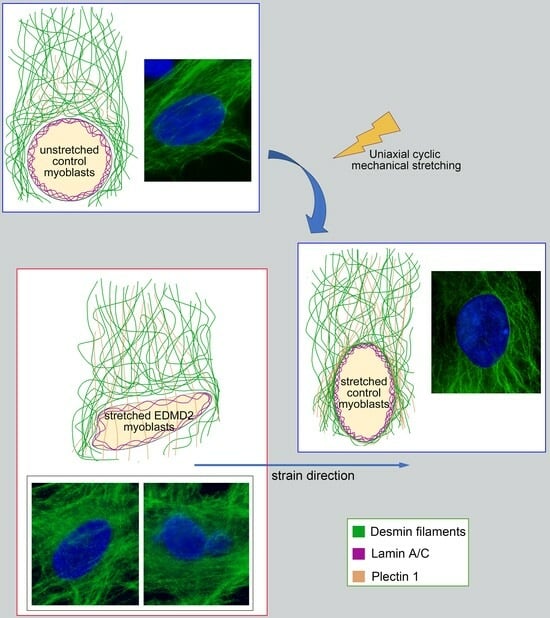
Desmin and Plectin Recruitment to the Nucleus and Nuclei Orientation Are Lost in Emery-Dreifuss Muscular Dystrophy Myoblasts Subjected to Mechanical Stimulation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture, Transfection and siRNA
2.2. Mechanical Stretching
2.3. Antibodies
2.4. Immunoprecipitation
2.5. In Situ Proximity Ligation Assay
2.6. Immunofluorescence Analysis
2.7. Imaging and SIM Analysis
2.8. Statistical Analysis
3. Results
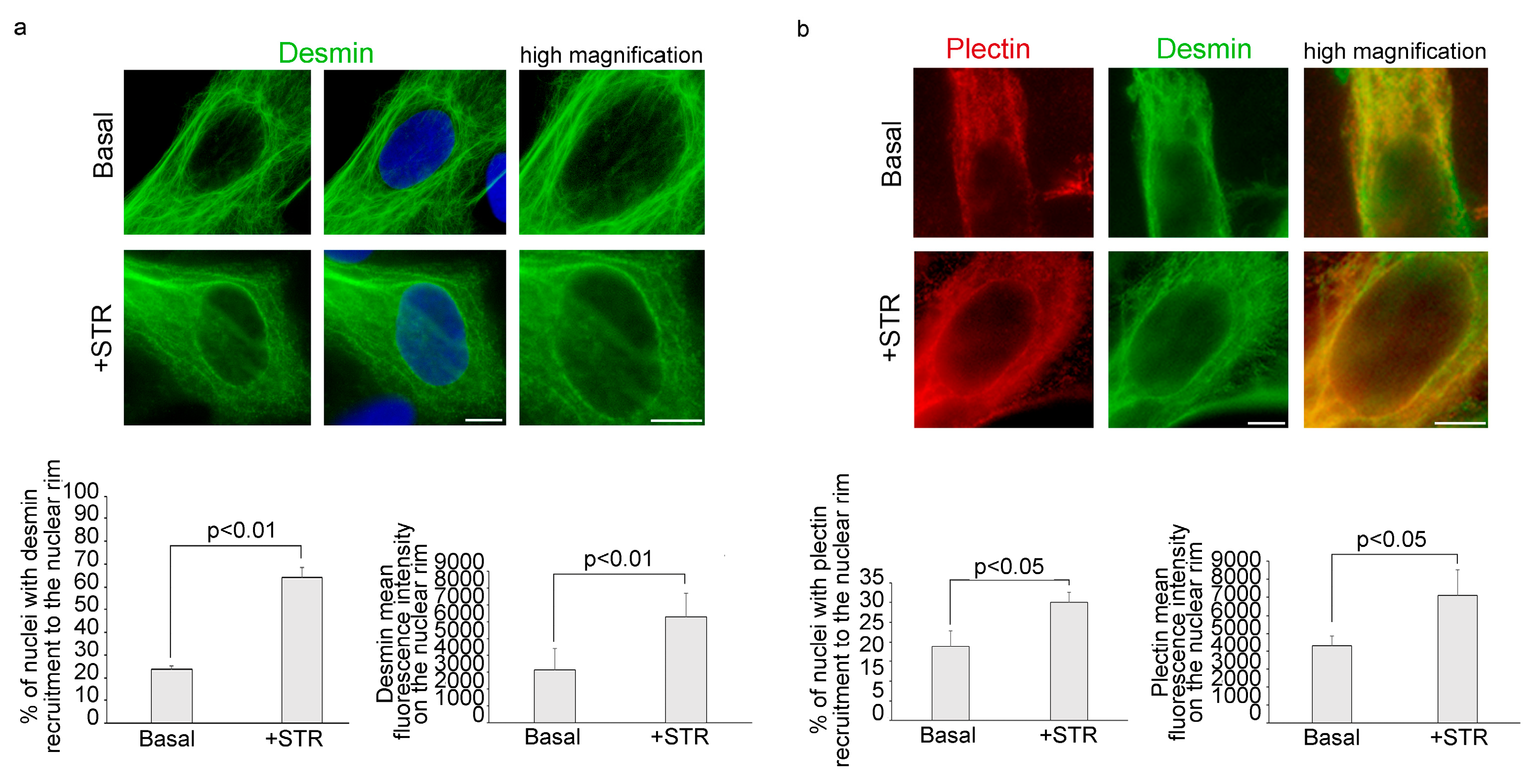
3.1. Mechanical Stretching Stimulates Desmin and Plectin 1 Translocation to the Nuclear Envelope in Human Myoblasts
3.2. Mechanical Stress Induces Desmin Recruitment to Lamina A/C in Human Myoblasts
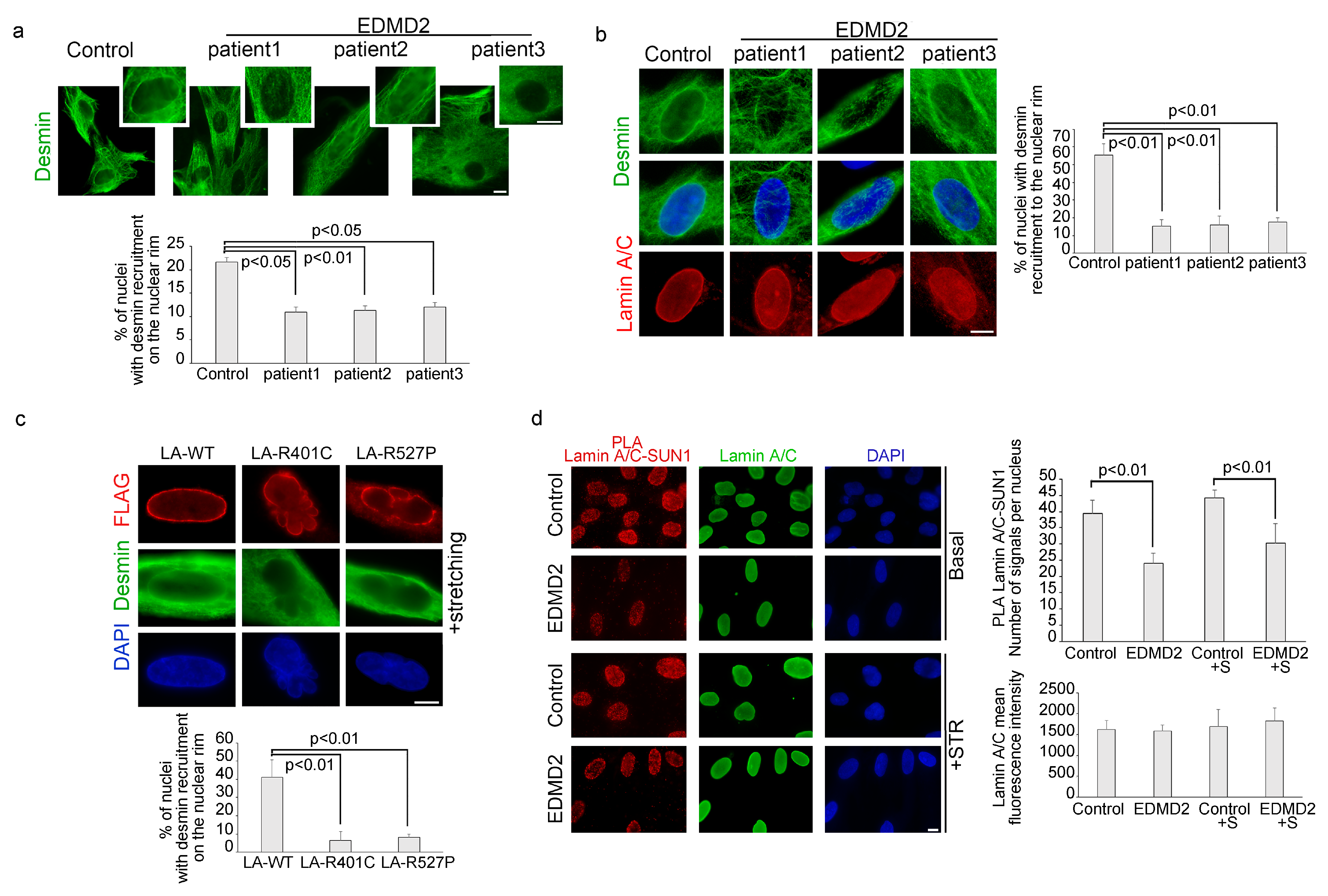
3.3. The Recruitment of Desmin to the Nuclear Envelope Is Altered in EDMD2 Myoblasts
3.4. LMNA-Mutated Myoblasts Subjected to Mechanical Stretching Show Reduced Recruitment of Plectin 1 to the Nuclear Envelope and Nuclei Reorientation Defects
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Osmanagic-Myers, S.; Dechat, T.; Foisner, R. Lamins at the crossroads of mechanosignaling. Genes Dev. 2015, 29, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, M.L.; Jaalouk, D.E.; Shanahan, C.M.; Burke, B.; Roux, K.J.; Lammerding, J. The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J. Biol. Chem. 2011, 286, 26743–26753. [Google Scholar] [CrossRef] [PubMed]
- Houben, F.; Willems, C.H.; Declercq, I.L.; Hochstenbach, K.; Kamps, M.A.; Snoeckx, L.H.; Ramaekers, F.C.; Broers, J.L. Disturbed nuclear orientation and cellular migration in A-type lamin deficient cells. Biochim. Biophys. Acta 2009, 1793, 312–324. [Google Scholar] [CrossRef]
