Linkage Map of a Gene Controlling Zero Tannins (zt-1) in Faba Bean (Vicia faba L.) with SSR and ISSR Markers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Tannin Measurement
2.3. DNA Extraction
2.4. Marker Analysis
2.5. Statistical Analysis and Genetic Mapping
3. Results
3.1. Phenotypic and Genetic Analyses
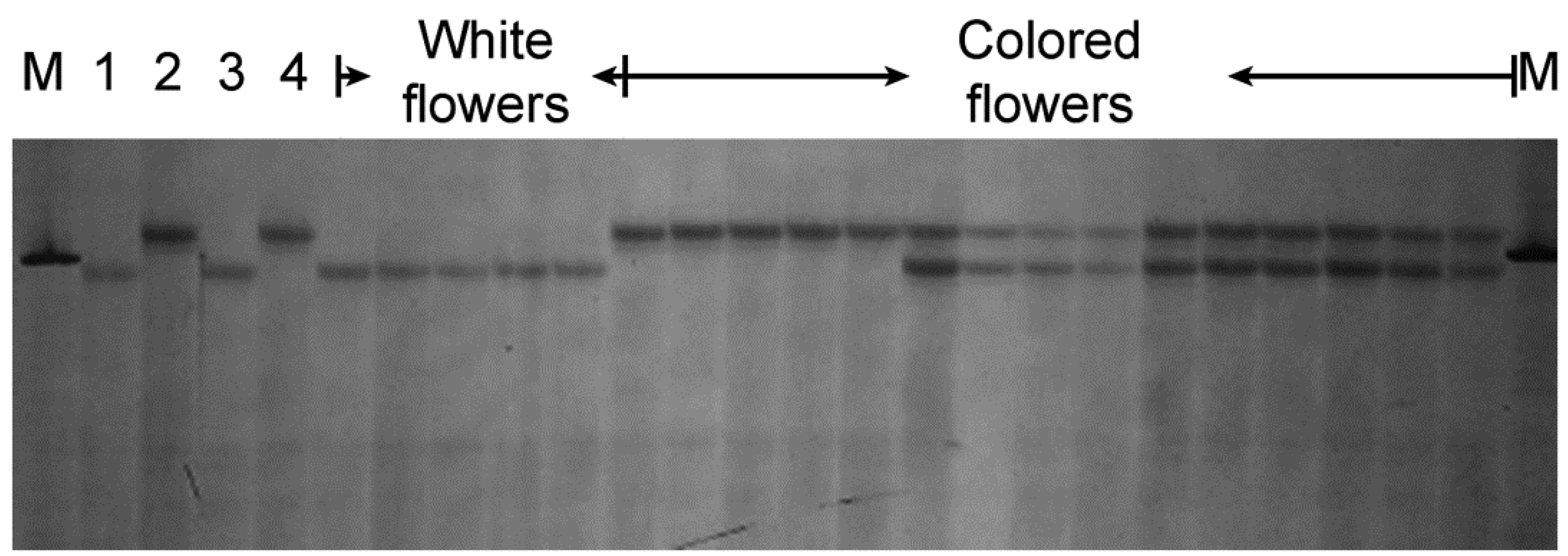
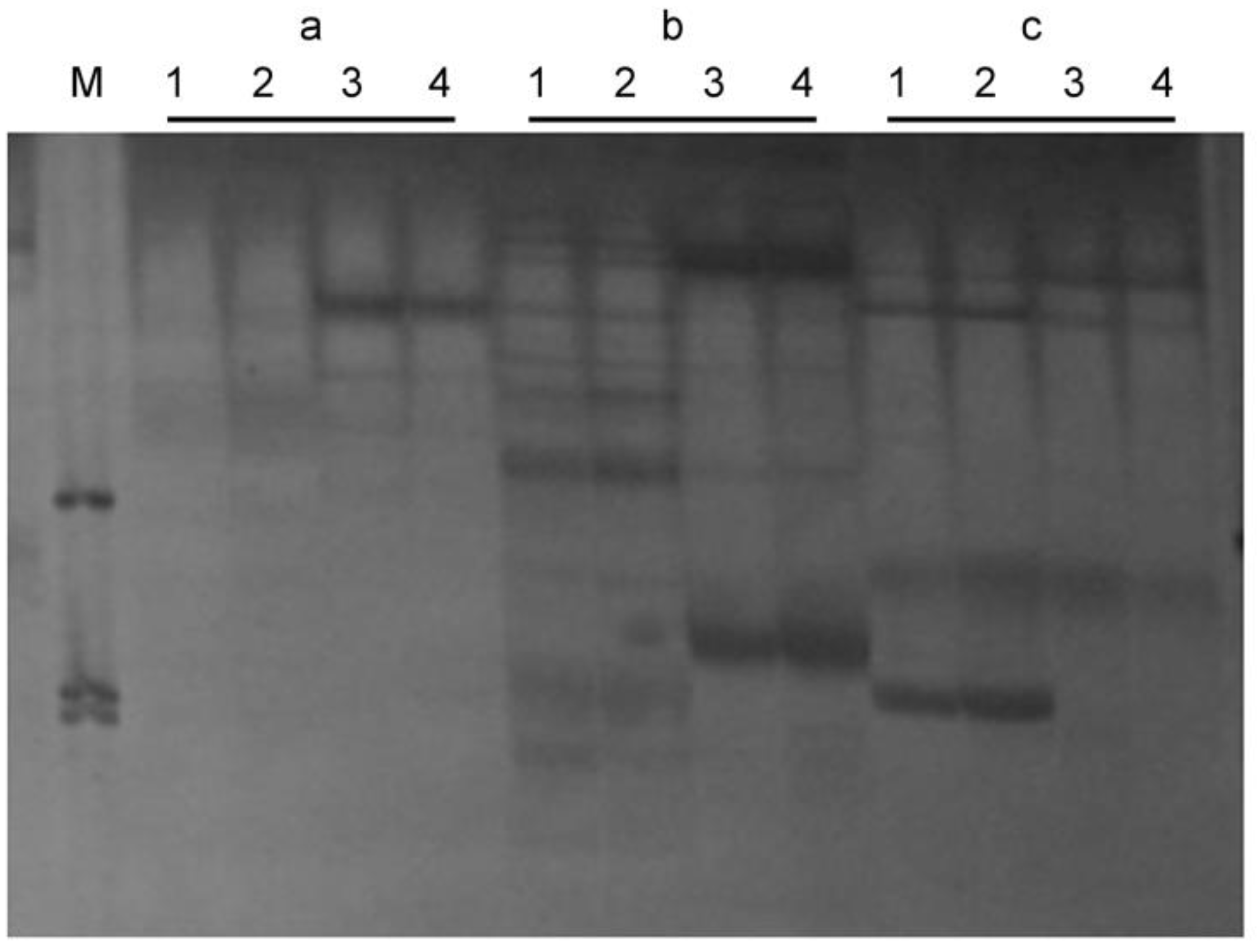
