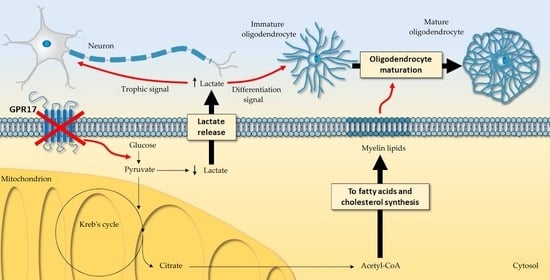
Rewiring of Glucose and Lipid Metabolism Induced by G Protein-Coupled Receptor 17 Silencing Enables the Transition of Oligodendrocyte Progenitors to Myelinating Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Care
2.2. Rat OPC Isolation
2.3. siRNA Transfection
2.4. Transcriptome Profiling and Microarray Analysis
2.5. Bioinformatic Analysis
2.6. Targeted Metabolomics and Lipidomics
2.7. Immunocytochemistry and Cell Counting
2.8. Treatment with AR-C155858
2.9. Total RNA Extraction, Retrotranscription and qPCR Analysis
2.10. Statistical Analysis
3. Results
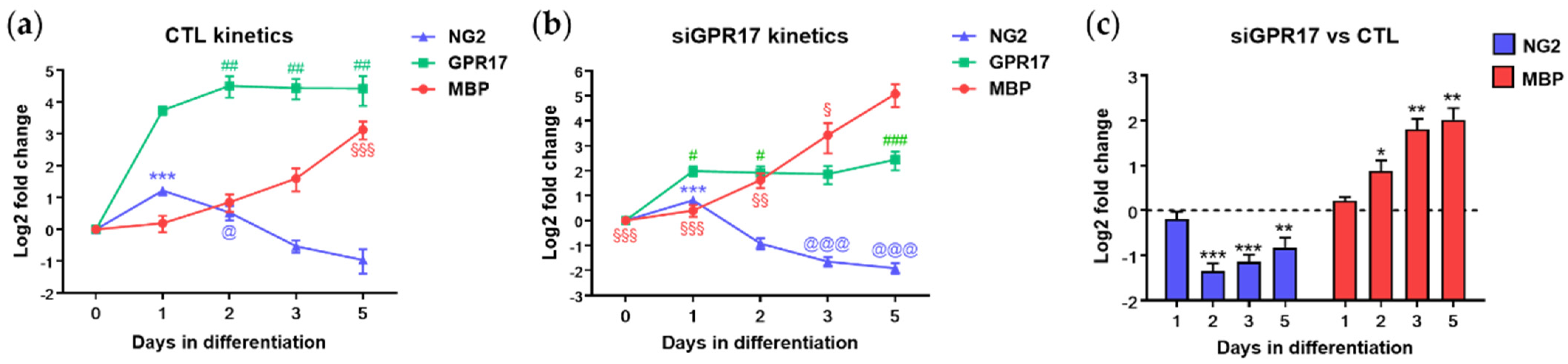
3.1. GPR17 Silencing in Differentiating OPCs Significantly Altered the Expression of Genes Involved in Lipid and Glucose Metabolism
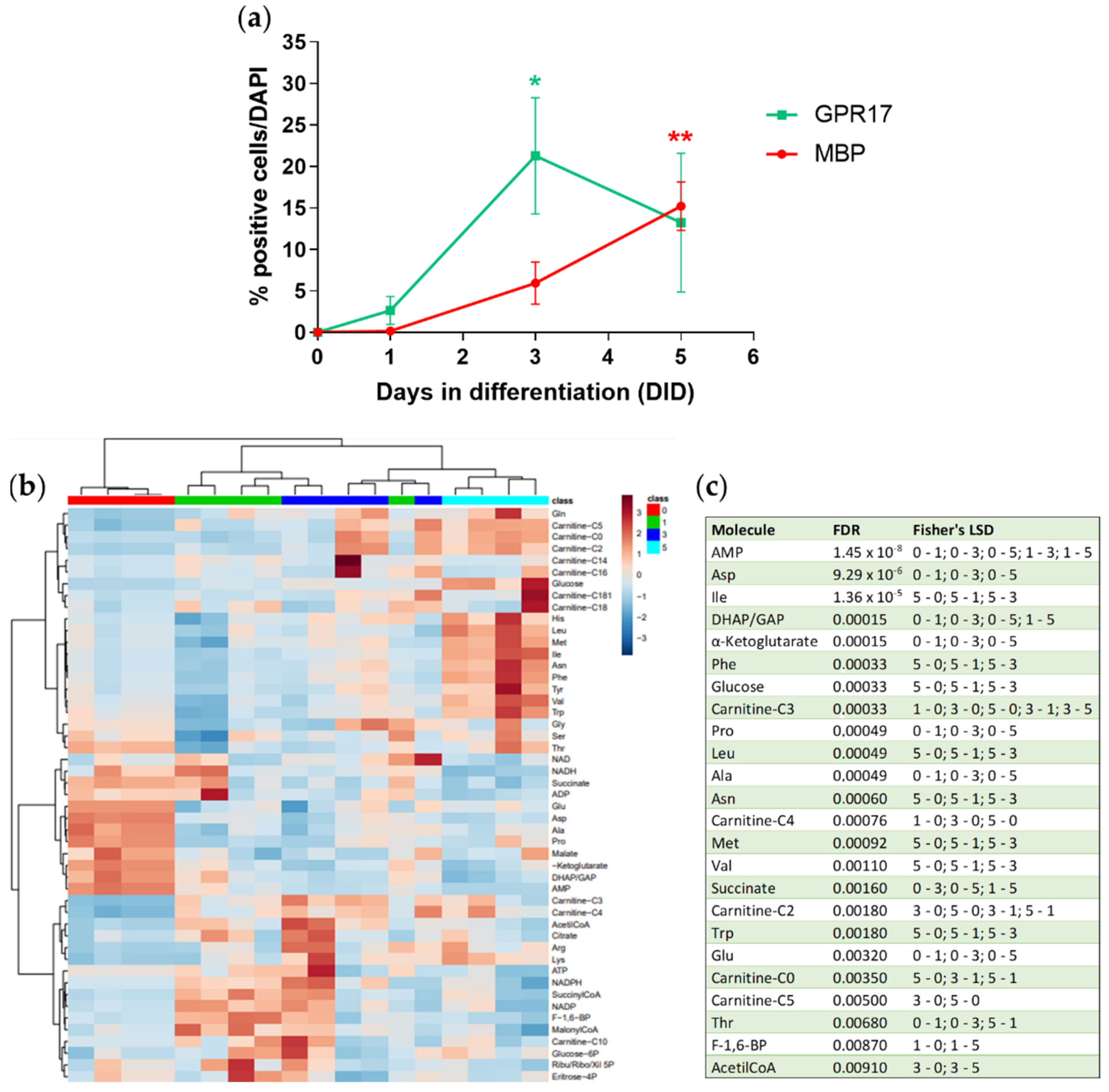
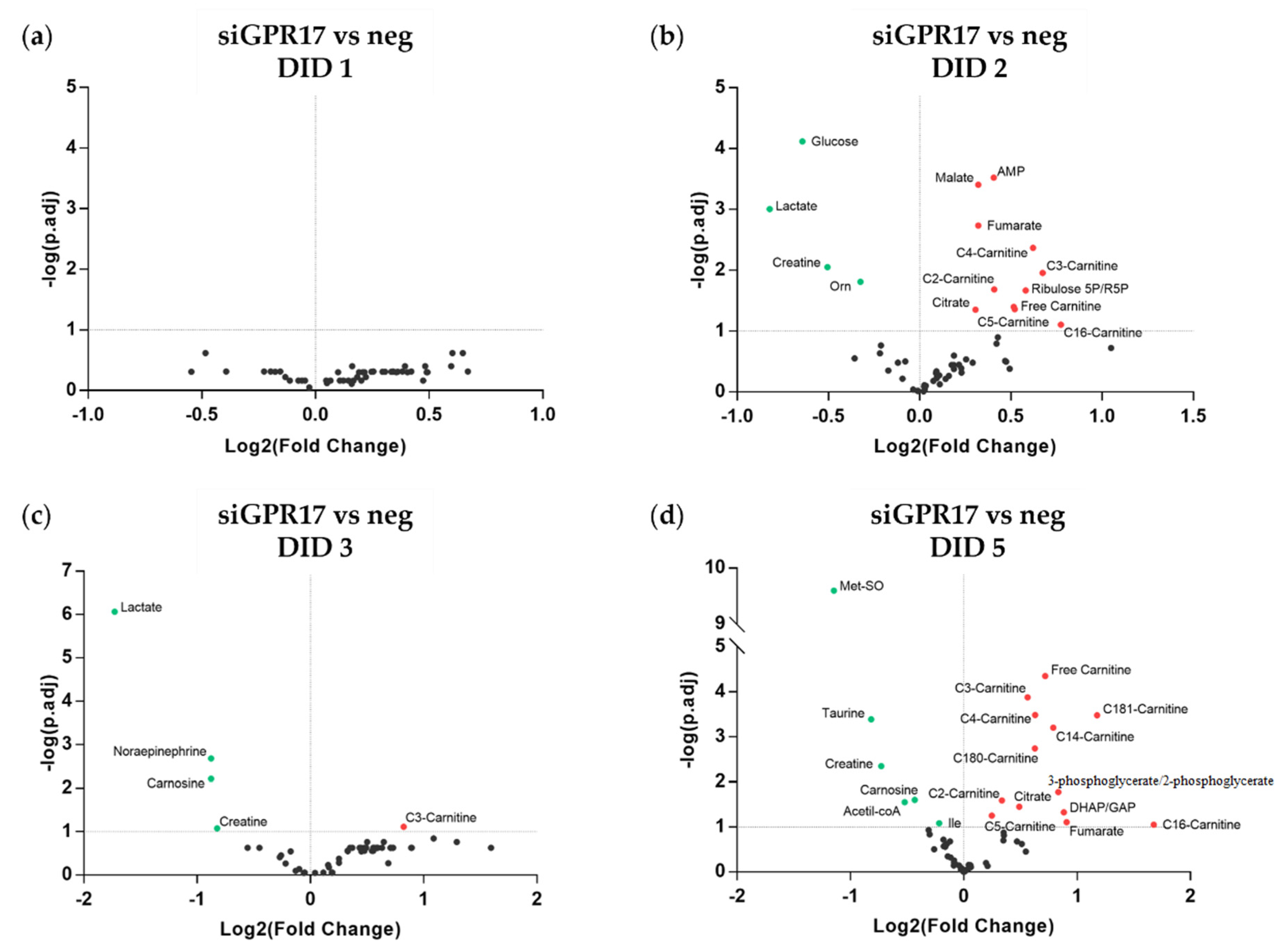
3.2. Metabolomic Analysis during OPC Differentiation In Vitro
3.3. Metabolomic Analysis after GPR17 Silencing during OPC Maturation In Vitro
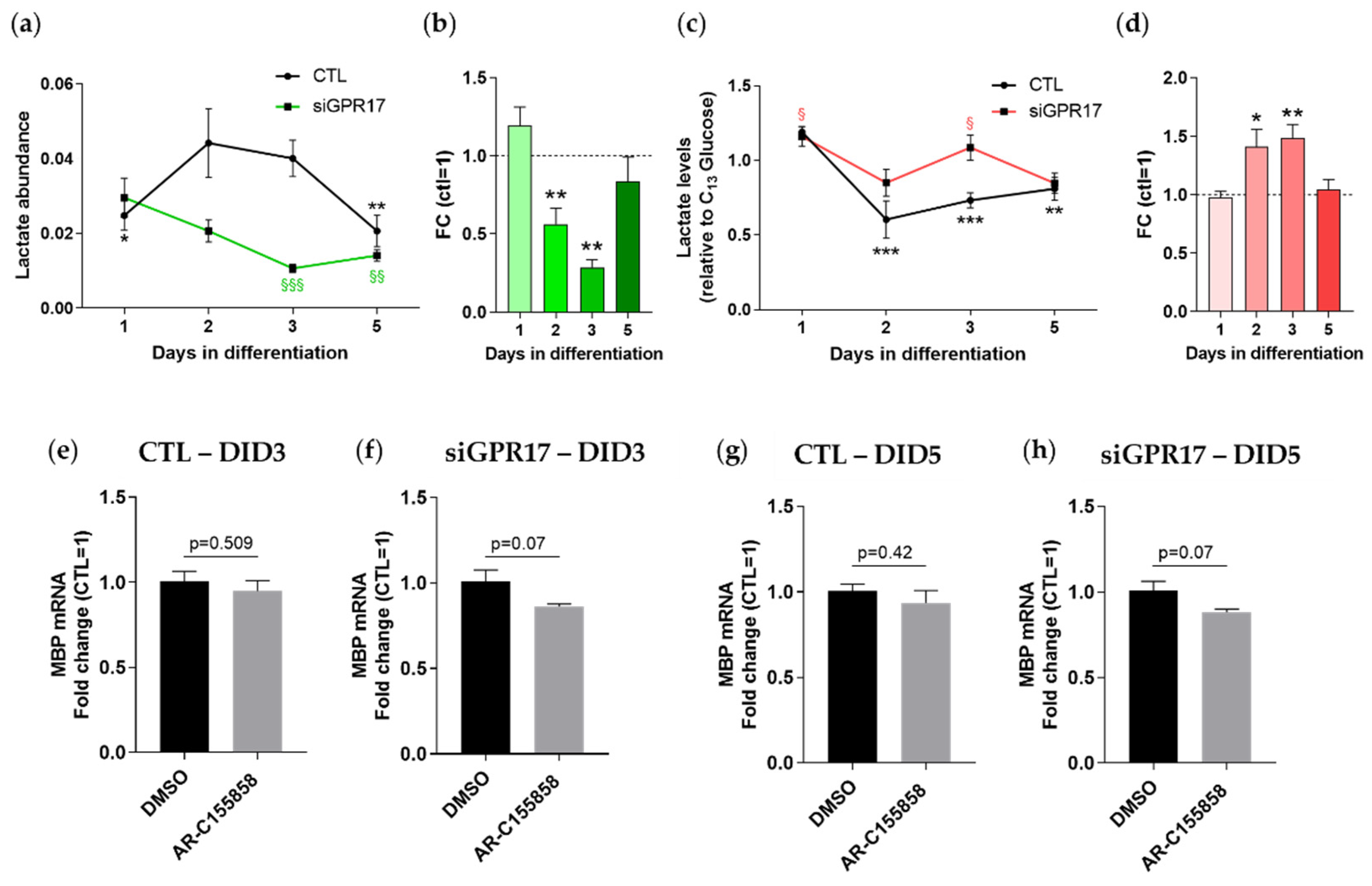
3.4. GPR17 Modulates Lactate Release during OPC Maturation
3.5. Lipidomic Analysis after GPR17 Silencing during OPC Maturation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Butt, A.M.; Papanikolaou, M.; Rivera, A. Physiology of Oligodendroglia. Adv. Exp. Med. Biol. 2019, 1175, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Nave, K.A.; Werner, H.B. Myelination of the nervous system: Mechanisms and functions. Annu. Rev. Cell Dev. Biol. 2014, 30, 503–533. [Google Scholar] [CrossRef] [PubMed]
- Narine, M.; Colognato, H. Current Insights Into Oligodendrocyte Metabolism and Its Power to Sculpt the Myelin Landscape. Front. Cell. Neurosci. 2022, 16, 892968. [Google Scholar] [CrossRef]
- Tepavcevic, V. Oligodendroglial Energy Metabolism and (re)Myelination. Life 2021, 11, 238. [Google Scholar] [CrossRef] [PubMed]
- Rao, V.T.S.; Khan, D.; Cui, Q.L.; Fuh, S.C.; Hossain, S.; Almazan, G.; Multhaup, G.; Healy, L.M.; Kennedy, T.E.; Antel, J.P. Distinct age and differentiation-state dependent metabolic profiles of oligodendrocytes under optimal and stress conditions. PLoS ONE 2017, 12, e0182372. [Google Scholar] [CrossRef] [Green Version]
- Schoenfeld, R.; Wong, A.; Silva, J.; Li, M.; Itoh, A.; Horiuchi, M.; Itoh, T.; Pleasure, D.; Cortopassi, G. Oligodendroglial differentiation induces mitochondrial genes and inhibition of mitochondrial function represses oligodendroglial differentiation. Mitochondrion 2010, 10, 143–150. [Google Scholar] [CrossRef] [Green Version]