- Mattioli, E.; Columbaro, M.; Capanni, C.; Maraldi, N.M.; Cenni, V.; Scotlandi, K.; Marino, M.T.; Merlini, L.; Squarzoni, S.; Lattanzi, G. Prelamin A-mediated recruitment of SUN1 to the nuclear envelope directs nuclear positioning in human muscle. Cell Death Differ. 2011, 18, 1305–1315. [Google Scholar] [CrossRef] [PubMed]
- Meinke, P.; Mattioli, E.; Haque, F.; Antoku, S.; Columbaro, M.; Straatman, K.R.; Worman, H.J.; Gundersen, G.G.; Lattanzi, G.; Wehnert, M.; et al. Muscular dystrophy-associated SUN1 and SUN2 variants disrupt nuclear-cytoskeletal connections and myonuclear organization. PLoS Genet. 2014, 10, e1004605. [Google Scholar] [CrossRef]
- Chang, W.; Antoku, S.; Ostlund, C.; Worman, H.J.; Gundersen, G.G. Linker of nucleoskeleton and cytoskeleton (LINC) complex-mediated actin-dependent nuclear positioning orients centrosomes in migrating myoblasts. Nucleus 2015, 6, 77–88. [Google Scholar] [CrossRef]
- Vahabikashi, A.; Sivagurunathan, S.; Nicdao, F.A.S.; Han, Y.L.; Park, C.Y.; Kittisopikul, M.; Wong, X.; Tran, J.R.; Gundersen, G.G.; Reddy, K.L.; et al. Nuclear lamin isoforms differentially contribute to LINC complex-dependent nucleocytoskeletal coupling and whole-cell mechanics. Proc. Natl. Acad. Sci. USA 2022, 119, e2121816119. [Google Scholar] [CrossRef]
- Kim, J.K.; Louhghalam, A.; Lee, G.; Schafer, B.W.; Wirtz, D.; Kim, D.H. Nuclear lamin A/C harnesses the perinuclear apical actin cables to protect nuclear morphology. Nat. Commun. 2017, 8, 2123. [Google Scholar] [CrossRef]
- Bertrand, A.T.; Ziaei, S.; Ehret, C.; Duchemin, H.; Mamchaoui, K.; Bigot, A.; Mayer, M.; Quijano-Roy, S.; Desguerre, I.; Laine, J.; et al. Cellular microenvironments reveal defective mechanosensing responses and elevated YAP signaling in LMNA-mutated muscle precursors. J. Cell Sci. 2014, 127, 2873–2884. [Google Scholar] [CrossRef]