3.2. Identification of the SSR and ISSR Markers
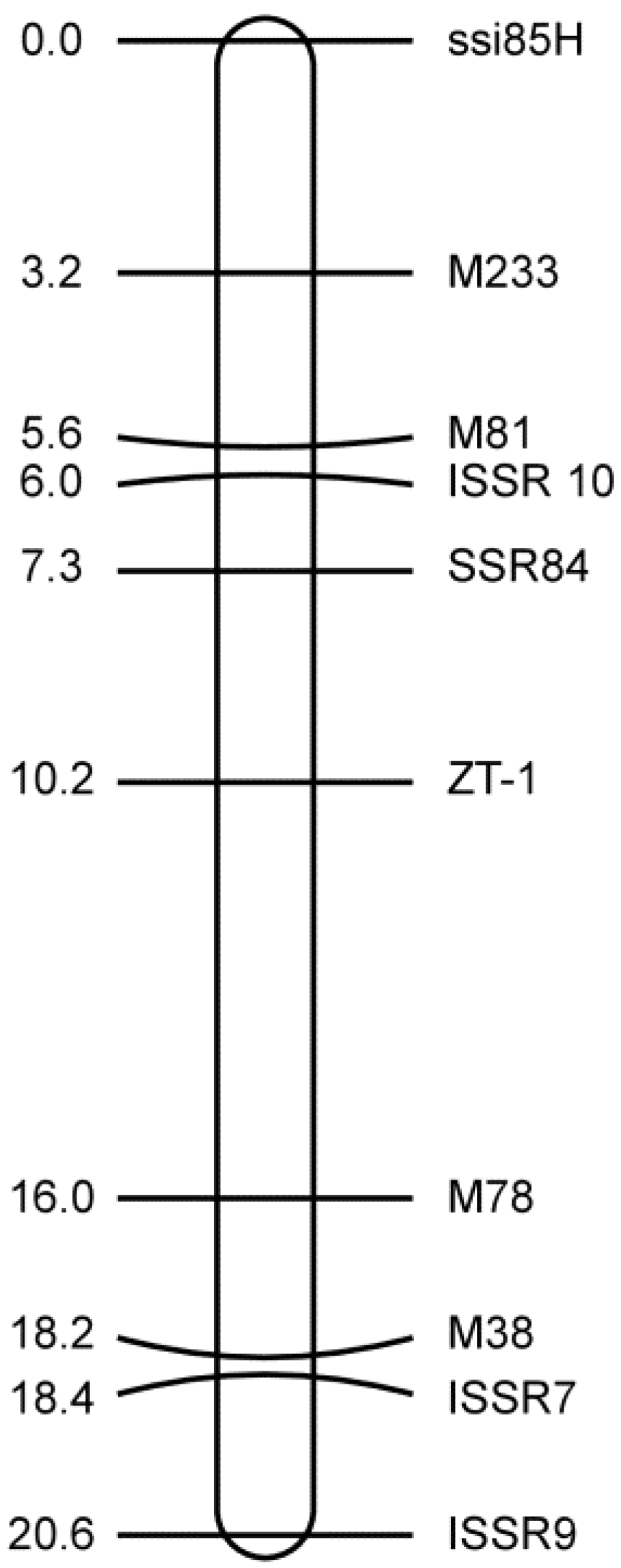
3.3. Mapping the SSR and ISSR Markers
3.4. Closely Linked Markers for the Marker-Assisted Selection of zt-1
4. Discussion
4.1. Zt-1 Gene in Faba Bean Variety M3290
4.2. SSR and ISSR Markers
4.3. Closely Linked Markers and Their Application in MAS
4.4. Faba Bean Breeding Program of Variety M3290
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Duc, G.; Bao, S.; Baumc, M.; Redden, B.; Sadiki, M. Diversity maintenance and use of Vicia faba L. genetic resources. Field Crop. Res. 2010, 115, 270–278. [Google Scholar] [CrossRef]
- FAO. United Nations. 2016. Available online: http://faostat3.fao.org/download/Q/QC/E (accessed on 10 October 2016).
