Major Histocompatibility Complex (MHC) Genes and Disease Resistance in Fish
Abstract
:1. Introduction
1.1. The Polymorphism of MHC Genes
1.2. MHC Variation and Resistance to Infectious Diseases in Tetrapod Species
1.3. MHC in Fish
1.4. Associations between MHC Variation and Differences in Disease Resistance in Fish
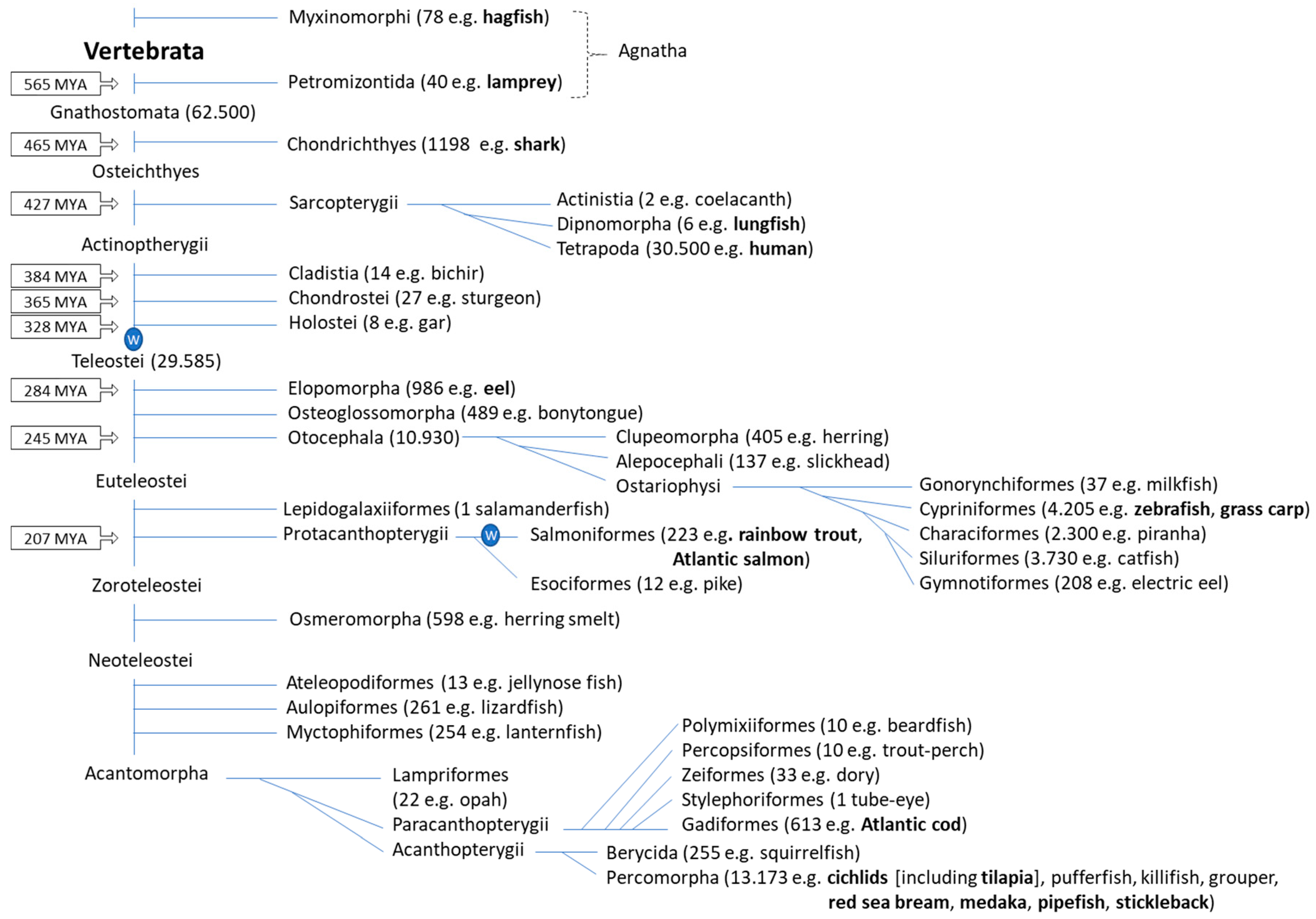
2. Fish Phylogeny
3. Classical and Nonclassical MHC Genes
3.1. Classical and Nonclassical MHC Class II
3.2. Classical and Nonclassical MHC Class I
4. Allelic Variation in Classical MHC Molecules in Fish
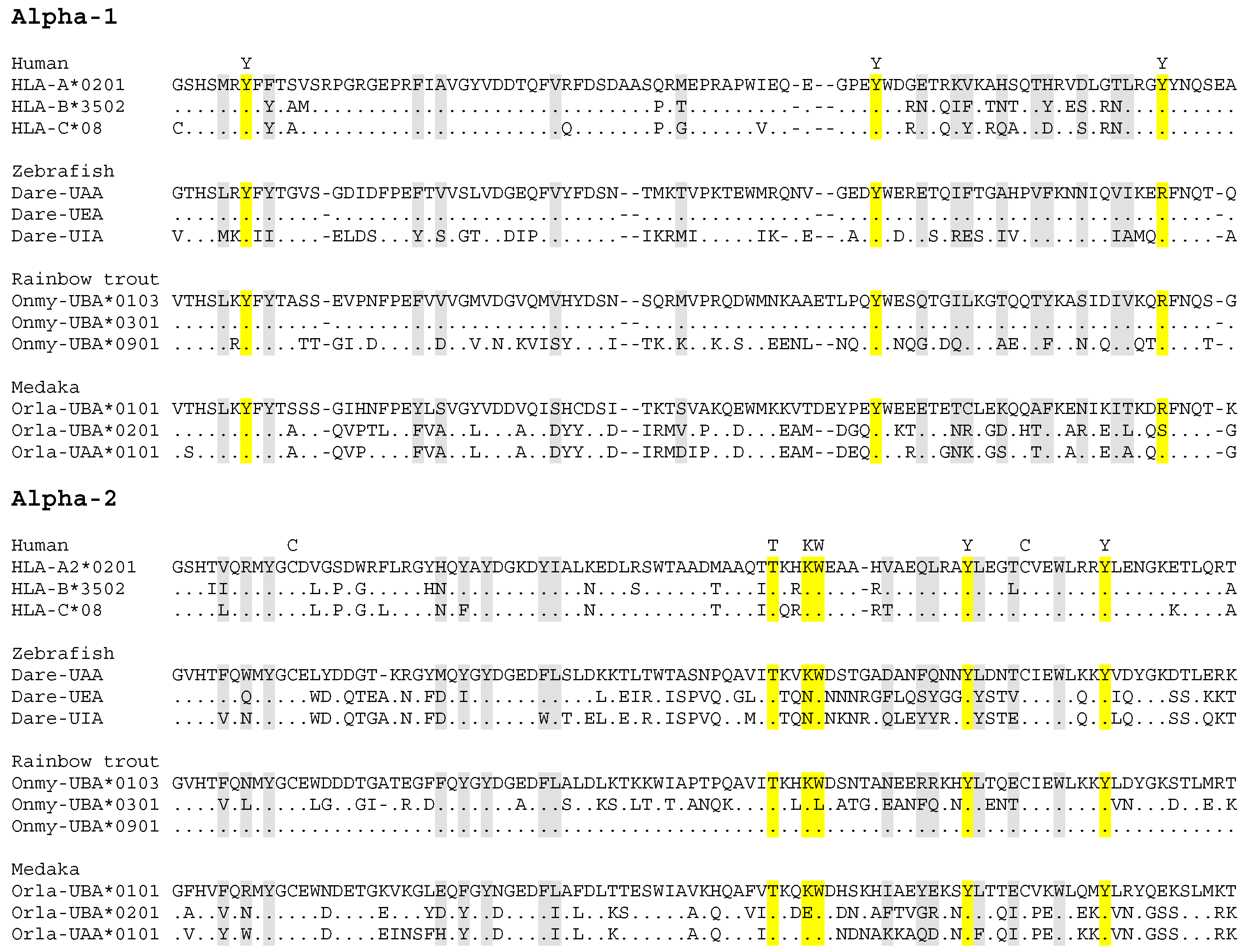
4.1. Allelic Variation in Fish Classical MHC Class II
4.2. Allelic Variation in Fish Classical MHC Class I
5. Functional Analyses of Fish Classical MHC Genes and Molecules
5.1. Expression Patterns of Classical MHC Class II
5.2. Expression Patterns of Classical MHC Class I
5.3. Binding of Peptide Ligands by Classical MHC Class I Molecules
5.4. MHC class I Restriction of Cell-Mediated Cytotoxicity by Lymphocytes
5.5. Additional, Indirect Indications for Classical MHC Functions in Fish
6. Genomic Organization/Haplotype Variation
6.1. Teleost Fish Mhc Allelic/Haplotype Sequence Variation in PSMB and TAP2 Genes
6.2. Copy Number Differences in MHC Class II Genes
6.3. Copy Number Differences in MHC Class I Genes
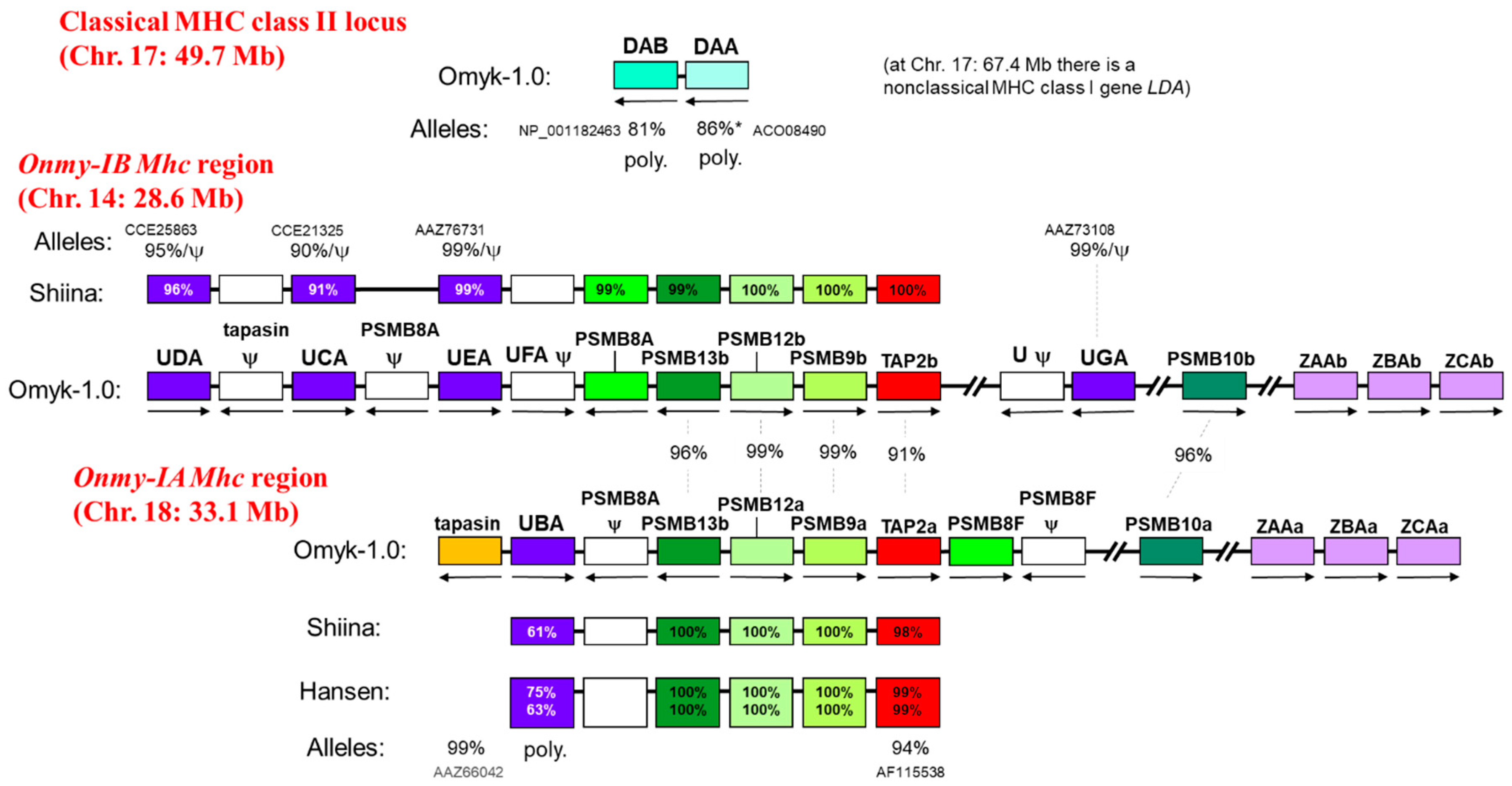
7. The Genomic Organization of the Classical MHC Gene Loci and the Duplicated Mhc Regions Onmy-IA and Onmy-IB in Rainbow Trout
8. Association of Teleost Fish MHC Genes with Disease Resistance
9. Association of Teleost Fish MHC Genes with Allograft Rejection
10. Association of Teleost Fish MHC Genes with Partner Selection
11. Association of Teleost Fish Classical MHC Class I with Behavior and Behavior-Related Growth
12. Discussion
Funding
Conflicts of Interest
References
- Kimura, M.; Ohta, T. The Average Number of Generations until Fixation of a Mutant Gene in a Finite Population. Genetics 1969, 61, 763–771. [Google Scholar] [PubMed]
- Kimura, M. The Neutral Theory of Molecular Evolution; Cambridge University Press: Cambridge, MA, USA, 1983. [Google Scholar]
- Hedrick, P.W.; Thomson, G. Evidence for balancing selection at HLA. Genetics 1983, 104, 449–456. [Google Scholar]
- Hughes, A.L.; Yeager, M. Natural selection and the evolutionary history of major histocompatibility complex loci. Front. Biosci. 1998, 3, d509–d516. [Google Scholar] [CrossRef]
- Mungall, A.J.; Palmer, S.A.; Sims, S.K.; Edwards, C.A.; Ashurst, J.L.; Wilming, L.; Jones, M.C.; Horton, R.; Hunt, S.E.; Scott, C.E.; et al. The DNA sequence and analysis of human chromosome 6. Nature 2003, 425, 805–811. [Google Scholar] [CrossRef]
- Norman, P.J.; Norberg, S.J.; Guethlein, L.A.; Nemat-Gorgani, N.; Royce, T.; Wroblewski, E.E.; Dunn, T.; Mann, T.; Alicata, C.; Hollenbach, J.A.; et al. Sequences of 95 human MHC haplotypes reveal extreme coding variation in genes other than highly polymorphic HLA class I and II. Genome Res. 2017, 27, 813–823. [Google Scholar] [CrossRef]
- Maccari, G.; Robinson, J.; Ballingall, K.; Guethlein, L.A.; Grimholt, U.; Kaufman, J.; Ho, C.S.; de Groot, N.G.; Flicek, P.; Bontrop, R.E.; et al. IPD-MHC 2.0: An improved inter-species database for the study of the major histocompatibility complex. Nucleic Acids Res. 2017, 45, D860–D864. [Google Scholar] [CrossRef] [PubMed]
- Kelly, A.; Trowsdale, J. Genetics of antigen processing and presentation. Immunogenetics 2019, 71, 161–170. [Google Scholar] [CrossRef]
- Germain, R.N. MHC-dependent antigen processing and peptide presentation: Providing ligands for T lymphocyte activation. Cell 1994, 76, 287–299. [Google Scholar] [CrossRef]
- Rammensee, H.G.; Friede, T.; Stevanoviíc, S. MHC ligands and peptide motifs: First listing. Immunogenetics 1995, 41, 178–228. [Google Scholar]
- Michalova, V.; Murray, B.W.; Sultmann, H.; Klein, J. A contig map of the Mhc class I genomic region in the zebrafish reveals ancient synteny. J. Immunol. 2000, 164, 5296–5305. [Google Scholar] [CrossRef] [PubMed]
- Satta, Y.; O’hUigin, C.; Takahata, N.; Klein, J. Intensity of natural selection at the major histocompatibility complex loci. Proc. Natl. Acad. Sci. USA 1994, 91, 7184–7188. [Google Scholar] [CrossRef]
- Kelly, A.; Trowsdale, J. Introduction: MHC/KIR and governance of specificity. Immunogenetics 2017, 69, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Neefjes, J.; Jongsma, M.L.; Paul, P.; Bakke, O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 2011, 11, 823–836. [Google Scholar] [CrossRef] [PubMed]
- Madden, D.R. The three-dimensional structure of peptide-MHC complexes. Annu. Rev. Immunol. 1995, 13, 587–622. [Google Scholar] [CrossRef]
- Hughes, A.L.; Nei, M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 1988, 335, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.L.; Nei, M. Nucleotide substitution at major histocompatibility complex class II loci: Evidence for overdominant selection. Proc. Natl. Acad. Sci. USA 1989, 86, 958–962. [Google Scholar] [CrossRef]
- Figueroa, F.; Günther, E.; Klein, J. MHC polymorphism pre-dating speciation. Nature 1988, 335, 265–267. [Google Scholar] [CrossRef] [PubMed]
- Parham, P.; Lawlor, D.A.; Lomen, C.E.; Ennis, P.D. Diversity and diversification of HLA-A,B,C alleles. J. Immunol. 1989, 142, 3937–3950. [Google Scholar] [PubMed]
- Kaufman, J. Generalists and Specialists: A New View of How MHC Class I Molecules Fight Infectious Pathogens. Trends Immunol. 2018, 39, 367–379. [Google Scholar] [CrossRef]
- Trowsdale, J.; Knight, J.C. Major histocompatibility complex genomics and human disease. Annu. Rev. Genomics Hum. Genet. 2013, 14, 301–323. [Google Scholar] [CrossRef] [PubMed]
- Goulder, P.J.; Walker, B.D. HIV and HLA class I: An evolving relationship. Immunity 2012, 37, 426–440. [Google Scholar] [CrossRef] [PubMed]
- Iwama, G.; Nakanishi, T. The Fish Immune System; Academic Press: San Diego, CA, USA, 1996. [Google Scholar]
- Hashimoto, K.; Nakanishi, T.; Kurosawa, Y. Isolation of carp genes encoding major histocompatibility complex antigens. Proc. Natl. Acad. Sci. USA 1990, 87, 6863–6867. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, K.; Nakanishi, T.; Kurosawa, Y. Identification of a shark sequence resembling the major histocompatibility complex class I alpha 3 domain. Proc. Natl. Acad. Sci. USA 1992, 89, 2209–2212. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, M.; Vazquez, M.; Sato, K.; McKinney, E.C.; Flajnik, M.F. Evolution of the major histocompatibility complex: Isolation of class II A cDNA clones from the cartilaginous fish. Proc. Natl. Acad. Sci. USA 1992, 89, 6688–6692. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, J.M.; Grimholt, U.; Leong, J.; Koop, B.F.; Hashimoto, K. Comprehensive analysis of MHC class II genes in teleost fish genomes reveals dispensability of the peptide-loading DM system in a large part of vertebrates. BMC Evol. Biol. 2013, 13, 260. [Google Scholar] [CrossRef] [PubMed]
- Betz, U.A.; Mayer, W.E.; Klein, J. Major histocompatibility complex class I genes of the coelacanth Latimeria chalumnae. Proc. Natl. Acad. Sci. USA 1994, 91, 11065–11069. [Google Scholar] [CrossRef]
- Sato, A.; Sultmann, H.; Mayer, W.E.; Klein, J. Mhc class I gene of African lungfish. Immunogenetics 2000, 51, 491–495. [Google Scholar] [CrossRef]
- Flajnik, M.F.; Kasahara, M. Origin and evolution of the adaptive immune system: Genetic events and selective pressures. Nat. Rev. Genet. 2010, 11, 47–59. [Google Scholar] [CrossRef]
- Star, B.; Nederbragt, A.J.; Jentoft, S.; Grimholt, U.; Malmstrøm, M.; Gregers, T.F.; Rounge, T.B.; Paulsen, J.; Solbakken, M.H.; Sharma, A.; et al. The genome sequence of Atlantic cod reveals a unique immune system. Nature 2011, 477, 207–210. [Google Scholar] [CrossRef]
- Malmstrøm, M.; Matschiner, M.; Tørresen, O.K.; Star, B.; Snipen, L.G.; Hansen, T.F.; Baalsrud, H.T.; Nederbragt, A.J.; Hanel, R.; Salzburger, W.; et al. Evolution of the immune system influences speciation rates in teleost fishes. Nat. Genet. 2016, 48, 1204–1210. [Google Scholar] [CrossRef]
- Haase, D.; Roth, O.; Kalbe, M.; Schmiedeskamp, G.; Scharsack, J.P.; Rosenstiel, P.; Reusch, T.B. Absence of major histocompatibility complex class II mediated immunity in pipefish, Syngnathus typhle: Evidence from deep transcriptome sequencing. Biol. Lett. 2013, 9, 20130044. [Google Scholar] [CrossRef] [PubMed]
- Ono, H.; Figueroa, F.; O’hUigin, C.; Klein, J. Cloning of the beta 2-microglobulin gene in the zebrafish. Immunogenetics 1993, 38, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sültmann, H.; Mayer, W.E.; Figueroa, F.; O’Huigin, C.; Klein, J. Organization of Mhc class II B genes in the zebrafish (Brachydanio rerio). Genomics 1994, 23, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Harstad, H.; Lukacs, M.F.; Bakke, H.G.; Grimholt, U. Multiple expressed MHC class II loci in salmonids; details of one non-classical region in Atlantic salmon (Salmo salar). BMC Genomics 2008, 9, 193. [Google Scholar] [CrossRef] [PubMed]
