.3390/cells8080887
Chicago/Turabian StyleHu, Guangfu, Mulan He, Wendy K. W. Ko, Cheng Ye, Qiongyao Hu, and Anderson O. L. Wong. 2019. "IGFs Potentiate TAC3-induced SLα Expression via Upregulation of TACR3 Expression in Grass Carp Pituitary Cells" Cells 8, no. 8: 887. https://doi.org/10.3390/cells8080887