- Stoddard, F.L.; Hovinen, S.; Kontturi, M.; Lindström, K.; Nykänen, A. Legumes in Finnish agriculture: history, present status and future prospects. Agric. Food Sci. 2009, 18, 191–205. [Google Scholar] [CrossRef]
- Friedman, M. Nutritional value of proteins from different food sources. A review. J. Agric. Food Chem. 1996, 44, 6–29. [Google Scholar] [CrossRef]
- Wiseman, J.; Cole, D.J.A. European legumes in diets for non-ruminants. In Recent Advances in Animal Nutrition; Haresign, W., Cole, D.J.A., Eds.; Butterworths: London, UK, 1988; pp. 13–37. [Google Scholar]
- Martin-Tanguy, H.; Guillaume, J.; Kossa, A. Condensed tannins in horse bean seeds: Chemical structure and apparent effects on poultry. J. Sci. Food Agric. 1977, 28, 757–765. [Google Scholar] [CrossRef]
- Cansfield, P.E.; Marquardt, R.R.; Campbell, L.D. Condensed proanthocyanidins of faba beans. J. Sci. Food Agric. 1980, 31, 802–812. [Google Scholar] [CrossRef] [PubMed]
- Vander Poel, A.F.B.; Dellaert, L.M.W.; Van Norel, A.; Helsper, J.P.F.G. The digestibility in piglets of faba bean (Vicia faba L.) as affected by breeding towards the absence of condensed tannins. Br. J. Nutr. 1992, 68, 793–800. [Google Scholar] [CrossRef]
- Picard, J. Apercu sur l’héreditédu caractére absence de tannins dans les graines de féverole (Vicia faba L.). Ann. Amelior. Plant. 1976, 26, 101–106. [Google Scholar]
- Bond, D.A. In vitro digestibility of the testa in tannin-free field beans (Vicia faba L.). J. Agric. Sci. Camb. 1976, 86, 561–566. [Google Scholar] [CrossRef]
- Cabrera, A.; Martin, A. Genetics of tannin content and its relationship with flower and testa colours in Vicia faba L. J. Agric. Sci. Camb. 1989, 113, 93–98. [Google Scholar] [CrossRef]
- Crofton, G.R.A.; Bond, D.A.; Duc, G. Potential seed multiplication problems arising from the existence of two genes for the absence of tannin in Vicia faba L. Plant Var. Seeds 2000, 13, 131–139. [Google Scholar]
- Alghamdi, S.S.; Migdadi, H.M.; Ammar, M.H.; Paull, J.G.; Siddique, K.H.M. Faba bean genomics: Current status and future prospects. Euphytica 2012, 186, 609–624. [Google Scholar] [CrossRef]
- Bennett, M.D.; Smith, J.B. Nuclear DNA amounts in angiosperms. Proc. R. Soc. Lond. B Biol. Sci. 1982, 216, 179–199. [Google Scholar] [CrossRef]
- Johnston, J.S.; Bennett, M.D.; Rayburn, A.L.; Galbraith, D.W.; Price, H.J. Reference standards for determination of DNA content of plant nuclei. Am. J. Bot. 1999, 86, 609–613. [Google Scholar] [CrossRef] [PubMed]