- Khatau, S.B.; Hale, C.M.; Stewart-Hutchinson, P.J.; Patel, M.S.; Stewart, C.L.; Searson, P.C.; Hodzic, D.; Wirtz, D. A perinuclear actin cap regulates nuclear shape. Proc. Natl. Acad. Sci. USA 2009, 106, 19017–19022. [Google Scholar] [CrossRef]
- Lammerding, J.; Schulze, P.C.; Takahashi, T.; Kozlov, S.; Sullivan, T.; Kamm, R.D.; Stewart, C.L.; Lee, R.T. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J. Clin. Investig. 2004, 113, 370–378. [Google Scholar] [CrossRef]
- Muchir, A.; Worman, H.J. Emery-Dreifuss muscular dystrophy: Focal point nuclear envelope. Curr. Opin. Neurol. 2019, 32, 728–734. [Google Scholar] [CrossRef]
- Maggi, L.; Mavroidis, M.; Psarras, S.; Capetanaki, Y.; Lattanzi, G. Skeletal and Cardiac Muscle Disorders Caused by Mutations in Genes Encoding Intermediate Filament Proteins. Int. J. Mol. Sci. 2021, 22, 4256. [Google Scholar] [CrossRef] [PubMed]
- Ripoll-Vera, T.; Zorio, E.; Gamez, J.M.; Molina, P.; Govea, N.; Cremer, D. Phenotypic Patterns of Cardiomyopathy Caused by Mutations in the Desmin Gene. A Clinical and Genetic Study in Two Inherited Heart Disease Units. Rev. Esp. Cardiol. (Engl. Ed.) 2015, 68, 1027–1029. [Google Scholar] [CrossRef]
- Agnetti, G.; Herrmann, H.; Cohen, S. New roles for desmin in the maintenance of muscle homeostasis. FEBS J. 2022, 289, 2755–2770. [Google Scholar] [CrossRef]
- Capetanaki, Y.; Papathanasiou, S.; Diokmetzidou, A.; Vatsellas, G.; Tsikitis, M. Desmin related disease: A matter of cell survival failure. Curr. Opin. Cell Biol. 2015, 32, 113–120. [Google Scholar] [CrossRef]
- Hol, E.M.; Capetanaki, Y. Type III Intermediate Filaments Desmin, Glial Fibrillary Acidic Protein (GFAP), Vimentin, and Peripherin. Cold Spring Harb. Perspect. Biol. 2017, 9, a021642. [Google Scholar] [CrossRef] [PubMed]