- Rone, M.B.; Cui, Q.L.; Fang, J.; Wang, L.C.; Zhang, J.; Khan, D.; Bedard, M.; Almazan, G.; Ludwin, S.K.; Jones, R.; et al. Oligodendrogliopathy in Multiple Sclerosis: Low Glycolytic Metabolic Rate Promotes Oligodendrocyte Survival. J. Neurosci. 2016, 36, 4698–4707. [Google Scholar] [CrossRef] [Green Version]
- Jackman, N.; Ishii, A.; Bansal, R. Oligodendrocyte development and myelin biogenesis: Parsing out the roles of glycosphingolipids. Physiology 2009, 24, 290–297. [Google Scholar] [CrossRef] [Green Version]
- Philips, T.; Rothstein, J.D. Oligodendroglia: Metabolic supporters of neurons. J. Clin. Investig. 2017, 127, 3271–3280. [Google Scholar] [CrossRef] [Green Version]
- Saab, A.S.; Nave, K.A. Myelin dynamics: Protecting and shaping neuronal functions. Curr. Opin. Neurobiol. 2017, 47, 104–112. [Google Scholar] [CrossRef]
- Herbert, A.L.; Monk, K.R. Advances in myelinating glial cell development. Curr. Opin. Neurobiol. 2017, 42, 53–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fumagalli, M.; Daniele, S.; Lecca, D.; Lee, P.R.; Parravicini, C.; Fields, R.D.; Rosa, P.; Antonucci, F.; Verderio, C.; Trincavelli, M.L.; et al. Phenotypic changes, signaling pathway, and functional correlates of GPR17-expressing neural precursor cells during oligodendrocyte differentiation. J. Biol. Chem. 2011, 286, 10593–10604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lecca, D.; Raffaele, S.; Abbracchio, M.P.; Fumagalli, M. Regulation and signaling of the GPR17 receptor in oligodendroglial cells. Glia 2020, 68, 1957–1967. [Google Scholar] [CrossRef] [PubMed]
- Lecca, D.; Trincavelli, M.L.; Gelosa, P.; Sironi, L.; Ciana, P.; Fumagalli, M.; Villa, G.; Verderio, C.; Grumelli, C.; Guerrini, U.; et al. The recently identified P2Y-like receptor GPR17 is a sensor of brain damage and a new target for brain repair. PLoS ONE 2008, 3, e3579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coppolino, G.T.; Marangon, D.; Negri, C.; Menichetti, G.; Fumagalli, M.; Gelosa, P.; Dimou, L.; Furlan, R.; Lecca, D.; Abbracchio, M.P. Differential local tissue permissiveness influences the final fate of GPR17-expressing oligodendrocyte precursors in two distinct models of demyelination. Glia 2018, 66, 1118–1130. [Google Scholar] [CrossRef] [PubMed]
- Nyamoya, S.; Leopold, P.; Becker, B.; Beyer, C.; Hustadt, F.; Schmitz, C.; Michel, A.; Kipp, M. G-Protein-Coupled Receptor Gpr17 Expression in Two Multiple Sclerosis Remyelination Models. Mol. Neurobiol. 2019, 56, 1109–1123. [Google Scholar] [CrossRef] [PubMed]
- Boda, E.; Vigano, F.; Rosa, P.; Fumagalli, M.; Labat-Gest, V.; Tempia, F.; Abbracchio, M.P.; Dimou, L.; Buffo, A. The GPR17 receptor in NG2 expressing cells: Focus on in vivo cell maturation and participation in acute trauma and chronic damage. Glia 2011, 59, 1958–1973. [Google Scholar] [CrossRef] [PubMed]