- Summers, K.; Roney, K.E.; da Silva, J.; Capraro, G.; Cuthbertson, B.J.; Kazianis, S.; Rosenthal, G.G.; Ryan, M.J.; McConnell, T.J. Divergent patterns of selection on the DAB and DXB MHC class II loci in Xiphophorus fishes. Genetica 2009, 135, 379–390. [Google Scholar] [CrossRef]
- Bannai, H.P.; Nonaka, M. Comprehensive analysis of medaka major histocompatibility complex (MHC) class II genes: Implications for evolution in teleosts. Immunogenetics 2013, 65, 883–895. [Google Scholar] [CrossRef] [PubMed]
- Ono, H.; Klein, D.; Vincek, V.; Figueroa, F.; O’hUigin, C.; Tichy, H.; Klein, J. Major histocompatibility complex class II genes of zebrafish. Proc. Natl. Acad. Sci. USA 1992, 89, 11886–11890. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, J.M.; Yamaguchi, T.; Grimholt, U. Conservation of sequence motifs suggests that the nonclassical MHC class I lineages CD1/PROCR and UT were established before the emergence of tetrapod species. Immunogenetics 2018, 70, 459–476. [Google Scholar] [CrossRef]
- Hashimoto, K.; Okamura, K.; Yamaguchi, H.; Ototake, M.; Nakanishi, T.; Kurosawa, Y. Conservation and diversification of MHC class I and its related molecules in vertebrates. Immunol. Rev. 1999, 167, 81–100. [Google Scholar] [CrossRef]
- Grimholt, U.; Hordvik, I.; Fosse, V.M.; Olsaker, I.; Endresen, C.; Lie, O. Molecular cloning of major histocompatibility complex class I cDNAs from Atlantic salmon (Salmo salar). Immunogenetics 1993, 37, 469–473. [Google Scholar] [CrossRef]
- Grimholt, U.; Tsukamoto, K.; Azuma, T.; Leong, J.; Koop, B.F.; Dijkstra, J.M. A comprehensive analysis of teleost MHC class I sequences. BMC Evol. Biol. 2015, 15, 32. [Google Scholar] [CrossRef]
- Kruiswijk, C.P.; Hermsen, T.T.; Westphal, A.H.; Savelkoul, H.F.; Stet, R.J. A novel functional class I lineage in zebrafish (Danio rerio), carp (Cyprinus carpio), and large barbus (Barbus intermedius) showing an unusual conservation of the peptide binding domains. J. Immunol. 2002, 169, 1936–1947. [Google Scholar] [CrossRef]
- Dijkstra, J.M.; Katagiri, T.; Hosomichi, K.; Yanagiya, K.; Inoko, H.; Ototake, M.; Aoki, T.; Hashimoto, K.; Shiina, T. A third broad lineage of major histocompatibility complex (MHC) class I in teleost fish; MHC class II linkage and processed genes. Immunogenetics 2007, 59, 305–321. [Google Scholar] [CrossRef]
- Shum, B.P.; Guethlein, L.; Flodin, L.R.; Adkison, M.A.; Hedrick, R.P.; Nehring, R.B.; Stet, R.J.; Secombes, C.; Parham, P. Modes of salmonid MHC class I and II evolution differ from the primate paradigm. J. Immunol. 2001, 166, 3297–3308. [Google Scholar] [CrossRef] [PubMed]
- Bingulac-Popovic, J.; Figueroa, F.; Sato, A.; Talbot, W.S.; Johnson, S.L.; Gates, M.; Postlethwait, J.H.; Klein, J. Mapping of Mhc class I and class II regions to different linkage groups in the zebrafish, Danio rerio. Immunogenetics 1997, 46, 129–134. [Google Scholar] [CrossRef]
- Sato, A.; Figueroa, F.; Murray, B.W.; Malaga-Trillo, E.; Zaleska-Rutczynska, Z.; Sultmann, H.; Toyosawa, S.; Wedekind, C.; Steck, N.; Klein, J. Nonlinkage of major histocompatibility complex class I and class II loci in bony fishes. Immunogenetics 2000, 51, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, K.; Hayashi, S.; Matsuo, M.Y.; Nonaka, M.I.; Kondo, M.; Shima, A.; Asakawa, S.; Shimizu, N.; Nonaka, M. Unprecedented intraspecific diversity of the MHC class I region of a teleost medaka, Oryzias latipes. Immunogenetics 2005, 57, 420–431. [Google Scholar] [CrossRef] [PubMed]
- Shiina, T.; Dijkstra, J.M.; Shimizu, S.; Watanabe, A.; Yanagiya, K.; Kiryu, I.; Fujiwara, A.; Nishida-Umehara, C.; Kaba, Y.; Hirono, I.; et al. Interchromosomal duplication of major histocompatibility complex class I regions in rainbow trout (Oncorhynchus mykiss), a species with a presumably recent tetraploid ancestry. Immunogenetics 2005, 56, 878–893. [Google Scholar] [CrossRef]
- Hansen, J.D.; Strassburger, P.; Thorgaard, G.H.; Young, W.P.; Du Pasquier, L. Expression, linkage, and polymorphism of MHC-related genes in rainbow trout, Oncorhynchus mykiss. J. Immunol. 1999, 163, 774–786. [Google Scholar]
- Clark, M.S.; Shaw, L.; Kelly, A.; Snell, P.; Elgar, G. Characterization of the MHC class I region of the Japanese pufferfish (Fugu rubripes). Immunogenetics 2001, 52, 174–185. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, K.; Takizawa, F.; Tokumaru, N.; Nakayasu, C.; Toda, H.; Fischer, U.; Moritomo, T.; Hashimoto, K.; Nakanishi, T.; Dijkstra, J.M. A molecule in teleost fish, related with human MHC-encoded G6F, has a cytoplasmic tail with ITAM and marks the surface of thrombocytes and in some fishes also of erythrocytes. Immunogenetics 2010, 62, 543–559. [Google Scholar] [CrossRef]
- McConnell, S.C.; Hernandez, K.M.; Wcisel, D.J.; Kettleborough, R.N.; Stemple, D.L.; Yoder, J.A.; Andrade, J.; de Jong, J.L. Alternative haplotypes of antigen processing genes in zebrafish diverged early in vertebrate evolution. Proc. Natl. Acad. Sci. USA 2016, 113, E5014–E5023. [Google Scholar] [CrossRef]
- Takami, K.; Zaleska-Rutczynska, Z.; Figueroa, F.; Klein, J. Linkage of LMP, TAP, and RING3 with Mhc class I rather than class II genes in the zebrafish. J. Immunol. 1997, 159, 6052–6060. [Google Scholar]
- Grimholt, U. Whole genome duplications have provided teleosts with many roads to peptide loaded MHC class I molecules. BMC Evol. Biol. 2018, 18, 25. [Google Scholar] [CrossRef]
- Murray, B.W.; Sültmann, H.; Klein, J. Analysis of a 26-kb region linked to the Mhc in zebrafish: Genomic organization of the proteasome component beta/transporter associated with antigen processing-2 gene cluster and identification of five new proteasome beta subunit genes. J. Immunol. 1999, 163, 2657–2666. [Google Scholar]
- Rodrigues, P.N.; Hermsen, T.T.; Rombout, J.H.; Egberts, E.; Stet, R.J. Detection of MHC class II transcripts in lymphoid tissues of the common carp (Cyprinus carpio, L.). Dev. Comp. Immunol. 1995, 19, 483–496. [Google Scholar] [CrossRef]
- Dijkstra, J.M.; Kiryu, I.; Kollner, B.; Yoshiura, Y.; Ototake, M. MHC class II invariant chain homologues in rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 2003, 15, 91–105. [Google Scholar] [CrossRef]
- Fischer, U.; Dijkstra, J.M.; Kollner, B.; Kiryu, I.; Koppang, E.O.; Hordvik, I.; Sawamoto, Y.; Ototake, M. The ontogeny of MHC class I expression in rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 2005, 18, 49–60. [Google Scholar] [CrossRef]
- Koppang, E.O.; Hordvik, I.; Bjerkås, I.; Torvund, J.; Aune, L.; Thevarajan, J.; Endresen, C. Production of rabbit antisera against recombinant MHC class II beta chain and identification of immunoreactive cells in Atlantic salmon (Salmo salar). Fish Shellfish Immunol. 2003, 14, 115–132. [Google Scholar] [CrossRef]
- Scharsack, J.P.; Kalbe, M.; Schaschl, H. Characterization of antisera raised against stickleback (Gasterosteus aculeatus) MHC class I and class II molecules. Fish Shellfish Immunol. 2007, 23, 991–1002. [Google Scholar] [CrossRef]
- Boudinot, P.; Blanco, M.; de Kinkelin, P.; Benmansour, A. Combined DNA immunization with the glycoprotein gene of viral hemorrhagic septicemia virus and infectious hematopoietic necrosis virus induces double-specific protective immunity and nonspecific response in rainbow trout. Virology 1998, 249, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.M.; Hu, Y.W.; Xue, N.N.; Ren, S.S.; Chen, S.N.; Nie, P.; Chang, M.X. Role of zebrafish NLRC5 in antiviral response and transcriptional regulation of MHC related genes. Dev. Comp. Immunol. 2017, 68, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Syed, M.; Vestrheim, O.; Mikkelsen, B.; Lundin, M. Isolation of the promoters of Atlantic salmon MHCII genes. Mar. Biotechnol. 2003, 5, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Cardwell, T.N.; Sheffer, R.J.; Hedrick, P.W. MHC variation and tissue transplantation in fish. J. Hered. 2001, 92, 305–308. [Google Scholar] [CrossRef]
- Dijkstra, J.M.; Yamaguchi, T. Ancient features of the MHC class II presentation pathway, and a model for the possible origin of MHC molecules. Immunogenetics 2019, 71, 233–249. [Google Scholar] [CrossRef]
- Boudinot, P.; Boubekeur, S.; Benmansour, A. Rhabdovirus infection induces public and private T cell responses in teleost fish. J. Immunol. 2001, 167, 6202–6209. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, J.M. TH2 and Treg candidate genes in elephant shark. Nature 2014, 511, E7–E9. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Takizawa, F.; Fischer, U.; Dijkstra, J.M. Along the Axis between Type 1 and Type 2 Immunity; Principles Conserved in Evolution from Fish to Mammals. Biology 2015, 17, 814–859. [Google Scholar] [CrossRef]