- Raina, S.N.; Ogihara, Y. Ribosomal DNA repeat unit polymorphism in 49 Vicia species. Theor. Appl. Genet. 1995, 90, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Zeid, M.; Schon, C.C.; Lin, W. Genetic diversity in recent elite faba bean lines using AFLP markers. Theor. Appl. Genet. 2003, 107, 1304–1314. [Google Scholar] [CrossRef] [PubMed]
- Zong, X.; Liu, X.; Guan, J.; Wang, S.; Liu, Q.; Paull, J.G.; Redden, R. Molecular variation among Chinese and global winter faba bean germplasm. Theor. Appl. Genet. 2009, 118, 971–978. [Google Scholar] [CrossRef] [PubMed]
- Zong, X.; Ren, J.; Guan, J.; Wang, S.; Liu, Q.; Paull, J.G.; Redden, R. Molecular variation among Chinese and global germplasm in spring faba bean areas. Plant Breed. 2010, 129, 508–513. [Google Scholar] [CrossRef]
- Gutierrez, N.; Avila, C.; Rodriguez-Suarez, C.; Moreno, M.; Torres, A. Development of SCAR markers linked to a gene controlling absence of tannins in faba bean. Mol. Breed. 2007, 19, 305–314. [Google Scholar] [CrossRef]
- Hagerman, A.E.; Butler, L.G. Protein precipitation method for the quantitative determination of tannins. J. Agric. Food Chem. 1978, 26, 809–812. [Google Scholar] [CrossRef]
- Song, W.N.; Langridge, P. Identification and mapping polymorphism in cereals based on polymerase chain reaction. Theor. Appl. Genet. 1991, 82, 209–213. [Google Scholar]
- Song, W.N.; Henry, R. Polymorphisms in the a-amy1 gene of wild and cultivated barley revealed by the polymerase chain reaction. Theor. Appl. Genet. 1994, 89, 509–512. [Google Scholar]
- Michelmore, R.W.; Paran, I.; Kesseli, R.V. Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 1991, 88, 9828–9832. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Yang, T.; Guan, S.; Wang, H.; Wang, X.; Zong, X. Development and characterization of 21 EST-derived microsatellite markers in Vicia faba (faba bean). Am. J. Bot. 2011, 98, 22–24. [Google Scholar] [CrossRef] [PubMed]
- El-Rodeny, W.; Kimura, M.; Hirakawa, H.; Sabah, A.; Shirasawa, K.; Sato, S. Development of EST-SSR markers and construction of a linkage map in faba bean (Vicia faba). Breed. Sci. 2014, 64, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Zietkiewicz, E.; Rafalski, A.; Labuda, D. Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 1994, 20, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Bassam, B.J.; Anolles, G.C.; Gresshoff, P.M. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal. Biochem. 1991, 196, 80–83. [Google Scholar] [CrossRef]