- Bonfanti, E.; Bonifacino, T.; Raffaele, S.; Milanese, M.; Morgante, E.; Bonanno, G.; Abbracchio, M.P.; Fumagalli, M. Abnormal Upregulation of GPR17 Receptor Contributes to Oligodendrocyte Dysfunction in SOD1 G93A Mice. Int. J. Mol. Sci. 2020, 21, 2395. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Wu, H.; Wang, S.; Koito, H.; Li, J.; Ye, F.; Hoang, J.; Escobar, S.S.; Gow, A.; Arnett, H.A.; et al. The oligodendrocyte-specific G protein-coupled receptor GPR17 is a cell-intrinsic timer of myelination. Nat. Neurosci. 2009, 12, 1398–1406. [Google Scholar] [CrossRef] [Green Version]
- Angelini, J.; Marangon, D.; Raffaele, S.; Lecca, D.; Abbracchio, M.P. The Distribution of GPR17-Expressing Cells Correlates with White Matter Inflammation Status in Brain Tissues of Multiple Sclerosis Patients. Int. J. Mol. Sci. 2021, 22, 4574. [Google Scholar] [CrossRef]
- Franke, H.; Parravicini, C.; Lecca, D.; Zanier, E.R.; Heine, C.; Bremicker, K.; Fumagalli, M.; Rosa, P.; Longhi, L.; Stocchetti, N.; et al. Changes of the GPR17 receptor, a new target for neurorepair, in neurons and glial cells in patients with traumatic brain injury. Purinergic. Signal. 2013, 9, 451–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satoh, J.I.; Kino, Y.; Yanaizu, M.; Tosaki, Y.; Sakai, K.; Ishida, T.; Saito, Y. Expression of GPR17, a regulator of oligodendrocyte differentiation and maturation, in Nasu-Hakola disease brains. Intractable Rare Dis. Res. 2017, 6, 50–54. [Google Scholar] [CrossRef] [Green Version]
- Han, R.; Liang, J.; Zhou, B. Glucose Metabolic Dysfunction in Neurodegenerative Diseases-New Mechanistic Insights and the Potential of Hypoxia as a Prospective Therapy Targeting Metabolic Reprogramming. Int. J. Mol. Sci. 2021, 22, 5887. [Google Scholar] [CrossRef] [PubMed]
- Pineda-Torra, I.; Siddique, S.; Waddington, K.E.; Farrell, R.; Jury, E.C. Disrupted Lipid Metabolism in Multiple Sclerosis: A Role for Liver X Receptors? Front. Endocrinol. 2021, 12, 639757. [Google Scholar] [CrossRef] [PubMed]
- Nijland, P.G.; Molenaar, R.J.; van der Pol, S.M.; van der Valk, P.; van Noorden, C.J.; de Vries, H.E.; van Horssen, J. Differential expression of glucose-metabolizing enzymes in multiple sclerosis lesions. Acta Neuropathol. Commun. 2015, 3, 79. [Google Scholar] [CrossRef] [Green Version]