- Chandar, S.; Yeo, L.S.; Leimena, C.; Tan, J.C.; Xiao, X.H.; Nikolova-Krstevski, V.; Yasuoka, Y.; Gardiner-Garden, M.; Wu, J.; Kesteven, S.; et al. Effects of mechanical stress and carvedilol in lamin A/C-deficient dilated cardiomyopathy. Circ. Res. 2010, 106, 573–582. [Google Scholar] [CrossRef]
- Nikolova, V.; Leimena, C.; McMahon, A.C.; Tan, J.C.; Chandar, S.; Jogia, D.; Kesteven, S.H.; Michalicek, J.; Otway, R.; Verheyen, F.; et al. Defects in nuclear structure and function promote dilated cardiomyopathy in lamin A/C-deficient mice. J. Clin. Investig. 2004, 113, 357–369. [Google Scholar] [CrossRef]
- Frock, R.L.; Kudlow, B.A.; Evans, A.M.; Jameson, S.A.; Hauschka, S.D.; Kennedy, B.K. Lamin A/C and emerin are critical for skeletal muscle satellite cell differentiation. Genes Dev. 2006, 20, 486–500. [Google Scholar] [CrossRef] [PubMed]
- Galata, Z.; Kloukina, I.; Kostavasili, I.; Varela, A.; Davos, C.H.; Makridakis, M.; Bonne, G.; Capetanaki, Y. Amelioration of desmin network defects by alphaB-crystallin overexpression confers cardioprotection in a mouse model of dilated cardiomyopathy caused by LMNA gene mutation. J. Mol. Cell. Cardiol. 2018, 125, 73–86. [Google Scholar] [CrossRef]
- Morioka, M.; Parameswaran, H.; Naruse, K.; Kondo, M.; Sokabe, M.; Hasegawa, Y.; Suki, B.; Ito, S. Microtubule dynamics regulate cyclic stretch-induced cell alignment in human airway smooth muscle cells. PLoS ONE 2011, 6, e26384. [Google Scholar] [CrossRef]
- Hoffman, L.M.; Smith, M.A.; Jensen, C.C.; Yoshigi, M.; Blankman, E.; Ullman, K.S.; Beckerle, M.C. Mechanical stress triggers nuclear remodeling and the formation of transmembrane actin nuclear lines with associated nuclear pore complexes. Mol. Biol. Cell 2020, 31, 1774–1787. [Google Scholar] [CrossRef]
- Jabre, S.; Hleihel, W.; Coirault, C. Nuclear Mechanotransduction in Skeletal Muscle. Cells 2021, 10, 318. [Google Scholar] [CrossRef]