- Lam, S.H.; Chua, H.L.; Gong, Z.; Wen, Z.; Lam, T.J.; Sin, Y.M. Morphologic transformation of the thymus in developing zebrafish. Dev. Dyn. 2002, 225, 87–94. [Google Scholar] [CrossRef]
- Langenau, D.M.; Zon, L.I. The zebrafish: A new model of T-cell and thymic development. Nat. Rev. Immunol. 2005, 5, 307–317. [Google Scholar] [CrossRef]
- Takizawa, F.; Dijkstra, J.M.; Kotterba, P.; Korytář, T.; Kock, H.; Köllner, B.; Jaureguiberry, B.; Nakanishi, T.; Fischer, U. The expression of CD8α discriminates distinct T cell subsets in teleost fish. Dev. Comp. Immunol. 2011, 35, 752–763. [Google Scholar] [CrossRef]
- Takizawa, F.; Magadan, S.; Parra, D.; Xu, Z.; Korytář, T.; Boudinot, P.; Sunyer, J.O. Novel Teleost CD4-Bearing Cell Populations Provide Insights into the Evolutionary Origins and Primordial Roles of CD4+ Lymphocytes and CD4+ Macrophages. J. Immunol. 2016, 196, 4522–4535. [Google Scholar] [CrossRef]
- Suetake, H.; Araki, K.; Suzuki, Y. Cloning, expression, and characterization of fugu CD4, the first ectothermic animal CD4. Immunogenetics 2004, 56, 368–374. [Google Scholar] [CrossRef]
- Dijkstra, J.M.; Somamoto, T.; Moore, L.; Hordvik, I.; Ototake, M.; Fischer, U. Identification and characterization of a second CD4-like gene in teleost fish. Mol. Immunol. 2006, 43, 410–419. [Google Scholar] [CrossRef]
- Laing, K.J.; Zou, J.J.; Purcell, M.K.; Phillips, R.; Secombes, C.J.; Hansen, J.D. Evolution of the CD4 family: teleost fish possess two divergent forms of CD4 in addition to lymphocyte activation gene-3. J. Immunol. 2006, 177, 3939–3951. [Google Scholar] [CrossRef] [PubMed]
- Taylor, E.B.; Wilson, M.; Bengten, E. The Src tyrosine kinase Lck binds to CD2, CD4–1, and CD4–2 T cell co-receptors in channel catfish, Ictalurus punctatus. Mol. Immunol. 2015, 66, 126–138. [Google Scholar] [CrossRef]
- Yoder, J.A.; Haire, R.N.; Litman, G.W. Cloning of two zebrafish cDNAs that share domains with the MHC class II-associated invariant chain. Immunogenetics 1999, 50, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Hansen, J.D.; Strassburger, P.; Du Pasquier, L. Conservation of an alpha 2 domain within the teleostean world, MHC class I from the rainbow trout Oncorhynchus mykiss. Dev. Comp. Immunol. 1996, 20, 417–425. [Google Scholar] [CrossRef]
- Dijkstra, J.M.; Kollner, B.; Aoyagi, K.; Sawamoto, Y.; Kuroda, A.; Ototake, M.; Nakanishi, T.; Fischer, U. The rainbow trout classical MHC class I molecule Onmy-UBA*501 is expressed in similar cell types as mammalian classical MHC class I molecules. Fish Shellfish Immunol. 2003, 14, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, J.M.; Yoshiura, Y.; Kiryu, I.; Aoyagi, K.; Kollner, B.; Fischer, U.; Nakanishi, T.; Ototake, M. The promoter of the classical MHC class I locus in rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 2003, 14, 177–185. [Google Scholar] [CrossRef]
- Rodrigues, P.N.; Dixon, B.; Roelofs, J.; Rombout, J.H.; Egberts, E.; Pohajdak, B.; Stet, R.J. Expression and temperature-dependent regulation of the beta2-microglobulin (Cyca-B2m) gene in a cold-blooded vertebrate, the common carp (Cyprinus carpio L.). Dev. Immunol. 1998, 5, 263–275. [Google Scholar] [CrossRef]
- Antao, A.B.; Chinchar, V.G.; McConnell, T.J.; Miller, N.W.; Clem, L.W.; Wilson, M.R. MHC class I genes of the channel catfish: Sequence analysis and expression. Immunogenetics 1999, 49, 303–311. [Google Scholar] [CrossRef]
- Chen, W.; Jia, Z.; Zhang, T.; Zhang, N.; Lin, C.; Gao, F.; Wang, L.; Li, X.; Jiang, Y.; Li, X.; et al. MHC class I presentation and regulation by IFN in bony fish determined by molecular analysis of the class I locus in grass carp. J. Immunol. 2010, 185, 2209–2221. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhang, N.; Qi, J.; Chen, R.; Dijkstra, J.M.; Li, X.; Wang, Z.; Wang, J.; Wu, Y.; Xia, C. The Structure of the MHC Class I Molecule of Bony Fishes Provides Insights into the Conserved Nature of the Antigen-Presenting System. J. Immunol. 2017, 199, 3668–3678. [Google Scholar] [CrossRef]
- Sarder, M.R.; Fischer, U.; Dijkstra, J.M.; Kiryu, I.; Yoshiura, Y.; Azuma, T.; Kollner, B.; Ototake, M. The MHC class I linkage group is a major determinant in the in vivo rejection of allogeneic erythrocytes in rainbow trout (Oncorhynchus mykiss). Immunogenetics 2003, 55, 315–324. [Google Scholar] [CrossRef]
- Quiniou, S.M.; Wilson, M.; Bengten, E.; Waldbieser, G.C.; Clem, L.W.; Miller, N.W. MHC RFLP analyses in channel catfish full-sibling families: Identification of the role of MHC molecules in spontaneous allogeneic cytotoxic responses. Dev. Comp. Immunol. 2005, 29, 457–467. [Google Scholar] [CrossRef]
- Somamoto, T.; Nakanishi, T.; Okamoto, N. Specific cell-mediated cytotoxicity against a virus-infected syngeneic cell line in isogeneic ginbuna crucian carp. Dev. Comp. Immunol. 2000, 24, 633–640. [Google Scholar] [CrossRef]
- Somamoto, T.; Nakanishi, T.; Okamoto, N. Role of specific cell-mediated cytotoxicity in protecting fish from viral infections. Virology 2002, 297, 120–127. [Google Scholar] [CrossRef]
- Somamoto, T.; Yoshiura, Y.; Sato, A.; Nakao, M.; Nakanishi, T.; Okamoto, N.; Ototake, M. Expression profiles of TCRbeta and CD8alpha mRNA correlate with virus-specific cell-mediated cytotoxic activity in ginbuna crucian carp. Virology 2006, 348, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Somamoto, T.; Nakanishi, T.; Nakao, M. Identification of anti-viral cytotoxic effector cells in the ginbuna crucian carp, Carassius auratus langsdorfii. Dev. Comp. Immunol. 2013, 39, 370–377. [Google Scholar] [CrossRef]
- Dijkstra, J.M.; Fischer, U.; Sawamoto, Y.; Ototake, M.; Nakanishi, T. Exogenous antigens and the stimulation of MHC class I restricted cell-mediated cytotoxicity: Possible strategies for fish vaccines. Fish Shellfish Immunol. 2001, 11, 437–458. [Google Scholar] [CrossRef] [PubMed]
- Utke, K.; Kock, H.; Schuetze, H.; Bergmann, S.M.; Lorenzen, N.; Einer-Jensen, K.; Köllner, B.; Dalmo, R.A.; Vesely, T.; Ototake, M.; et al. Cell-mediated immune responses in rainbow trout after DNA immunization against the viral hemorrhagic septicemia virus. Dev. Comp. Immunol. 2008, 32, 239–252. [Google Scholar] [CrossRef]
- Chang, Y.T.; Kai, Y.H.; Chi, S.C.; Song, Y.L. Cytotoxic CD8α+ leucocytes have heterogeneous features in antigen recognition and class I MHC restriction in grouper. Fish Shellfish Immunol. 2011, 30, 1283–1293. [Google Scholar] [CrossRef]
- Nelson, J.S.; Grande, T.C.; Wilson, M.V.H. Fishes of the World, 5th ed.; John Wiley & Sons: Hoboken, NJ, USA, 2016. [Google Scholar]
- Kumar, S.; Hedges, S.B. A molecular timescale for vertebrate evolution. Nature 1998, 392, 917–920. [Google Scholar] [CrossRef] [PubMed]
- Broughton, R.E.; Betancur-R, R.; Li, C.; Arratia, G.; Ortí, G. Multi-locus phylogenetic analysis reveals the pattern and tempo of bony fish evolution. PLoS Curr. 2013, 5. [Google Scholar] [CrossRef]
- Jaillon, O.; Aury, J.M.; Brunet, F.; Petit, J.L.; Stange-Thomann, N.; Mauceli, E.; Bouneau, L.; Fischer, C.; Ozouf-Costaz, C.; Bernot, A.; et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 2004, 431, 946–957. [Google Scholar] [CrossRef]
- Painter, C.A.; Stern, L.J. Conformational variation in structures of classical and non-classical MHCII proteins and functional implications. Immunol. Rev. 2012, 250, 144–157. [Google Scholar] [CrossRef]
- Kasahara, M.; McKinney, E.C.; Flajnik, M.F.; Ishibashi, T. The evolutionary origin of the major histocompatibility complex: Polymorphism of class II alpha chain genes in the cartilaginous fish. Eur. J. Immunol. 1993, 23, 2160–2165. [Google Scholar] [CrossRef]
- Hughes, A.L.; Nei, M. Evolution of the major histocompatibility complex: Independent origin of nonclassical class I genes in different groups of mammals. Mol. Biol. Evol. 1989, 6, 559–579. [Google Scholar]
- Adams, E.J.; Luoma, A.M. The adaptable major histocompatibility complex (MHC) fold: Structure and function of nonclassical and MHC class I-like molecules. Annu. Rev. Immunol. 2013, 31, 529–561. [Google Scholar] [CrossRef]
- Lukacs, M.F.; Harstad, H.; Grimholt, U.; Beetz-Sargent, M.; Cooper, G.A.; Reid, L.; Bakke, H.G.; Phillips, R.B.; Miller, K.M.; Davidson, W.S.; et al. Genomic organization of duplicated major histocompatibility complex class I regions in Atlantic salmon (Salmo salar). BMC Genomics 2007, 8, 251. [Google Scholar] [CrossRef]
- Dirscherl, H.; McConnell, S.C.; Yoder, J.A.; de Jong, J.L. The MHC class I genes of zebrafish. Dev. Comp. Immunol. 2014, 46, 11–23. [Google Scholar] [CrossRef]