- Vallee, A.; Lecarpentier, Y.; Guillevin, R.; Vallee, J.N. Demyelination in Multiple Sclerosis: Reprogramming Energy Metabolism and Potential PPARgamma Agonist Treatment Approaches. Int. J. Mol. Sci. 2018, 19, 1212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marangon, D.; Abbracchio, M.P.; Lecca, D. Pathway-Focused Profiling of Oligodendrocytes Over-Expressing miR-125a-3p Reveals Alteration of Wnt and Cell-to-Cell Signaling. Cell. Mol. Neurobiol. 2021, 41, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Marangon, D.; Boda, E.; Parolisi, R.; Negri, C.; Giorgi, C.; Montarolo, F.; Perga, S.; Bertolotto, A.; Buffo, A.; Abbracchio, M.P.; et al. In vivo silencing of miR-125a-3p promotes myelin repair in models of white matter demyelination. Glia 2020, 68, 2001–2014. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009, 37, W305–W311. [Google Scholar] [CrossRef]
- Brivio, P.; Audano, M.; Gallo, M.T.; Gruca, P.; Lason, M.; Litwa, E.; Fumagalli, F.; Papp, M.; Mitro, N.; Calabrese, F. Metabolomic signature and mitochondrial dynamics outline the difference between vulnerability and resilience to chronic stress. Transl. Psychiatry 2022, 12, 87. [Google Scholar] [CrossRef] [PubMed]
- Macchi, C.; Moregola, A.; Greco, M.F.; Svecla, M.; Bonacina, F.; Dhup, S.; Dadhich, R.K.; Audano, M.; Sonveaux, P.; Mauro, C.; et al. Monocarboxylate transporter 1 deficiency impacts CD8(+) T lymphocytes proliferation and recruitment to adipose tissue during obesity. iScience 2022, 25, 104435. [Google Scholar] [CrossRef] [PubMed]
- Cermenati, G.; Audano, M.; Giatti, S.; Carozzi, V.; Porretta-Serapiglia, C.; Pettinato, E.; Ferri, C.; D’Antonio, M.; De Fabiani, E.; Crestani, M.; et al. Lack of sterol regulatory element binding factor-1c imposes glial Fatty Acid utilization leading to peripheral neuropathy. Cell Metab. 2015, 21, 571–583. [Google Scholar] [CrossRef] [Green Version]
- Hoxha, E.; Balbo, I.; Parolisi, R.; Audano, M.; Montarolo, F.; Ravera, F.; Guglielmotto, M.; Muratori, L.; Raimondo, S.; DiGregorio, E.; et al. Elovl5 is required for proper action potential conduction along peripheral myelinated fibers. Glia 2021, 69, 2419–2428. [Google Scholar] [CrossRef] [PubMed]
- Ghaffari, M.H.; Jahanbekam, A.; Sadri, H.; Schuh, K.; Dusel, G.; Prehn, C.; Adamski, J.; Koch, C.; Sauerwein, H. Metabolomics meets machine learning: Longitudinal metabolite profiling in serum of normal versus overconditioned cows and pathway analysis. J. Dairy Sci. 2019, 102, 11561–11585. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, R.A.; Hoefsloot, H.C.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef] [Green Version]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef]
- Fumagalli, M.; Bonfanti, E.; Daniele, S.; Zappelli, E.; Lecca, D.; Martini, C.; Trincavelli, M.L.; Abbracchio, M.P. The ubiquitin ligase Mdm2 controls oligodendrocyte maturation by intertwining mTOR with G protein-coupled receptor kinase 2 in the regulation of GPR17 receptor desensitization. Glia 2015, 63, 2327–2339. [Google Scholar] [CrossRef]