- Wiche, G. Plectin-Mediated Intermediate Filament Functions: Why Isoforms Matter. Cells 2021, 10, 2154. [Google Scholar] [CrossRef]
- Staszewska, I.; Fischer, I.; Wiche, G. Plectin isoform 1-dependent nuclear docking of desmin networks affects myonuclear architecture and expression of mechanotransducers. Hum. Mol. Genet. 2015, 24, 7373–7389. [Google Scholar] [CrossRef]
- Konieczny, P.; Fuchs, P.; Reipert, S.; Kunz, W.S.; Zeold, A.; Fischer, I.; Paulin, D.; Schroder, R.; Wiche, G. Myofiber integrity depends on desmin network targeting to Z-disks and costameres via distinct plectin isoforms. J. Cell Biol. 2008, 181, 667–681. [Google Scholar] [CrossRef]
- Prechova, M.; Adamova, Z.; Schweizer, A.L.; Maninova, M.; Bauer, A.; Kah, D.; Meier-Menches, S.M.; Wiche, G.; Fabry, B.; Gregor, M. Plectin-mediated cytoskeletal crosstalk controls cell tension and cohesion in epithelial sheets. J. Cell Biol. 2022, 221, e202105146. [Google Scholar] [CrossRef]
- Zrelski, M.M.; Kustermann, M.; Winter, L. Muscle-Related Plectinopathies. Cells 2021, 10, 2480. [Google Scholar] [CrossRef]
- Cenni, V.; Bertacchini, J.; Beretti, F.; Lattanzi, G.; Bavelloni, A.; Riccio, M.; Ruzzene, M.; Marin, O.; Arrigoni, G.; Parnaik, V.; et al. Lamin A Ser404 is a nuclear target of Akt phosphorylation in C2C12 cells. J. Proteome Res. 2008, 7, 4727–4735. [Google Scholar] [CrossRef]
- Santi, S.; Cenni, V.; Capanni, C.; Lattanzi, G.; Mattioli, E. PCAF Involvement in Lamin A/C-HDAC2 Interplay during the Early Phase of Muscle Differentiation. Cells 2020, 9, 1735. [Google Scholar] [CrossRef] [PubMed]
- Gregor, M.; Zeold, A.; Oehler, S.; Marobela, K.A.; Fuchs, P.; Weigel, G.; Hardie, D.G.; Wiche, G. Plectin scaffolds recruit energy-controlling AMP-activated protein kinase (AMPK) in differentiated myofibres. J. Cell Sci. 2006, 119, 1864–1875. [Google Scholar] [CrossRef] [PubMed]