- Shum, B.P.; Rajalingam, R.; Magor, K.E.; Azumi, K.; Carr, W.H.; Dixon, B.; Stet, R.J.; Adkison, M.A.; Hedrick, R.P.; Parham, P. A divergent non-classical class I gene conserved in salmonids. Immunogenetics 1999, 49, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Dirscherl, H.; Yoder, J.A. Characterization of the Z lineage Major histocompatability complex class I genes in zebrafish. Immunogenetics 2014, 66, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Nonaka, M.I.; Aizawa, K.; Mitani, H.; Bannai, H.P.; Nonaka, M. Retained orthologous relationships of the MHC Class I genes during euteleost evolution. Mol. Biol. Evol. 2011, 28, 3099–3112. [Google Scholar] [CrossRef] [PubMed]
- Gómez, D.; Conejeros, P.; Marshall, S.H.; Consuegra, S. MHC evolution in three salmonid species: A comparison between class II alpha and beta genes. Immunogenetics 2010, 62, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Sun, W.X.; Hu, J.F.; Hong, D.W.; Chen, S.L. Molecular characterization, polymorphism and expression analysis of swamp eel major histocompatibility complex class II B gene, after infection by Aeromonas Hydrophilia. J. Anim. Plant Sci. 2014, 24, 481–491. [Google Scholar]
- Stet, R.J.; de Vries, B.; Mudde, K.; Hermsen, T.; van Heerwaarden, J.; Shum, B.P.; Grimholt, U. Unique haplotypes of co-segregating major histocompatibility class II A and class II B alleles in Atlantic salmon (Salmo salar) give rise to diverse class II genotypes. Immunogenetics 2002, 54, 320–331. [Google Scholar] [CrossRef]
- Okamura, K.; Ototake, M.; Nakanishi, T.; Kurosawa, Y.; Hashimoto, K. The most primitive vertebrates with jaws possess highly polymorphic MHC class I genes comparable to those of humans. Immunity 1997, 7, 777–790. [Google Scholar] [CrossRef]
- Aoyagi, K.; Dijkstra, J.M.; Xia, C.; Denda, I.; Ototake, M.; Hashimoto, K.; Nakanishi, T. Classical MHC class I genes composed of highly divergent sequence lineages share a single locus in rainbow trout (Oncorhynchus mykiss). J. Immunol. 2002, 168, 260–273. [Google Scholar] [CrossRef]
- Kiryu, I.; Dijkstra, J.M.; Sarder, R.I.; Fujiwara, A.; Yoshiura, Y.; Ototake, M. New MHC class Ia domain lineages in rainbow trout (Oncorhynchus mykiss) which are shared with other fish species. Fish Shellfish Immunol. 2005, 18, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Nonaka, M.I.; Nonaka, M. Evolutionary analysis of two classical MHC class I loci of the medaka fish, Oryzias latipes: Haplotype-specific genomic diversity, locus-specific polymorphisms, and interlocus homogenization. Immunogenetics 2010, 62, 319–332. [Google Scholar] [CrossRef]
- McConnell, S.C.; Restaino, A.C.; de Jong, J.L. Multiple divergent haplotypes express completely distinct sets of class I MHC genes in zebrafish. Immunogenetics 2014, 66, 199–213. [Google Scholar] [CrossRef]
- Dijkstra, J.M.; Grimholt, U. Major histocompatibility complex (MHC) fragment numbers alone—In Atlantic cod and in general—Do not represent functional variability. Version 2. F1000Research 2018, 7, 963. [Google Scholar] [CrossRef]
- Vogel, T.U.; Evans, D.T.; Urvater, J.A.; O’Connor, D.H.; Hughes, A.L.; Watkins, D.I. Major histocompatibility complex class I genes in primates: Co-evolution with pathogens. Immunol. Rev. 1999, 167, 327–337. [Google Scholar] [CrossRef] [PubMed]
- De Groot, N.G.; Otting, N.; Doxiadis, G.G.; Balla-Jhagjhoorsingh, S.S.; Heeney, J.L.; van Rood, J.J.; Gagneux, P.; Bontrop, R.E. Evidence for an ancient selective sweep in the MHC class I gene repertoire of chimpanzees. Proc. Natl. Acad. Sci. USA 2002, 99, 11748–11753. [Google Scholar] [CrossRef]
- Lafont, B.A.; Buckler-White, A.; Plishka, R.; Buckler, C.; Martin, M.A. Characterization of pig-tailed macaque classical MHC class I genes: Implications for MHC evolution and antigen presentation in macaques. J. Immunol. 2003, 171, 875–885. [Google Scholar] [CrossRef] [PubMed]
- Meredith, R.W.; Janečka, J.E.; Gatesy, J.; Ryder, O.A.; Fisher, C.A.; Teeling, E.C.; Goodbla, A.; Eizirik, E.; Simão, T.L.; Stadler, T.; et al. Impacts of the Cretaceous Terrestrial Revolution and KPg extinction on mammal diversification. Science 2011, 334, 521–524. [Google Scholar] [CrossRef]
- Barber, L.D.; Percival, L.; Valiante, N.M.; Chen, L.; Lee, C.; Gumperz, J.E.; Phillips, J.H.; Lanier, L.L.; Bigge, J.C.; Parekh, R.B.; et al. The inter-locus recombinant HLA-B*4601 has high selectivity in peptide binding and functions characteristic of HLA-C. J. Exp. Med. 1996, 184, 735–740. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.; Kiryu, I.; Dijkstra, J.M.; Azuma, T.; Nakanishi, T.; Ototake, M. Differences in MHC class I genes between strains of rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 2002, 12, 287–301. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.M.; Li, S.; Ming, T.J.; Kaukinen, K.H.; Schulze, A.D. The salmonid MHC class I: More ancient loci uncovered. Immunogenetics 2006, 58, 571–589. [Google Scholar] [CrossRef] [PubMed]
- Grimholt, U.; Drablos, F.; Jorgensen, S.M.; Hoyheim, B.; Stet, R.J. The major histocompatibility class I locus in Atlantic salmon (Salmo salar, L.): Polymorphism, linkage analysis and protein modelling. Immunogenetics 2002, 54, 570–581. [Google Scholar] [CrossRef]
- Consuegra, S.; Megens, H.J.; Schaschl, H.; Leon, K.; Stet, R.J.; Jordan, W.C. Rapid evolution of the MH class I locus results in different allelic compositions in recently diverged populations of Atlantic salmon. Mol. Biol. Evol. 2005, 22, 1095–1106. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, H.; Figueroa, F.; O’Huigin, C.; Klein, J. Cloning and characterization of class I Mhc genes of the zebrafish, Brachydanio rerio. Immunogenetics 1995, 42, 77–84. [Google Scholar] [CrossRef]
- Graser, R.; Vincek, V.; Takami, K.; Klein, J. Analysis of zebrafish Mhc using BAC clones. Immunogenetics 1998, 47, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Sültmann, H.; Murray, B.W.; Klein, J. Identification of seven genes in the major histocompatibility complex class I region of the zebrafish. Scand. J. Immunol. 2000, 51, 577–585. [Google Scholar] [CrossRef]
- Van Erp, S.H.; Dixon, B.; Figueroa, F.; Egberts, E.; Stet, R.J. Identification and characterization of a new major histocompatibility complex class I gene in carp (Cyprinus carpio, L.). Immunogenetics 1996, 44, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, M.; Fremont, D.H.; Peterson, P.A.; Wilson, I.A. Emerging principles for the recognition of peptide antigens by MHC class I molecules. Science 1992, 257, 927–934. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, M.Y.; Asakawa, S.; Shimizu, N.; Kimura, H.; Nonaka, M. Nucleotide sequence of the MHC class I genomic region of a teleost, the medaka (Oryzias latipes). Immunogenetics 2002, 53, 930–940. [Google Scholar]
- Persson, A.C.; Stet, R.J.; Pilström, L. Characterization of MHC class I and beta(2)-microglobulin sequences in Atlantic cod reveals an unusually high number of expressed class I genes. Immunogenetics 1999, 50, 49–59. [Google Scholar] [CrossRef]
- Miller, K.M.; Kaukinen, K.H.; Schulze, A.D. Expansion and contraction of major histocompatibility complex genes: A teleostean example. Immunogenetics 2002, 53, 941–963. [Google Scholar]
- Schaschl, H.; Wegner, K.M. Contrasting mode of evolution between the MHC class I genomic region and class II region in the three-spined stickleback (Gasterosteus aculeatus, L.; Gasterosteidae: Teleostei). Immunogenetics 2007, 59, 295–304. [Google Scholar] [CrossRef]
- Tørresen, O.K.; Brieuc, M.S.O.; Solbakken, M.H.; Sørhus, E.; Nederbragt, A.J.; Jakobsen, K.S.; Meier, S.; Edvardsen, R.B.; Jentoft, S. Genomic architecture of haddock (Melanogrammus aeglefinus) shows expansions of innate immune genes and short tandem repeats. BMC Genomics 2018, 19, 240. [Google Scholar] [CrossRef] [PubMed]
- Bartl, S.; Baish, M.A.; Flajnik, M.F.; Ohta, Y. Identification of class I genes in cartilaginous fish, the most ancient group of vertebrates displaying an adaptive immune response. J. Immunol. 1997, 159, 6097–6104. [Google Scholar]
- Koppang, E.O.; Dannevig, B.H.; Lie, O.; Ronningen, K.; Press, C.M.L. Expression of MHC class I and II mRNA in a macrophage-like cell line (SHK-1) derived from Atlantic salmon, Salmo salar, L., head kidney. Fish Shellfish Immunol. 1999, 9, 473–489. [Google Scholar] [CrossRef]
- Jørgensen, S.M.; Syvertsen, B.L.; Lukacs, M.; Grimholt, U.; Gjøen, T. Expression of MHC class I pathway genes in response to infectious salmon anaemia virus in Atlantic salmon (Salmo salar, L.) cells. Fish Shellfish Immunol. 2006, 21, 548–560. [Google Scholar] [CrossRef]
- Antao, A.B.; Wilson, M.; Wang, J.; Bengten, E.; Miller, N.W.; Clem, L.W.; Chinchar, V.G. Genomic organization and differential expression of channel catfish MHC class I genes. Dev. Comp. Immunol. 2001, 25, 579–595. [Google Scholar] [CrossRef]
- Stet, R.J.; Kruiswijk, C.P.; Dixon, B. Major histocompatibility lineages and immune gene function in teleost fishes: The road not taken. Crit. Rev. Immunol. 2003, 23, 441–471. [Google Scholar] [CrossRef]