- Van Ooijen, J.W. JoinMap4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations; Kyazma BV: Wageningen, The Netherlands, 2006. [Google Scholar]
- Kosambi, D.D. The estimation of map distances from recombination values. Ann. Eugen. 1944, 12, 172–175. [Google Scholar] [CrossRef]
- Kelly, J.D. Use of random amplified polymorphic DNA markers in breeding for major gene resistance to plant pathogens. HortScience 1995, 30, 461–465. [Google Scholar]
- Terzopoulos, P.J.; Bebeli, P.J. Genetic diversity analysis of Mediterranean faba bean (Vicia faba L.) with ISSR markers. Field Crop. Res. 2008, 108, 39–44. [Google Scholar] [CrossRef]
- Link, W.; Ederer, W.; Metz, P.; Buiel, H.; Melchinger, A.E. Genotypic and environmental variation for degree of cross-fertilization in faba bean. Crop Sci. 1994, 34, 960–964. [Google Scholar] [CrossRef]
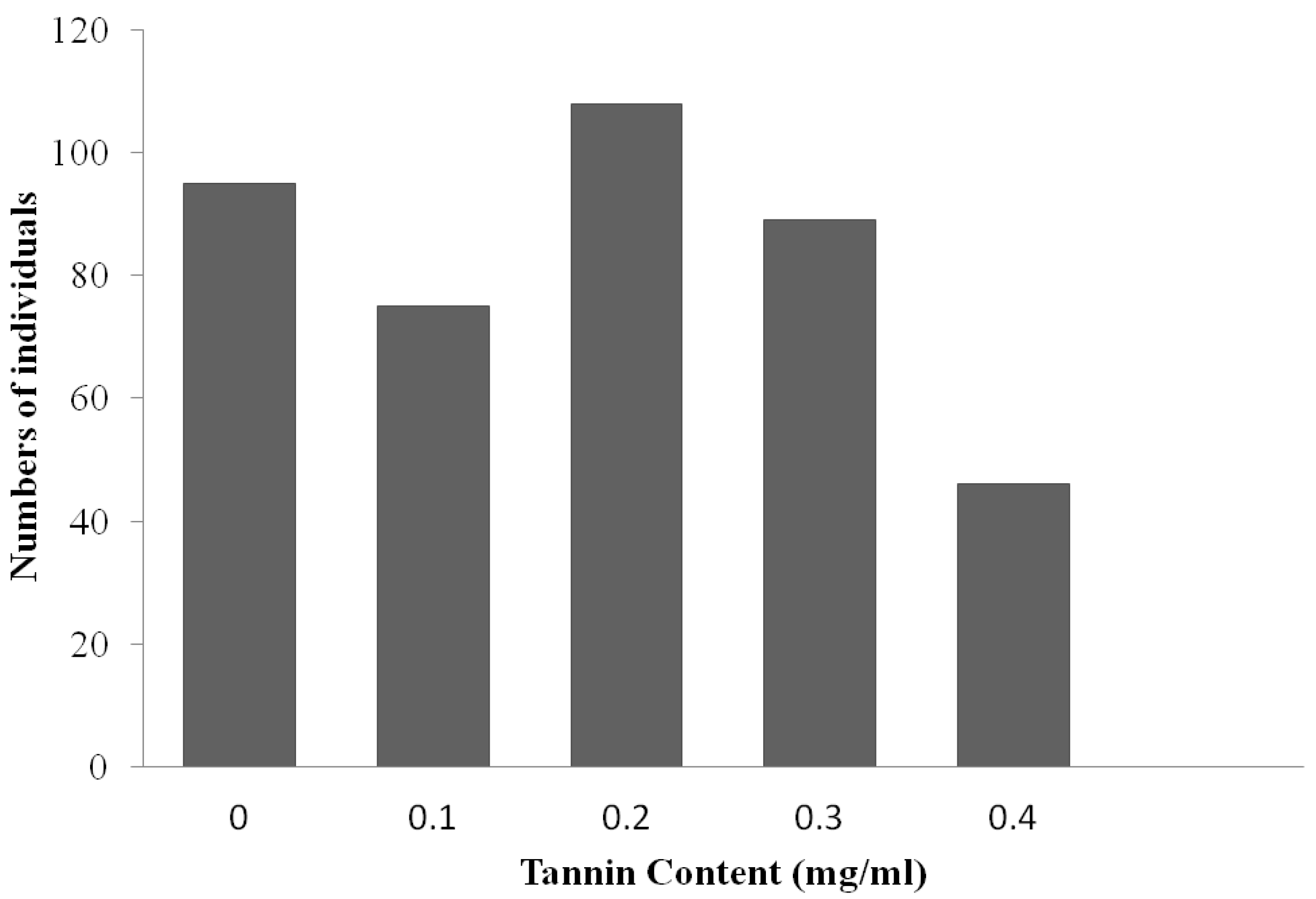
| CrossProgeny | Observed Number of Plants or Lines | Expected Ratio | χ2 | P | ||
|---|---|---|---|---|---|---|
| W | Seg | P | ||||
| M3290 | 15 | 0 | 0 | |||
| Qinghai12 | 0 | 0 | 15 | |||
| F1 | 15 | - | - | 1:0 | ||
| F2 | 95 | - | 318 | 1:3 | 0.879 | 0.348 |
| F3 | 95 | 205 | 113 | 1:2:1 | 1.59 | 0.451 |
| Name | Marker Type | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Annealing Temperature (°C) |
|---|---|---|---|---|
| SSR84 | SSR | TCTGAAAACGAGTTCAGTGGA | CTGGTGCCGAACTAACCAGT | 52 |
| M38 | SSR | GCTACTGGAGGAGGCTTTCA | GCCTTCTACACAACGGCTTC | 53 |
| M78 | SSR | GTCAAATCGAGTGGCGAAAG | TTGGGATATGGAAGTAGCTTCAG | 52 |
| M81 | SSR | CCTCATGCCATTCCTCTGAT | TTCCGCGTGGTAAATTCTATG | 55 |
| M233 | SSR | CATCCCAACAATATACCGGC | CTGGGGTACCACCGTAACTC | 51 |
| ssi85H | SSR | AACAACTACGTAATGCCAGAC | ACATGAGGGGCCAAGTAT | 52 |
| ISSR7 | ISSR | AGA GAG AGA GAG AGA GT | 53 | |
| ISSR9 | ISSR | AGA GAG AGA GAG AGA GG | 50 | |
| ISSR10 | ISSR | GAG AGA GAG AGA GAG AT | 51 |
| No. | Variety | Province | Flower a | SSR84 b | M78 b | ||
|---|---|---|---|---|---|---|---|
| 900 bp | 1100 bp | 400 bp | 420 bp | ||||
| 1 | Qinghai11 | Qinghai | Purple | - c | + c | - | + |
| 2 | Qinghai12 | Qinghai | Purple | - | + | - | + |
| 3 | Qinghai13 | Qinghai | Purple | - | + | - | + |
| 4 | Qingcan14 | Qinghai | Purple | - | + | - | + |