- Folker, E.S.; Ostlund, C.; Luxton, G.W.; Worman, H.J.; Gundersen, G.G. Lamin A variants that cause striated muscle disease are defective in anchoring transmembrane actin-associated nuclear lines for nuclear movement. Proc. Natl. Acad. Sci. USA 2011, 108, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Broers, J.L.; Hutchison, C.J.; Ramaekers, F.C. Laminopathies. J. Pathol. 2004, 204, 478–488. [Google Scholar] [CrossRef]
- Ueda, N.; Maekawa, M.; Matsui, T.S.; Deguchi, S.; Takata, T.; Katahira, J.; Higashiyama, S.; Hieda, M. Inner Nuclear Membrane Protein, SUN1, is Required for Cytoskeletal Force Generation and Focal Adhesion Maturation. Front. Cell Dev. Biol. 2022, 10, 885859. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cenni, V.; Evangelisti, C.; Santi, S.; Sabatelli, P.; Neri, S.; Cavallo, M.; Lattanzi, G.; Mattioli, E. Desmin and Plectin Recruitment to the Nucleus and Nuclei Orientation Are Lost in Emery-Dreifuss Muscular Dystrophy Myoblasts Subjected to Mechanical Stimulation. Cells 2024, 13, 162. https://doi.org/10.3390/cells13020162
Cenni V, Evangelisti C, Santi S, Sabatelli P, Neri S, Cavallo M, Lattanzi G, Mattioli E. Desmin and Plectin Recruitment to the Nucleus and Nuclei Orientation Are Lost in Emery-Dreifuss Muscular Dystrophy Myoblasts Subjected to Mechanical Stimulation. Cells. 2024; 13(2):162. https://doi.org/10.3390/cells13020162
Chicago/Turabian StyleCenni, Vittoria, Camilla Evangelisti, Spartaco Santi, Patrizia Sabatelli, Simona Neri, Marco Cavallo, Giovanna Lattanzi, and Elisabetta Mattioli. 2024. "Desmin and Plectin Recruitment to the Nucleus and Nuclei Orientation Are Lost in Emery-Dreifuss Muscular Dystrophy Myoblasts Subjected to Mechanical Stimulation" Cells 13, no. 2: 162. https://doi.org/10.3390/cells13020162