- Bly, J.E.; Clem, L.W. Temperature-mediated processes in teleost immunity: In vitro immunosuppression induced by in vivo low temperature in channel catfish. Vet. Immunol. Immunopathol. 1991, 28, 365–377. [Google Scholar] [CrossRef]
- Abram, Q.H.; Dixon, B.; Katzenback, B.A. Impacts of Low Temperature on the Teleost Immune System. Biology 2017, 6, 39. [Google Scholar] [CrossRef] [PubMed]
- Sever, L.; Vo, N.T.K.; Bols, N.C.; Dixon, B. Tapasin’s protein interactions in the rainbow trout peptide-loading complex. Dev. Comp. Immunol. 2018, 81, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Criscitiello, M.F.; Ohta, Y.; Saltis, M.; McKinney, E.C.; Flajnik, M.F. Evolutionarily conserved TCR binding sites, identification of T cells in primary lymphoid tissues, and surprising trans-rearrangements in nurse shark. J. Immunol. 2010, 184, 6950–6960. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, N.; Wang, Z.; Yanan, W.; Zhang, L.; Xia, C. Structural insights into the evolution feature of a bony fish CD8αα homodimer. Mol. Immunol. 2018, 97, 109–116. [Google Scholar] [CrossRef]
- Hansen, J.D.; Strassburger, P. Description of an ectothermic TCR coreceptor, CD8 alpha, in rainbow trout. J. Immunol. 2000, 164, 3132–3139. [Google Scholar] [CrossRef]
- Moore, L.J.; Somamoto, T.; Lie, K.K.; Dijkstra, J.M.; Hordvik, I. Characterisation of salmon and trout CD8alpha and CD8beta. Mol. Immunol. 2005, 42, 1225–1234. [Google Scholar] [CrossRef]
- Suetake, H.; Araki, K.; Akatsu, K.; Somamoto, T.; Dijkstra, J.M.; Yoshiura, Y.; Kikuchi, K.; Suzuki, Y. Genomic organization and expression of CD8alpha and CD8beta genes in fugu Takifugu rubripes. Fish Shellfish Immunol. 2007, 23, 1107–1118. [Google Scholar] [CrossRef]
- Hansen, J.D.; Farrugia, T.J.; Woodson, J.; Laing, K.J. Description of an elasmobranch TCR coreceptor: CD8α from Rhinobatos productus. Dev. Comp. Immunol. 2011, 35, 452–460. [Google Scholar] [CrossRef]
- Venkatesh, B.; Lee, A.P.; Ravi, V.; Maurya, A.K.; Lian, M.M.; Swann, J.B.; Ohta, Y.; Flajnik, M.F.; Sutoh, Y.; Kasahara, M.; et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature 2014, 505, 174–179. [Google Scholar] [CrossRef]
- Hayashi, N.; Takeuchi, M.; Nakanishi, T.; Hashimoto, K.; Dijkstra, J.M. Zinc-dependent binding between peptides derived from rainbow trout CD8alpha and LCK. Fish Shellfish Immunol. 2010, 28, 72–76. [Google Scholar] [CrossRef]
- Criscitiello, M.F.; Ohta, Y.; Graham, M.D.; Eubanks, J.O.; Chen, P.L.; Flajnik, M.F. Shark class II invariant chain reveals ancient conserved relationships with cathepsins and MHC class II. Dev. Comp. Immunol. 2012, 36, 521–533. [Google Scholar] [CrossRef]
- Fujiki, K.; Smith, C.M.; Liu, L.; Sundick, R.S.; Dixon, B. Alternate forms of MHC class II-associated invariant chain are not produced by alternative splicing in rainbow trout (Oncorhynchus mykiss) but are encoded by separate genes. Dev. Comp. Immunol. 2003, 27, 377–391. [Google Scholar] [CrossRef]
- Wilson, A.B. MHC and adaptive immunity in teleost fishes. Immunogenetics 2017, 69, 521–528. [Google Scholar] [CrossRef] [PubMed]
- Kulski, J.K.; Shiina, T.; Anzai, T.; Kohara, S.; Inoko, H. Comparative genomic analysis of the MHC: The evolution of class I duplication blocks, diversity and complexity from shark to man. Immunol. Rev. 2002, 190, 95–122. [Google Scholar] [CrossRef]
- Ohta, Y.; Okamura, K.; McKinney, E.C.; Bartl, S.; Hashimoto, K.; Flajnik, M.F. Primitive synteny of vertebrate major histocompatibility complex class I and class II genes. Proc. Natl. Acad. Sci. USA 2000, 97, 4712–4717. [Google Scholar] [CrossRef] [PubMed]
- Ohta, Y.; McKinney, E.C.; Criscitiello, M.F.; Flajnik, M.F. Proteasome, transporter associated with antigen processing, and class I genes in the nurse shark Ginglymostoma cirratum: Evidence for a stable class I region and MHC haplotype lineages. J. Immunol. 2002, 168, 771–781. [Google Scholar] [CrossRef]
- Ohta, Y.; Shiina, T.; Lohr, R.L.; Hosomichi, K.; Pollin, T.I.; Heist, E.J.; Suzuki, S.; Inoko, H.; Flajnik, M.F. Primordial linkage of β2-microglobulin to the MHC. J. Immunol. 2011, 186, 3563–3571. [Google Scholar] [CrossRef] [PubMed]
- Blees, A.; Januliene, D.; Hofmann, T.; Koller, N.; Schmidt, C.; Trowitzsch, S.; Moeller, A.; Tampé, R. Structure of the human MHC-I peptide-loading complex. Nature 2017, 551, 525–528. [Google Scholar] [CrossRef]
- Murata, S.; Takahama, Y.; Kasahara, M.; Tanaka, K. The immunoproteasome and thymoproteasome: Functions, evolution and human disease. Nat. Immunol. 2018, 19, 923–931. [Google Scholar] [CrossRef] [PubMed]
- Robledo, D.; Taggart, J.B.; Ireland, J.H.; McAndrew, B.J.; Starkey, W.G.; Haley, C.S.; Hamilton, A.; Guy, D.R.; Mota-Velasco, J.C.; Gheyas, A.A.; et al. Gene expression comparison of resistant and susceptible Atlantic salmon fry challenged with Infectious Pancreatic Necrosis virus reveals a marked contrast in immune response. BMC Genomics 2016, 17, 279. [Google Scholar] [CrossRef] [PubMed]
- Hansen, J.D.; La Patra, S. Induction of the rainbow trout MHC class I pathway during acute IHNV infection. Immunogenetics 2002, 54, 654–661. [Google Scholar] [CrossRef]
- Martin, S.A.; Zou, J.; Houlihan, D.F.; Secombes, C.J. Directional responses following recombinant cytokine stimulation of rainbow trout (Oncorhynchus mykiss) RTS-11 macrophage cells as revealed by transcriptome profiling. BMC Genomics 2007, 8, 150. [Google Scholar] [CrossRef]
- Kasthuri, S.R.; Umasuthan, N.; Whang, I.; Lim, B.S.; Jung, H.B.; Oh, M.J.; Jung, S.J.; Yeo, S.Y.; Kim, S.Y.; Lee, J. Molecular characterization and expressional affirmation of the beta proteasome subunit cluster in rock bream immune defense. Mol. Biol. Rep. 2014, 41, 5413–5427. [Google Scholar] [CrossRef] [PubMed]
- Momburg, F.; Roelse, J.; Howard, J.C.; Butcher, G.W.; Hämmerling, G.J.; Neefjes, J.J. Selectivity of MHC-encoded peptide transporters from human, mouse and rat. Nature 1994, 367, 648–651. [Google Scholar] [CrossRef]
- Powis, S.J.; Young, L.L.; Joly, E.; Barker, P.J.; Richardson, L.; Brandt, R.P.; Melief, C.J.; Howard, J.C.; Butcher, G.W. The rat cim effect: TAP allele-dependent changes in a class I MHC anchor motif and evidence against C-terminal trimming of peptides in the ER. Immunity 1996, 4, 159–165. [Google Scholar] [CrossRef]
- Joly, E.; Le Rolle, A.F.; González, A.L.; Mehling, B.; Stevens, J.; Coadwell, W.J.; Hünig, T.; Howard, J.C.; Butcher, G.W. Co-evolution of rat TAP transporters and MHC class I RT1-A molecules. Curr. Biol. 1998, 8, 169–172. [Google Scholar] [CrossRef]
- Walker, B.A.; Hunt, L.G.; Sowa, A.K.; Skjødt, K.; Göbel, T.W.; Lehner, P.J.; Kaufman, J. The dominantly expressed class I molecule of the chicken MHC is explained by coevolution with the polymorphic peptide transporter (TAP) genes. Proc. Natl. Acad. Sci. USA 2011, 108, 8396–8401. [Google Scholar] [CrossRef] [PubMed]
- Van Hateren, A.; Carter, R.; Bailey, A.; Kontouli, N.; Williams, A.P.; Kaufman, J.; Elliott, T. A mechanistic basis for the co-evolution of chicken tapasin and major histocompatibility complex class I (MHC I) proteins. J. Biol. Chem. 2013, 288, 32797–32808. [Google Scholar] [CrossRef]
- Tregaskes, C.A.; Harrison, M.; Sowa, A.K.; van Hateren, A.; Hunt, L.G.; Vainio, O.; Kaufman, J. Surface expression, peptide repertoire, and thermostability of chicken class I molecules correlate with peptide transporter specificity. Proc. Natl. Acad. Sci. USA 2016, 113, 692–697. [Google Scholar] [CrossRef] [PubMed]
- Namikawa, C.; Salter-Cid, L.; Flajnik, M.F.; Kato, Y.; Nonaka, M.; Sasaki, M. Isolation of Xenopus LMP-7 homologues. Striking allelic diversity and linkage to MHC. J. Immunol. 1995, 155, 1964–1971. [Google Scholar]
- Ohta, Y.; Powis, S.J.; Lohr, R.L.; Nonaka, M.; Pasquier, L.D.; Flajnik, M.F. Two highly divergent ancient allelic lineages of the transporter associated with antigen processing (TAP) gene in Xenopus: Further evidence for co-evolution among MHC class I region genes. Eur. J. Immunol. 2003, 33, 3017–3027. [Google Scholar] [CrossRef]
- Noro, M.; Nonaka, M. Evolution of dimorphisms of the proteasome subunit beta type 8 gene (PSMB8) in basal ray-finned fish. Immunogenetics 2014, 66, 325–334. [Google Scholar] [CrossRef]
- Kandil, E.; Namikawa, C.; Nonaka, M.; Greenberg, A.S.; Flajnik, M.F.; Ishibashi, T.; Kasahara, M. Isolation of low molecular mass polypeptide complementary DNA clones from primitive vertebrates. Implications for the origin of MHC class I-restricted antigen presentation. J. Immunol. 1996, 156, 4245–4253. [Google Scholar] [PubMed]
- Tsukamoto, K.; Miura, F.; Fujito, N.T.; Yoshizaki, G.; Nonaka, M. Long-lived dichotomous lineages of the proteasome subunit beta type 8 (PSMB8) gene surviving more than 500 million years as alleles or paralogs. Mol. Biol. Evol. 2012, 29, 3071–3079. [Google Scholar] [CrossRef] [PubMed]