- Audano, M.; Pedretti, S.; Cermenati, G.; Brioschi, E.; Diaferia, G.R.; Ghisletti, S.; Cuomo, A.; Bonaldi, T.; Salerno, F.; Mora, M.; et al. Zc3h10 is a novel mitochondrial regulator. EMBO Rep. 2018, 19, e45531. [Google Scholar] [CrossRef]
- Ichihara, Y.; Doi, T.; Ryu, Y.; Nagao, M.; Sawada, Y.; Ogata, T. Oligodendrocyte Progenitor Cells Directly Utilize Lactate for Promoting Cell Cycling and Differentiation. J. Cell. Physiol. 2017, 232, 986–995. [Google Scholar] [CrossRef]
- Fratangeli, A.; Parmigiani, E.; Fumagalli, M.; Lecca, D.; Benfante, R.; Passafaro, M.; Buffo, A.; Abbracchio, M.P.; Rosa, P. The regulated expression, intracellular trafficking, and membrane recycling of the P2Y-like receptor GPR17 in Oli-neu oligodendroglial cells. J. Biol. Chem. 2013, 288, 5241–5256. [Google Scholar] [CrossRef] [Green Version]
- Ou, Z.; Sun, Y.; Lin, L.; You, N.; Liu, X.; Li, H.; Ma, Y.; Cao, L.; Han, Y.; Liu, M.; et al. Olig2-Targeted G-Protein-Coupled Receptor Gpr17 Regulates Oligodendrocyte Survival in Response to Lysolecithin-Induced Demyelination. J. Neurosci. 2016, 36, 10560–10573. [Google Scholar] [CrossRef] [Green Version]
- Bauernfeind, A.L.; Barks, S.K.; Duka, T.; Grossman, L.I.; Hof, P.R.; Sherwood, C.C. Aerobic glycolysis in the primate brain: Reconsidering the implications for growth and maintenance. Brain Struct. Funct. 2014, 219, 1149–1167. [Google Scholar] [CrossRef]
- Simon, K.; Hennen, S.; Merten, N.; Blattermann, S.; Gillard, M.; Kostenis, E.; Gomeza, J. The Orphan G Protein-coupled Receptor GPR17 Negatively Regulates Oligodendrocyte Differentiation via Galphai/o and Its Downstream Effector Molecules. J. Biol. Chem. 2016, 291, 705–718. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.J.; Vainshtein, A.; Maik-Rachline, G.; Peles, E. G protein-coupled receptor 37 is a negative regulator of oligodendrocyte differentiation and myelination. Nat. Commun. 2016, 7, 10884. [Google Scholar] [CrossRef] [Green Version]
- Beyer, B.A.; Fang, M.; Sadrian, B.; Montenegro-Burke, J.R.; Plaisted, W.C.; Kok, B.P.C.; Saez, E.; Kondo, T.; Siuzdak, G.; Lairson, L.L. Metabolomics-based discovery of a metabolite that enhances oligodendrocyte maturation. Nat. Chem. Biol. 2018, 14, 22–28. [Google Scholar] [CrossRef] [Green Version]
- Chamberlain, K.A.; Chapey, K.S.; Nanescu, S.E.; Huang, J.K. Creatine Enhances Mitochondrial-Mediated Oligodendrocyte Survival After Demyelinating Injury. J. Neurosci. 2017, 37, 1479–1492. [Google Scholar] [CrossRef] [Green Version]