| 5 | Qingcan15 | Qinghai | Purple | - | + | - | + |
| 6 | M3290 | Qinghai | White | + | - | + | - |
| 7 | TF26 | Qinghai | White | + | - | + | - |
| 8 | TF29 | Qinghai | White | + | - | - | + |
| 9 | TF34 | Qinghai | White | + | - | - | + |
| 10 | 2005-00 | Qinghai | White | + | - | - | + |
| 11 | Lincan6 | Gansu | Purple | - | + | - | + |
| 12 | Lincan7 | Gansu | Purple | - | + | - | + |
| 13 | Lincan8 | Gansu | Purple | - | + | - | + |
| 14 | Yangyandou | Gansu | Purple | - | + | - | + |
| 15 | Yundou7 | Yunnan | Purple | - | + | - | + |
| 16 | Yundou8 | Yunnan | Purple | - | + | - | + |
| 17 | Yundou9 | Yunnan | Purple | - | + | - | + |
| 18 | Touxinlv | Yunnan | Purple | - | + | - | + |
| 19 | Dahudou | Sichuan | Purple | - | + | - | + |
| 20 | Xiaohudou | Sichuan | Purple | - | + | - | + |
| 21 | Honghudou | Sichuan | Purple | - | + | - | + |
| 22 | Chenghu9 | Sichuan | Purple | - | + | - | + |
| 23 | Chenghu10 | Sichuan | Purple | - | + | - | + |
| 24 | Chenghu11 | Sichuan | Purple | - | + | - | + |
| 25 | Tongcanxian7 | Jiangsu | Purple | - | + | - | + |
| 26 | Tongcanxian8 | Jiangsu | Purple | - | + | - | + |
| 27 | Qidongbaipi | Jiangsu | Purple | - | + | - | + |
| 28 | Haimendabaipi | Jiangsu | Purple | - | + | - | + |
| 29 | Tongcan5 | Jiangsu | Purple | - | + | - | + |
| 30 | Nantongsanbai | Jiangsu | Purple | - | + | - | + |
| 31 | DAqingpi | Jiangsu | Purple | - | + | - | + |
| 32 | Lvpidou | Zhejiang | Purple | - | + | - | + |
| 33 | Luohandou | Zhejiang | Purple | - | + | - | + |
| 34 | Xiaoqingdou | Zhejiang | Purple | - | + | - | + |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, W.; Zhang, X.; Yan, Q.; Li, P.; Sha, W.; Tian, Y.; Liu, Y. Linkage Map of a Gene Controlling Zero Tannins (zt-1) in Faba Bean (Vicia faba L.) with SSR and ISSR Markers. Agronomy 2018, 8, 80. https://doi.org/10.3390/agronomy8060080
Hou W, Zhang X, Yan Q, Li P, Sha W, Tian Y, Liu Y. Linkage Map of a Gene Controlling Zero Tannins (zt-1) in Faba Bean (Vicia faba L.) with SSR and ISSR Markers. Agronomy. 2018; 8(6):80. https://doi.org/10.3390/agronomy8060080
Chicago/Turabian StyleHou, Wanwei, Xiaojuan Zhang, Qingbiao Yan, Ping Li, Weichao Sha, Yingying Tian, and Yujiao Liu. 2018. "Linkage Map of a Gene Controlling Zero Tannins (zt-1) in Faba Bean (Vicia faba L.) with SSR and ISSR Markers" Agronomy 8, no. 6: 80. https://doi.org/10.3390/agronomy8060080