- Roos, C.; Walter, L. Considerable haplotypic diversity in the RT1-CE class I gene region of the rat major histocompatibility complex. Immunogenetics 2005, 56, 773–777. [Google Scholar] [CrossRef]
- Hosomichi, K.; Shiina, T.; Suzuki, S.; Tanaka, M.; Shimizu, S.; Iwamoto, S.; Hara, H.; Yoshida, Y.; Kulski, J.K.; Inoko, H.; et al. The major histocompatibility complex (Mhc) class IIB region has greater genomic structural flexibility and diversity in the quail than the chicken. BMC Genomics 2006, 7, 322. [Google Scholar] [CrossRef] [PubMed]
- Flajnik, M.F.; Kasahara, M.; Shum, B.P.; Salter-Cid, L.; Taylor, E.; Du Pasquier, L. A novel type of class I gene organization in vertebrates: A large family of non-MHC-linked class I genes is expressed at the RNA level in the amphibian Xenopus. EMBO J. 1993, 12, 4385–4396. [Google Scholar] [CrossRef] [PubMed]
- Bartl, S.; Weissman, I.L. Isolation and characterization of major histocompatibility complex class IIB genes from the nurse shark. Proc. Natl. Acad. Sci. USA 1994, 91, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Málaga-Trillo, E.; Zaleska-Rutczynska, Z.; McAndrew, B.; Vincek, V.; Figueroa, F.; Sültmann, H.; Klein, J. Linkage relationships and haplotype polymorphism among cichlid Mhc class II B loci. Genetics 1998, 149, 1527–1537. [Google Scholar] [PubMed]
- Murray, B.W.; Shintani, S.; Sültmann, H.; Klein, J. Major histocompatibility complex class II A genes in cichlid fishes: Identification, expression, linkage relationships, and haplotype variation. Immunogenetics 2000, 51, 576–586. [Google Scholar] [CrossRef]
- Dirscherl, H.; Yoder, J.A. A nonclassical MHC class I U lineage locus in zebrafish with a null haplotypic variant. Immunogenetics 2015, 67, 501–513. [Google Scholar] [CrossRef]
- Sato, A.; Klein, D.; Sültmann, H.; Figueroa, F.; O’hUigin, C.; Klein, J. Class I mhc genes of cichlid fishes: Identification, expression, and polymorphism. Immunogenetics 1997, 46, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Murray, B.W.; Nilsson, P.; Zaleska-Rutczynska, Z.; Sültmann, H.; Klein, J. Linkage Relationships and Haplotype Variation of the Major Histocompatibility Complex Class I A Genes in the Cichlid Fish Oreochromis niloticus. Mar. Biotechnol 2000, 2, 437–448. [Google Scholar] [PubMed]
- Anderson, C.K.; Brossay, L. The role of MHC class Ib-restricted T cells during infection. Immunogenetics 2016, 68, 677–691. [Google Scholar] [CrossRef]
- Treiner, E.; Duban, L.; Bahram, S.; Radosavljevic, M.; Wanner, V.; Tilloy, F.; Affaticati, P.; Gilfillan, S.; Lantz, O. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature 2003, 422, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.H.; Chiu, N.M.; Mandal, M.; Wang, N.; Wang, C.R. Impaired NK1+ T cell development and early IL-4 production in CD1-deficient mice. Immunity 1997, 6, 459–467. [Google Scholar] [CrossRef]
- Allendorf, F.W.; Thorgaard, G.H. Tetraploidy and the evolution of salmonid fishes. In Evolutionary Genetics of Fishes; Turner, B.J., Ed.; Plenum Press: New York, NY, USA, 1984; pp. 1–53. [Google Scholar]
- Berthelot, C.; Brunet, F.; Chalopin, D.; Juanchich, A.; Bernard, M.; Noël, B.; Bento, P.; Da Silva, C.; Labadie, K.; Alberti, A.; et al. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat. Commun. 2014, 5, 3657. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, J.M.; Kiryu, I.; Yoshiura, Y.; Kumanovics, A.; Kohara, M.; Hayashi, N.; Ototake, M. Polymorphism of two very similar MHC class Ib loci in rainbow trout (Oncorhynchus mykiss). Immunogenetics 2006, 58, 152–167. [Google Scholar] [CrossRef]
- Landis, E.D.; Purcell, M.K.; Thorgaard, G.H.; Wheeler, P.A.; Hansen, J.D. Transcriptional profiling of MHC class I genes in rainbow trout infected with infectious hematopoietic necrosis virus. Mol. Immunol. 2008, 45, 1646–1657. [Google Scholar] [CrossRef]
- Pond, L.; Kuhn, L.A.; Teyton, L.; Schutze, M.P.; Tainer, J.A.; Jackson, M.R.; Peterson, P.A. A role for acidic residues in di-leucine motif-based targeting to the endocytic pathway. J. Biol. Chem. 1995, 270, 19989–19997. [Google Scholar] [CrossRef]
- Grimholt, U. MHC and Evolution in Teleosts. Biology 2016, 5, 6. [Google Scholar] [CrossRef]
- Lien, S.; Koop, B.F.; Sandve, S.R.; Miller, J.R.; Kent, M.P.; Nome, T.; Hvidsten, T.R.; Leong, J.S.; Minkley, D.R.; Zimin, A.; et al. The Atlantic salmon genome provides insights into rediploidization. Nature 2016, 533, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Phillips, R.B.; Nichols, K.M.; DeKoning, J.J.; Morasch, M.R.; Keatley, K.A.; Rexroad, C.E., 3rd; Gahr, S.A.; Danzmann, R.G.; Drew, R.E.; Thorgaard, G.H. Assignment of rainbow trout linkage groups to specific chromosomes. Genetics 2006, 174, 1661–1670. [Google Scholar] [CrossRef]
- Palti, Y.; Genet, C.; Luo, M.C.; Charlet, A.; Gao, G.; Hu, Y.; Castaño-Sánchez, C.; Tabet-Canale, K.; Krieg, F.; Yao, J.; et al. A first generation integrated map of the rainbow trout genome. BMC Genomics 2011, 12, 180. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, A.; Kiryu, I.; Dijkstra, J.M.; Yoshiura, Y.; Nishida-Umehara, C.; Ototake, M. Chromosome mapping of MHC class I in rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 2003, 14, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Phillips, R.B.; Zimmerman, A.; Noakes, M.A.; Palti, Y.; Morasch, M.R.; Eiben, L.; Ristow, S.S.; Thorgaard, G.H.; Hansen, J.D. Physical and genetic mapping of the rainbow trout major histocompatibility regions: evidence for duplication of the class I region. Immunogenetics 2003, 55, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, A.; Sakamoto, T.; Khoo, S.; Nakamura, K.; Coimbra, M.R.; Akutsu, T.; Okamoto, N. Quantitative trait loci (QTLs) associated with resistance/susceptibility to infectious pancreatic necrosis virus (IPNV) in rainbow trout (Oncorhynchus mykiss). Mol. Genet. Genomics 2001, 265, 23–31. [Google Scholar]
- Ozaki, A.; Khoo, S.; Yoshiura, Y.; Ototake, M.; Sakamoto, T.; Dijkstra, J.M.; Okamoto, N. Identification of Additional Quantitative Trait Loci (QTL) Responsible for Susceptibility to Infectious Pancreatic Necrosis Virus in Rainbow Trout. Fish Pathol. 2007, 42, 131–140. [Google Scholar] [CrossRef]
- Palti, Y.; Vallejo, R.L.; Gao, G.; Liu, S.; Hernandez, A.G.; Rexroad, C.E., 3rd; Wiens, G.D. Detection and Validation of QTL Affecting Bacterial Cold Water Disease Resistance in Rainbow Trout Using Restriction-Site Associated DNA Sequencing. PLoS ONE 2015, 10, e0138435. [Google Scholar] [CrossRef]
- Moen, T.; Baranski, M.; Sonesson, A.K.; Kjøglum, S. Confirmation and fine-mapping of a major QTL for resistance to infectious pancreatic necrosis in Atlantic salmon (Salmo salar): Population-level associations between markers and trait. BMC Genomics 2009, 10, 368. [Google Scholar] [CrossRef]
- Gonen, S.; Baranski, M.; Thorland, I.; Norris, A.; Grove, H.; Arnesen, P.; Bakke, H.; Lien, S.; Bishop, S.C.; Houston, R.D. Mapping and validation of a major QTL affecting resistance to pancreas disease (salmonid alphavirus) in Atlantic salmon (Salmo salar). Heredity 2015, 115, 405–414. [Google Scholar] [CrossRef]
- Khoo, S.K.; Ozaki, A.; Nakamura, F.; Arakawa, T.; Ichimoto, S.; Nickolov, R.; Sakamoto, T.; Akutsu, T.; Mochizuki, M.; Denda, I.; et al. Identification of a novel chromosomal region associated with infectious hematopoietic necrosis (IHN) resistance in rainbow trout Oncorhynchus mykiss. Fish Pathol. 2004, 39, 95–101. [Google Scholar] [CrossRef]
- Fraslin, C.; Dechamp, N.; Bernard, M.; Krieg, F.; Hervet, C.; Guyomard, R.; Esquerré, D.; Barbieri, J.; Kuchly, C.; Duchaud, E.; et al. Quantitative trait loci for resistance to Flavobacterium psychrophilum in rainbow trout: effect of the mode of infection and evidence of epistatic interactions. Genet. Sel. Evol. 2018, 50, 60. [Google Scholar] [CrossRef]
- Verrier, E.R.; Dorson, M.; Mauger, S.; Torhy, C.; Ciobotaru, C.; Hervet, C.; Dechamp, N.; Genet, C.; Boudinot, P.; Quillet, E. Resistance to a rhabdovirus (VHSV) in rainbow trout: Identification of a major QTL related to innate mechanisms. PLoS ONE 2013, 8, e55302. [Google Scholar] [CrossRef]
- Vallejo, R.L.; Liu, S.; Gao, G.; Fragomeni, B.O.; Hernandez, A.G.; Leeds, T.D.; Parsons, J.E.; Martin, K.E.; Evenhuis, J.P.; Welch, T.J.; et al. Similar Genetic Architecture with Shared and Unique Quantitative Trait Loci for Bacterial Cold Water Disease Resistance in Two Rainbow Trout Breeding Populations. Front. Genet. 2017, 8, 156. [Google Scholar] [CrossRef]
- Liu, S.; Vallejo, R.L.; Palti, Y.; Gao, G.; Marancik, D.P.; Hernandez, A.G.; Wiens, G.D. Identification of single nucleotide polymorphism markers associated with bacterial cold water disease resistance and spleen size in rainbow trout. Front. Genet. 2015, 6, 298. [Google Scholar] [CrossRef] [PubMed]