- Rinholm, J.E.; Hamilton, N.B.; Kessaris, N.; Richardson, W.D.; Bergersen, L.H.; Attwell, D. Regulation of oligodendrocyte development and myelination by glucose and lactate. J. Neurosci. 2011, 31, 538–548. [Google Scholar] [CrossRef] [Green Version]
- Jha, M.K.; Morrison, B.M. Lactate Transporters Mediate Glia-Neuron Metabolic Crosstalk in Homeostasis and Disease. Front. Cell. Neurosci. 2020, 14, 589582. [Google Scholar] [CrossRef]
- Ou, Z.; Ma, Y.; Sun, Y.; Zheng, G.; Wang, S.; Xing, R.; Chen, X.; Han, Y.; Wang, J.; Lu, Q.R.; et al. A GPR17-cAMP-Lactate Signaling Axis in Oligodendrocytes Regulates Whole-Body Metabolism. Cell Rep. 2019, 26, 2984–2997.e2984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker, S.A.; Gajera, C.R.; Wawro, A.M.; Corces, M.R.; Montine, T.J. GATM and GAMT synthesize creatine locally throughout the mammalian body and within oligodendrocytes of the brain. Brain Res. 2021, 1770, 147627. [Google Scholar] [CrossRef]
- Vitvitsky, V.; Garg, S.K.; Banerjee, R. Taurine biosynthesis by neurons and astrocytes. J. Biol. Chem. 2011, 286, 32002–32010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.; Morrison, B.M.; Li, Y.; Lengacher, S.; Farah, M.H.; Hoffman, P.N.; Liu, Y.; Tsingalia, A.; Jin, L.; Zhang, P.W.; et al. Oligodendroglia metabolically support axons and contribute to neurodegeneration. Nature 2012, 487, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Philips, T.; Mironova, Y.A.; Jouroukhin, Y.; Chew, J.; Vidensky, S.; Farah, M.H.; Pletnikov, M.V.; Bergles, D.E.; Morrison, B.M.; Rothstein, J.D. MCT1 Deletion in Oligodendrocyte Lineage Cells Causes Late-Onset Hypomyelination and Axonal Degeneration. Cell Rep. 2021, 34, 108610. [Google Scholar] [CrossRef] [PubMed]
- Meffre, D.; Shackleford, G.; Hichor, M.; Gorgievski, V.; Tzavara, E.T.; Trousson, A.; Ghoumari, A.M.; Deboux, C.; Nait Oumesmar, B.; Liere, P.; et al. Liver X receptors alpha and beta promote myelination and remyelination in the cerebellum. Proc. Natl. Acad. Sci. USA 2015, 112, 7587–7592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sensi, C.; Daniele, S.; Parravicini, C.; Zappelli, E.; Russo, V.; Trincavelli, M.L.; Martini, C.; Abbracchio, M.P.; Eberini, I. Oxysterols act as promiscuous ligands of class-A GPCRs: In silico molecular modeling and in vitro validation. Cell. Signal. 2014, 26, 2614–2620. [Google Scholar] [CrossRef] [PubMed]
- Elbaz, B.; Popko, B. Molecular Control of Oligodendrocyte Development. Trends Neurosci. 2019, 42, 263–277. [Google Scholar] [CrossRef] [PubMed]
- Polito, A.; Reynolds, R. NG2-expressing cells as oligodendrocyte progenitors in the normal and demyelinated adult central nervous system. J. Anat. 2005, 207, 707–716. [Google Scholar] [CrossRef]
- Imgrund, S.; Hartmann, D.; Farwanah, H.; Eckhardt, M.; Sandhoff, R.; Degen, J.; Gieselmann, V.; Sandhoff, K.; Willecke, K. Adult ceramide synthase 2 (CERS2)-deficient mice exhibit myelin sheath defects, cerebellar degeneration, and hepatocarcinomas. J. Biol. Chem. 2009, 284, 33549–33560. [Google Scholar] [CrossRef] [Green Version]
- Marangon, D.; Boccazzi, M.; Lecca, D.; Fumagalli, M. Regulation of Oligodendrocyte Functions: Targeting Lipid Metabolism and Extracellular Matrix for Myelin Repair. J. Clin. Med. 2020, 9, 470. [Google Scholar] [CrossRef] [Green Version]
- Podbielska, M.; Banik, N.L.; Kurowska, E.; Hogan, E.L. Myelin recovery in multiple sclerosis: The challenge of remyelination. Brain Sci. 2013, 3, 1282–1324. [Google Scholar] [CrossRef] [Green Version]
- Gopalakrishnan, G.; Awasthi, A.; Belkaid, W.; De Faria, O., Jr.; Liazoghli, D.; Colman, D.R.; Dhaunchak, A.S. Lipidome and proteome map of myelin membranes. J. Neurosci. Res. 2013, 91, 321–334. [Google Scholar] [CrossRef]
- Cermenati, G.; Giatti, S.; Audano, M.; Pesaresi, M.; Spezzano, R.; Caruso, D.; Mitro, N.; Melcangi, R.C. Diabetes alters myelin lipid profile in rat cerebral cortex: Protective effects of dihydroprogesterone. J. Steroid. Biochem. Mol. Biol. 2017, 168, 60–70. [Google Scholar] [CrossRef]
- Sindelar, P.J.; Guan, Z.; Dallner, G.; Ernster, L. The protective role of plasmalogens in iron-induced lipid peroxidation. Free Radic. Biol. Med. 1999, 26, 318–324. [Google Scholar] [CrossRef]
- Broniec, A.; Klosinski, R.; Pawlak, A.; Wrona-Krol, M.; Thompson, D.; Sarna, T. Interactions of plasmalogens and their diacyl analogs with singlet oxygen in selected model systems. Free Radic. Biol. Med. 2011, 50, 892–898. [Google Scholar] [CrossRef] [Green Version]
- Glaser, P.E.; Gross, R.W. Plasmenylethanolamine facilitates rapid membrane fusion: A stopped-flow kinetic investigation correlating the propensity of a major plasma membrane constituent to adopt an HII phase with its ability to promote membrane fusion. Biochemistry 1994, 33, 5805–5812. [Google Scholar] [CrossRef]
- Glaser, P.E.; Gross, R.W. Rapid plasmenylethanolamine-selective fusion of membrane bilayers catalyzed by an isoform of glyceraldehyde-3-phosphate dehydrogenase: Discrimination between glycolytic and fusogenic roles of individual isoforms. Biochemistry 1995, 34, 12193–12203. [Google Scholar] [CrossRef]
- Bechtold, S.; Blaschek, A.; Raile, K.; Dost, A.; Freiberg, C.; Askenas, M.; Frohlich-Reiterer, E.; Molz, E.; Holl, R.W. Higher relative risk for multiple sclerosis in a pediatric and adolescent diabetic population: Analysis from DPV database. Diabetes Care 2014, 37, 96–101. [Google Scholar] [CrossRef] [Green Version]