- Vallejo, R.L.; Palti, Y.; Liu, S.; Evenhuis, J.P.; Gao, G.; Rexroad, C.E., 3rd; Wiens, G.D. Detection of QTL in rainbow trout affecting survival when challenged with Flavobacterium psychrophilum. Mar. Biotechnol. 2014, 16, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Wiens, G.D.; Vallejo, R.L.; Leeds, T.D.; Palti, Y.; Hadidi, S.; Liu, S.; Evenhuis, J.P.; Welch, T.J.; Rexroad, C.E., 3rd. Assessment of genetic correlation between bacterial cold water disease resistance and spleen index in a domesticated population of rainbow trout: identification of QTL on chromosome Omy19. PLoS ONE 2013, 8, e75749. [Google Scholar] [CrossRef]
- Nichols, K.M.; Bartholomew, J.; Thorgaard, G.H. Mapping multiple genetic loci associated with Ceratomyxa shasta resistance in Oncorhynchus mykiss. Dis. Aquat. Organ. 2003, 56, 145–154. [Google Scholar] [CrossRef]
- Baerwald, M.R.; Petersen, J.L.; Hedrick, R.P.; Schisler, G.J.; May, B. A major effect quantitative trait locus for whirling disease resistance identified in rainbow trout (Oncorhynchus mykiss). Heredity 2011, 106, 920–926. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, A.M.; Evenhuis, J.P.; Thorgaard, G.H.; Ristow, S.S. A single major chromosomal region controls natural killer cell-like activity in rainbow trout. Immunogenetics 2004, 55, 825–835. [Google Scholar]
- Houston, R.D.; Haley, C.S.; Hamilton, A.; Guy, D.R.; Mota-Velasco, J.C.; Gheyas, A.A.; Tinch, A.E.; Taggart, J.B.; Bron, J.E.; Starkey, W.G.; et al. The susceptibility of Atlantic salmon fry to freshwater infectious pancreatic necrosis is largely explained by a major QTL. Heredity 2010, 105, 318–327. [Google Scholar] [CrossRef]
- Houston, R.D.; Haley, C.S.; Hamilton, A.; Guy, D.R.; Tinch, A.E.; Taggart, J.B.; McAndrew, B.J.; Bishop, S.C. Major quantitative trait loci affect resistance to infectious pancreatic necrosis in Atlantic salmon (Salmo salar). Genetics 2008, 178, 1109–1115. [Google Scholar] [CrossRef] [PubMed]
- Moen, T.; Fjalestad, K.T.; Munck, H.; Gomez-Raya, L. A multistage testing strategy for detection of quantitative trait Loci affecting disease resistance in Atlantic salmon. Genetics. 2004, 167, 851–858. [Google Scholar] [CrossRef]
- Gilbey, J.; Verspoor, E.; Mo, T.A.; Sterud, E.; Olstad, K.; Hytterød, S.; Jones, C.; Noble, L. Identification of genetic markers associated with Gyrodactylus salaris resistance in Atlantic salmon Salmo salar. Dis. Aquat. Organ. 2006, 25, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Sawayama, E.; Tanizawa, S.; Kitamura, S.I.; Nakayama, K.; Ohta, K.; Ozaki, A.; Takagi, M. Identification of Quantitative Trait Loci for Resistance to RSIVD in Red Sea Bream (Pagrus major). Mar. Biotechnol. 2017, 19, 601–613. [Google Scholar] [CrossRef]
- Miller, K.M.; Winton, J.R.; Schulze, A.D.; Purcell, M.K.; Ming, T.J. Major histocompatibility complex loci are associated with susceptibility of Atlantic salmon to infectious hematopoietic necrosis virus. Environ. Biol. Fishes 2004, 69, 307–316. [Google Scholar] [CrossRef]
- Johnson, N.A.; Vallejo, R.L.; Silverstein, J.T.; Welch, T.J.; Wiens, G.D.; Hallerman, E.M.; Palti, Y. Suggestive association of major histocompatibility IB genetic markers with resistance to bacterial cold water disease in rainbow trout (Oncorhynchus mykiss). Mar. Biotechnol. 2008, 10, 429–437. [Google Scholar] [CrossRef]
- Fischer, U.; Utke, K.; Ototake, M.; Dijkstra, J.M.; Kollner, B. Adaptive cell-mediated cytotoxicity against allogeneic targets by CD8-positive lymphocytes of rainbow trout (Oncorhynchus mykiss). Dev. Comp. Immunol. 2003, 27, 323–337. [Google Scholar] [CrossRef]
- Pearse, A.M.; Swift, K. Allograft theory: Transmission of devil facial-tumour disease. Nature 2006, 439, 549. [Google Scholar] [CrossRef]
- Murgia, C.; Pritchard, J.K.; Kim, S.Y.; Fassati, A.; Weiss, R.A. Clonal origin and evolution of a transmissible cancer. Cell 2006, 126, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Metzger, M.J.; Villalba, A.; Carballal, M.J.; Iglesias, D.; Sherry, J.; Reinisch, C.; Muttray, A.F.; Baldwin, S.A.; Goff, S.P. Widespread transmission of independent cancer lineages within multiple bivalve species. Nature 2016, 534, 705–709. [Google Scholar] [CrossRef]
- Qin, Q.W.; Ototake, M.; Nagoya, H.; Nakanishi, T. Graft-versus-host reaction (GVHR) in clonal amago salmon, Oncorhynchus rhodurus. Vet. Immunol. Immunopathol. 2002, 89, 83–89. [Google Scholar] [CrossRef]
- Griesemer, A.; Yamada, K.; Sykes, M. Xenotransplantation: Immunological hurdles and progress toward tolerance. Immunol. Rev. 2014, 258, 241–258. [Google Scholar] [CrossRef]
- Singh, P.B. Chemosensation and genetic individuality. Reproduction 2001, 121, 529–539. [Google Scholar] [CrossRef]
- Bernatchez, L.; Landry, C. MHC studies in nonmodel vertebrates: What have we learned about natural selection in 15 years? J. Evol. Biol. 2003, 16, 363–377. [Google Scholar] [CrossRef] [PubMed]
- Overath, P.; Sturm, T.; Rammensee, H.G. Of volatiles and peptides: In search for MHC-dependent olfactory signals in social communication. Cell Mol. Life Sci. 2014, 71, 2429–2442. [Google Scholar] [CrossRef]
- Lobmaier, J.S.; Fischbacher, U.; Probst, F.; Wirthmüller, U.; Knoch, D. Accumulating evidence suggests that men do not find body odours of human leucocyte antigen-dissimilar women more attractive. Proc. Biol. Sci. 2018, 285, 20180566. [Google Scholar] [CrossRef]
- Reusch, T.B.; Haberli, M.A.; Aeschlimann, P.B.; Milinski, M. Female sticklebacks count alleles in a strategy of sexual selection explaining MHC polymorphism. Nature 2001, 414, 300–302. [Google Scholar] [CrossRef]
- Bay, R.A.; Arnegard, M.E.; Conte, G.L.; Best, J.; Bedford, N.L.; McCann, S.R.; Dubin, M.E.; Chan, Y.F.; Jones, F.C.; Kingsley, D.M.; et al. Genetic Coupling of Female Mate Choice with Polygenic Ecological Divergence Facilitates Stickleback Speciation. Curr. Biol. 2017, 27, 3344–3349. [Google Scholar] [CrossRef]
- Glazer, A.M.; Killingbeck, E.E.; Mitros, T.; Rokhsar, D.S.; Miller, C.T. Genome Assembly Improvement and Mapping Convergently Evolved Skeletal Traits in Sticklebacks with Genotyping-by-Sequencing. G3 2015, 5, 1463–1472. [Google Scholar] [CrossRef]
- Corriveau, R.A.; Huh, G.S.; Shatz, C.J. Regulation of class I MHC expression in the developing and mature CNS by Neural activity. Neuron 1998, 21, 505–520. [Google Scholar] [CrossRef]
- Huh, G.S.; Boulanger, L.M.; Du, H.; Riquelme, P.A.; Brotz, T.M.; Shatz, C.J. Functional requirement for class I MHC in CNS development and plasticity. Science 2000, 290, 2155–2159. [Google Scholar] [CrossRef]
- Loconto, J.; Papes, F.; Chang, E.; Stowers, L.; Jones, E.P.; Takada, T.; Kumanovics, A.; Fischer Lindahl, K.; Dulac, C. Functional expression of murine V2R pheromone receptors involves selective association with the M10 and M1 families of MHC class Ib molecules. Cell 2003, 112, 607–618. [Google Scholar] [CrossRef]
- Oliveira, A.L.; Thams, S.; Lidman, O.; Piehl, F.; Hokfelt, T.; Karre, K.; Linda, H.; Cullheim, S. A role for MHC class I molecules in synaptic plasticity and regeneration of neurons after axotomy. Proc. Natl. Acad. Sci. USA 2004, 101, 17843–17848. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Brott, B.K.; Kirkby, L.A.; Adelson, J.D.; Cheng, S.; Feller, M.B.; Datwani, A.; Shatz, C.J. Synapse elimination and learning rules co-regulated by MHC class I H2-Db. Nature 2014, 509, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Adelson, J.D.; Sapp, R.W.; Brott, B.K.; Lee, H.; Miyamichi, K.; Luo, L.; Cheng, S.; Djurisic, M.; Shatz, C.J. Developmental Sculpting of Intracortical Circuits by MHC Class I H2-Db and H2-Kb. Cereb. Cortex 2016, 26, 1453–1463. [Google Scholar] [CrossRef] [PubMed]
- Shatz, C.J. MHC class I: An unexpected role in neuronal plasticity. Neuron 2009, 64, 40–45. [Google Scholar] [CrossRef]
- Elmer, B.M.; McAllister, A.K. Major histocompatibility complex class I proteins in brain development and plasticity. Trends Neurosci. 2012, 35, 660–670. [Google Scholar] [CrossRef]
- Misra, M.K.; Damotte, V.; Hollenbach, J.A. The Immunogenetics of neurological disease. Immunology 2018, 153, 399–414. [Google Scholar] [CrossRef]
- Benus, R.F.; Bohus, B.; Koolhaas, J.M.; van Oortmerssen, G.A. Heritable variation for aggression as a reflection of individual coping strategies. Experientia 1991, 47, 1008–1019. [Google Scholar] [CrossRef] [PubMed]
- Sokolowski, M.B.; Pereira, H.S.; Hughes, K. Evolution of foraging behavior in Drosophila by density-dependent selection. Proc. Natl. Acad. Sci. USA 1997, 94, 7373–7377. [Google Scholar] [CrossRef]
- Campbell, J.M.; Carter, P.A.; Wheeler, P.A.; Thorgaard, G.H. Aggressive behavior.; brain size and domestication in clonal rainbow trout lines. Behav. Genet. 2015, 45, 245–254. [Google Scholar] [CrossRef]
- Azuma, T.; Dijkstra, J.M.; Kiryu, I.; Sekiguchi, T.; Terada, Y.; Asahina, K.; Fischer, U.; Ototake, M. Growth and behavioral traits in Donaldson rainbow trout (Oncorhynchus mykiss) cosegregate with classical major histocompatibility complex (MHC) class I genotype. Behav. Genet. 2005, 35, 463–478. [Google Scholar] [CrossRef]
- Wright, D.; Nakamichi, R.; Krause, J.; Butlin, R.K. QTL analysis of behavioral and morphological differentiation between wild and laboratory zebrafish (Danio rerio). Behav. Genet. 2006, 36, 271–284. [Google Scholar] [CrossRef]
- Liu, S.; Vallejo, R.L.; Gao, G.; Palti, Y.; Weber, G.M.; Hernandez, A.; Rexroad, C.E., 3rd. Identification of single-nucleotide polymorphism markers associated with cortisol response to crowding in rainbow trout. Mar. Biotechnol. 2015, 17, 328–337. [Google Scholar] [CrossRef]
- Cnaani, A.; Zilberman, N.; Tinman, S.; Hulata, G.; Ron, M. Genome-scan analysis for quantitative trait loci in an F2 tilapia hybrid. Mol. Genet. Genomics 2004, 272, 162–172. [Google Scholar] [CrossRef]
- Phillips, R.B.; Ventura, A.B.; Dekoning, J.J.; Nichols, K.M. Mapping rainbow trout immune genes involved in inflammation reveals conserved blocks of immune genes in teleosts. Anim. Genet. 2013, 44, 107–113. [Google Scholar] [CrossRef]
| MHC molecule types MHC class I versus class II Like mammals, most fish possess MHC class I, MHC class IIα, MHC class IIβ and β2-m molecules. However, some teleost fish lost MHC class II genes in their evolution. [24,31,32,33,34] Classical versus nonclassical MHC class II As in mammals, in fish both polymorphic classical and nonpolymorphic nonclassical MHC class II are found. The nonclassical MHC class II lineages in teleost fish are not the same as in tetrapods, and, unlike the DM lineage distribution in tetrapods, they are not stably inherited throughout those teleosts that do possess an MHC class II system. [27,35,36,37,38,39] Classical versus nonclassical MHC class I Like mammals, all investigated teleost fish possess at least one polymorphic classical MHC class I gene and a number of nonpolymorphic nonclassical ones. The conclassical MHC class I lineages found in tetrapods and fish are not the same. Different from mammals is that throughout ray-finned fish members of an ancient nonclassical MHC class I lineage named "Z" are conserved which show highly conserved features for presumably binding a (modified) peptide ligand, while the Z lineage was lost in tetrapods. [24,40,41,42,43,44,45,46] Genomic organization Genomic location of classical MHC class II genes Unlike in mammals, teleost fish classical MHC class II genes are not situated in typical Mhc regions and are not linked with the classical MHC class I genes. Even between teleost fish species differences in genomic locations of the classical MHC class II genes can be observed, highlighting the relative plasticity of the teleost fish MHC class II system. [27,47,48] Genomic location of classical MHC class I genes As in mammals, fish classical MHC class I genes are linked together with TAP, tapasin, PSMB and other conserved "framework" genes in a typical Mhc region. [11,49,50,51,52,53] PSMB, TAP and tapasin genes in the Mhc region As in mammals, PSMB, TAP and tapasin genes are located in the teleost fish Mhc region, but their number and lineages differ. Unlike in mammals, teleost fish TAP1 is not linked with the Mhc region. Compared to mammals, teleost fish have additional ancient gene copies of the PSMB6/9/12 and PSMB7/10/13 lineages. In some but not all teleost fish, different from humans, considerable allelic or haplotype variation can be found for the Mhc-situated PSMB and TAP2 genes. [54,55,56,57] Functions of classical MHC class II Expression of classical MHC class II As in mammals, teleost fish classical MHC class II transcripts and molecules appear to be expressed in professional antigen presenting cells like for example B lymphocytes. Furthermore, as in mammals, their expression is upregulated after immune stimulation, which agrees with conservation of promoter motifs similar to those in mammals. [58,59,60,61,62,63,64,65] Peptide presentation by classical MHC class II For fish MHC class II, the structure or functions in peptide presentation have not been determined yet. However, fish classical MHC class II molecules have a set of conserved residues which suggest a similar mode of peptide ligand binding as known in mammals. [27] Allograft rejection As in mammals, teleost fish classical MHC class II genes have been found linked with allograft rejection. [66] Indirect evidence for classical MHC class II function Although fragmentary, there is abundant evidence for helper and regulatory T cell functions in teleost fish similar to as in mammals. Furthermore, teleost fish have CD4, LAG-3 and CD74 molecules that probably all participate in the MHC class II system as in mammals. [31,32,59,60,67,68,69,70,71,72,73,74,75,76,77,78,79] Functions of classical MHC class I Expression of classical MHC class I As in mammals, teleost fish classical MHC class I transcripts and molecules are ubiquitously expressed and show the highest expression in epithelial and lymphoid tissues. Furthermore, as in mammals, their expression is upregulated after immune stimulation, which agrees with conservation of promoter motifs similar to those in mammals. Different from mammals is that in teleost fish, which are ectotherm species, the levels of MHC class I can be temperature dependent. [62,80,81,82,83,84] Peptide presentation As in mammals, teleost fish classical MHC class I molecules form heterotrimer complexes with β2-m and peptide ligands of ~9 aa. X-ray crystallography analysis revealed a similar complex structure as known in mammals. [85,86] Allograft rejection As in mammals, teleost fish classical MHC class I genes have been found linked with allograft rejection. [87,88] MHC class I restriction of cell-mediated cytotoxicity by T cells Although conclusive experiments have not been performed yet, there are several lines of evidence that together suggest that specific cell-mediated cytotoxicity by teleost fish CD8+ T cells requires classical MHC class I matching of the target cells as known in mammals. [89,90,91,92,93,94,95] |
| A) Disease Resistance/Immunity Related QTL Studies in Rainbow Trout | ||
| Resistance against | QTL encoding linkage group | Reference |
| Infectious pancreatic necrosis (IPN) virus | Chr. 3, Chr. 7, Chr. 8, Chr. 14 (IB), Chr. 16, Chr-17 (II), Chr. 20, Chr. 24, Chr. 27 | Ozaki et al. 2007 [201] |
| Chr. 14 (IB), Chr. 16 | Ozaki et al. 2001 [200] | |
| Infectious hematopoietic necrosis (IHN) virus | Chr. 17 (II) | Khoo et al. 2004 [205] |
| Viral hemorrhagic septicemia (VHS) virus | Chr. 2, Chr. 3, Chr. 4, Chr. 5, Chr. 17 (II), Chr. 24 | Verrier et al. 2013 [207] |
| Cold water disease Flavobacterium psychrophilum (bacterium) | Chr. 2, Chr. 3, Chr. 7, Chr. 10, Chr. 17 (II), Chr. 21, Chr. 24, Chr. 25, Chr. 26, Chr. 29 | Fraslin et al. 2018 [206] |
| Chr. 3, Chr. 5, Chr. 8, Chr. 10, Chr. 13, Chr. 15, Chr. 25 | Vallejo et al. 2017 [208] | |
| Chr. 8, Chr. 19, Chr. 25 | Liu et al. 2015 [209] | |
| Chr. 1, Chr. 6, Chr. 7, Chr. 8, Chr. 11, Chr. 12, Chr. 14 (IB), Chr. 25 | Palti et al. 2015 [202] | |
| Chr. 2, Chr. 3, Chr. 6, Chr. 8, Chr. 12, Chr. 13, Chr. 20 | Vallejo et al. 2014 [210] | |
| Chr. 5, Chr. 16, Chr. 19 | Wiens et al. 2013 [211] | |
| Ceratomyxa shasta (parasite) | Chr. 9, Chr. 16, Chr. 20, Chr. 22, Chr. 29 | Nichols et al. 2003 [212] |
| Whirling disease Myxobolus cerebralis (parasite) | Chr. 9 | Baerwald et al. 2011 [213] |
| YAC-1 cells (murine tumor cell line) | Chr. 3 | Zimmerman et al. 2004 [214] |
| B) Disease Resistance/Immunity Related QTL Studies in Atlantic Salmon | ||
| Infectious pancreatic necrosis (IPN) virus | Chr. 26 | Houston et al. 2010 [215] |
| Chr. 4, Chr. 8, Chr. 26 | Houston et al. 2008 [216] | |
| Chr. 1, Chr. 3, Chr. 4, Chr. 5, Chr. 6, Chr. 7, Chr. 9, Chr.10, Chr. 14 (IB), Chr. 17, Chr. 18, Chr. 19, Chr. 20, Chr. 26 | Moen et al 2009 [203] | |
| Pancreas disease Salmonid alphavirus (SAV) | Chr. 2, Chr. 3, Chr. 4, Chr. 7, Chr. 14 (IB), Chr. 26 | Gonen et al. 2015 [204] |
| Infectious Salmon Anaemia (ISA) virus | Chr. 15 (maybe additional weaker QTL, but difficult to interpret) | Moen et al. 2004 [217] |
| Gyrodactylus salaris (parasite) | Chr. 4, Chr. 5, Chr. 6, Chr. 10, Chr. 13, Chr. 15, Chr. 16, Chr. 17, Chr. 23, Chr. 24 | Gilbey et al. 2006 [218] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamaguchi, T.; Dijkstra, J.M. Major Histocompatibility Complex (MHC) Genes and Disease Resistance in Fish. Cells 2019, 8, 378. https://doi.org/10.3390/cells8040378
Yamaguchi T, Dijkstra JM. Major Histocompatibility Complex (MHC) Genes and Disease Resistance in Fish. Cells. 2019; 8(4):378. https://doi.org/10.3390/cells8040378
Chicago/Turabian StyleYamaguchi, Takuya, and Johannes M. Dijkstra. 2019. "Major Histocompatibility Complex (MHC) Genes and Disease Resistance in Fish" Cells 8, no. 4: 378. https://doi.org/10.3390/cells8040378