- Tettey, P.; Simpson, S., Jr.; Taylor, B.; Blizzard, L.; Ponsonby, A.L.; Dwyer, T.; Kostner, K.; van der Mei, I. An adverse lipid profile is associated with disability and progression in disability, in people with MS. Mult. Scler. 2014, 20, 1737–1744. [Google Scholar] [CrossRef]
- Wheeler, D.; Bandaru, V.V.; Calabresi, P.A.; Nath, A.; Haughey, N.J. A defect of sphingolipid metabolism modifies the properties of normal appearing white matter in multiple sclerosis. Brain 2008, 131, 3092–3102. [Google Scholar] [CrossRef] [Green Version]
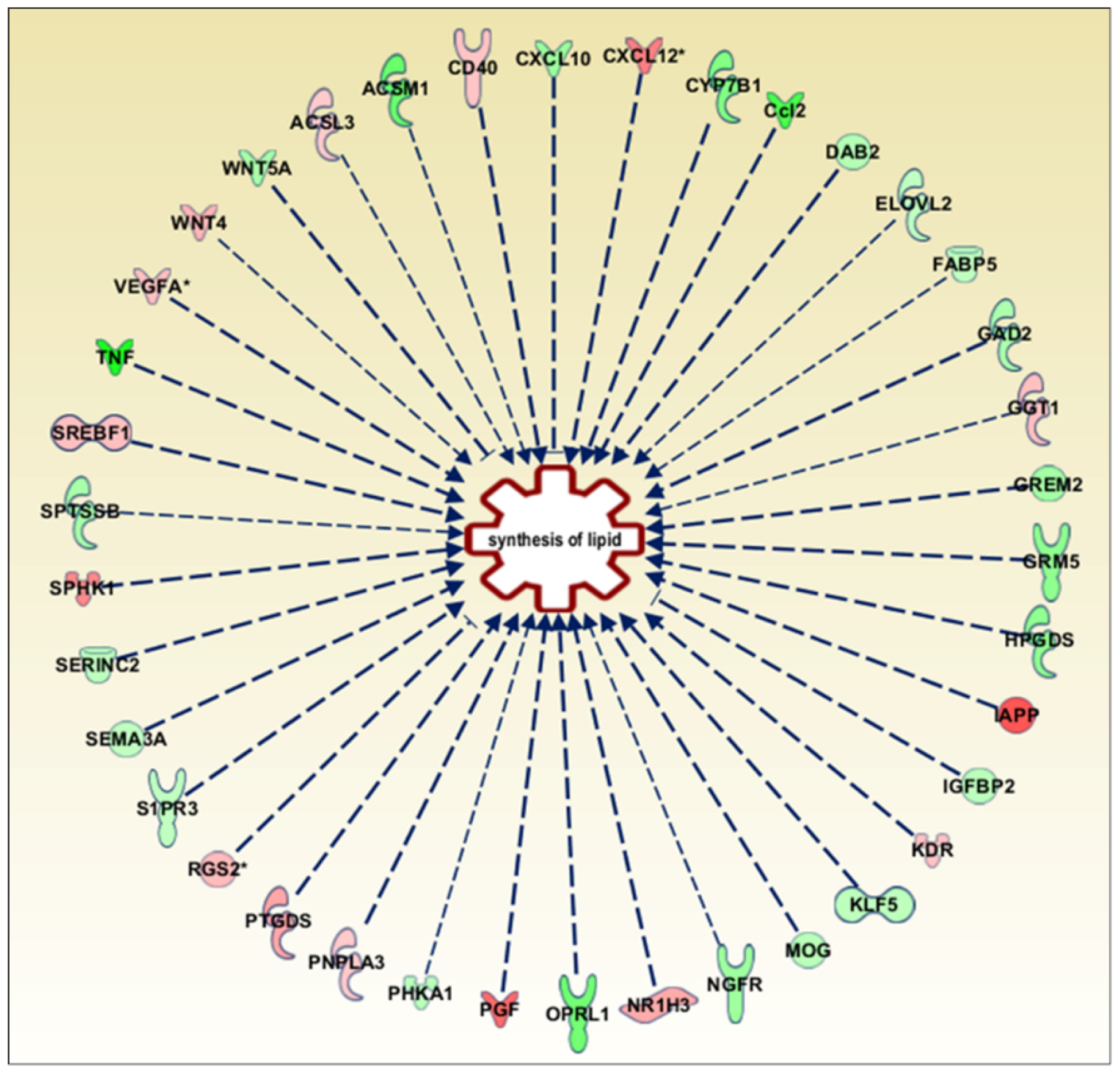
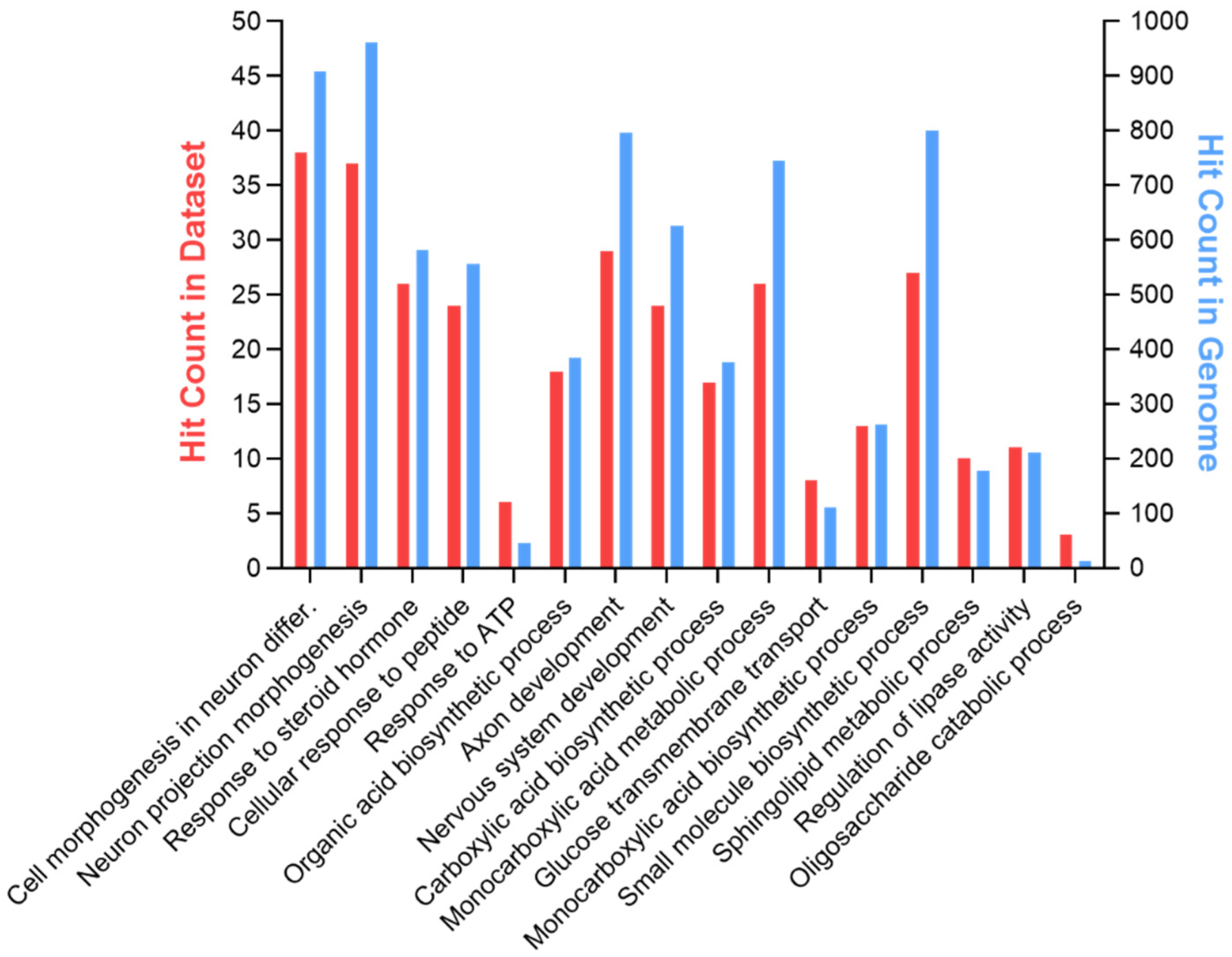


| Pathways | Total Genes | p-Value | FDR | Genes in Dataset |
|---|---|---|---|---|
| mTORC1 downstream signaling | 61 | 6.05 × 10−5 | 2.50 × 10−2 | 7 |
| TGF, WNT and cytoskeleton remodelling | 111 | 8.73 × 10−5 | 2.50 × 10−2 | 9 |
| IL-6 signaling in multiple myeloma | 51 | 1.80 × 10−4 | 3.27 × 10−2 | 6 |
| Regulation of lipid metabolism | 19 | 2.29 × 10−4 | 3.27 × 10−2 | 4 |
| Regulation of endothelial progenitor cell differentiation from adult stem cells | 60 | 4.42 × 10−4 | 4.87 × 10−2 | 6 |
| Regulation of lung epithelial progenitor cell differentiation | 41 | 5.34 × 10−4 | 4.87 × 10−2 | 5 |
| Regulation of tissue factor signaling in cancer | 43 | 6.68 × 10−4 | 4.87 × 10−2 | 5 |
| Role of IL-8 in angiogenesis | 65 | 6.81 × 10−4 | 4.87 × 10−2 | 6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marangon, D.; Audano, M.; Pedretti, S.; Fumagalli, M.; Mitro, N.; Lecca, D.; Caruso, D.; Abbracchio, M.P. Rewiring of Glucose and Lipid Metabolism Induced by G Protein-Coupled Receptor 17 Silencing Enables the Transition of Oligodendrocyte Progenitors to Myelinating Cells. Cells 2022, 11, 2369. https://doi.org/10.3390/cells11152369
Marangon D, Audano M, Pedretti S, Fumagalli M, Mitro N, Lecca D, Caruso D, Abbracchio MP. Rewiring of Glucose and Lipid Metabolism Induced by G Protein-Coupled Receptor 17 Silencing Enables the Transition of Oligodendrocyte Progenitors to Myelinating Cells. Cells. 2022; 11(15):2369. https://doi.org/10.3390/cells11152369
Chicago/Turabian StyleMarangon, Davide, Matteo Audano, Silvia Pedretti, Marta Fumagalli, Nico Mitro, Davide Lecca, Donatella Caruso, and Maria P. Abbracchio. 2022. "Rewiring of Glucose and Lipid Metabolism Induced by G Protein-Coupled Receptor 17 Silencing Enables the Transition of Oligodendrocyte Progenitors to Myelinating Cells" Cells 11, no. 15: 2369. https://doi.org/10.3390/cells11152369