Existing and Potential Statistical and Computational Approaches for the Analysis of 3D CT Images of Plant Roots
Abstract
:1. Introduction
2. X-ray CT Imaging
3. Existing Approaches
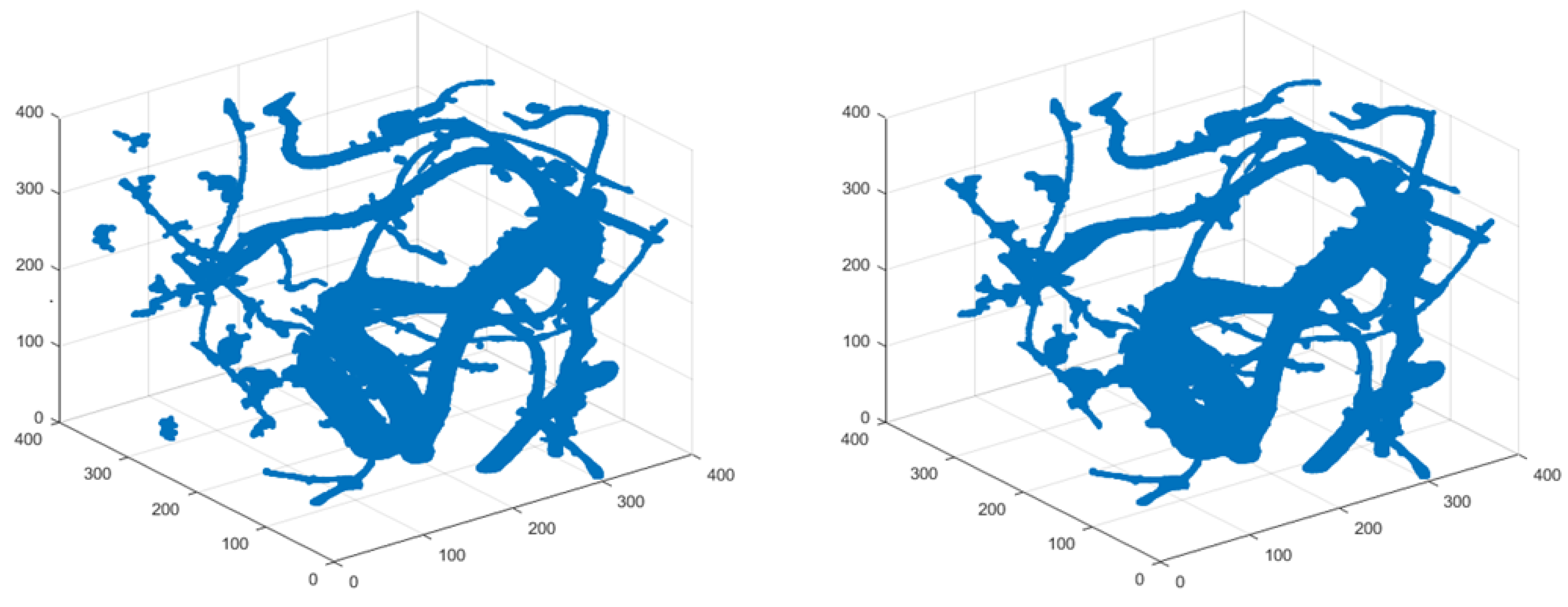
3.1. Statistical Branch Tracking Based on Vesselness Measures
3.2. Bayesian Single Branch Tracking
| Algorithm 1 Single Branch Tracking |
|
| Algorithm 2 Branching Detection |
|
3.3. Bayesian Tracking with Overlap of Intensities
3.4. Tracking Based on A Constrained Geometric Model for Tabular Structures
4. 2D to 3D Structure Determination
4.1. cryoSPARC for Cyro-EM 3D Structure Estimation
4.2. KNOSSOS and ElektroNN2
5. Other Recent Approaches
5.1. Deep Learning
5.2. Graph-Based Segmentation
6. Available Computational Tools
6.1. ImageJ and Root1
6.2. RooTrak
6.3. VG Studio Max
6.4. Volume Player Plus (VPP) and Modular Algorithms for Volume Images (MAVI)
6.5. Digital Imaging of Root Traits (DIRT)
7. Discussion and Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| 2D | Two Dimension |
| 3D | Three Dimension |
| ANN | Artificial Neural Network |
| CNN | Convolutional Neural Network |
| COPD | Chronic Obstructive Pulmonary Disease |
| CT | Computed Topology |
| CTF | Contrast Transfer Function |
| CTN | Computed Topology Number |
| DIRT | Digital Imaging of Root Traits |
| DNN | Deep Neural Network |
| EM (algorithm) | Expected-Maximum (algorithm) |
| EM (image) | Electronic Microscopic (image) |
| MAP | Maximum A Posteriori |
| MAVI | Modular Algorithms for Volume Images |
| MCMC | Markov Chain Monte Carlo |
| MRI | Magnetic Resonance Imaging |
| NIH | National Institute of Health |
| NN | Nearest Neighbor |
| RGB (image) | Red Green Blue (image) |
| RSA | Root Structure Architecture |
| SGD | Stochastic Gradient Descent |
| TACC | Texas Advanced Computing Center |
| VPP | Volume Player Plus |
Appendix A. Methodological Details
Appendix A.1. Statistical Branch Tracking Based on Vesselness Measures
Appendix A.2. Bayesian Single Branch Tracking
Appendix B. Bayesian Tracking with Overlap of Intensities
Appendix B.1. Tracking Based on A Constrained Geometric Model for Tabular Structures
Appendix B.2. cryoSPARC for cryo-EM 3D Structure Estimation
References
- Tollner, E.; Verma, B.; Cheshire, J. Observing soil-tool interactions and soil organisms using X-ray computer tomography. Trans. Am. Soc. Agric. Eng. 1987, 30, 1605–1610. [Google Scholar] [CrossRef]
- Dutilleul, P.; Lafond, J. Branching and Rooting Out with a CT Scanner: The Why, the How, and the Outcomes, Present and Possibly Future; Frontiers Media: Lusanne, Switzerland, 2016. [Google Scholar]
- Trachsel, S.; Kaeppler, S.; Brown, K.; Lynch, J. Shovelomics: High throughput phenotyping of maize (Zea mays L.) root architecture in the field. Plant Soil 2010, 341, 75–87. [Google Scholar] [CrossRef]
- Chen, X.; Ding, Q.; Blaszkiewicz, Z.; Sun, J.; Sun, Q.; He, R.; Li, Y. Phenotyping for the dynamics of field wheat root system architecture. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Downie, H.; Adu, M.; Schmidt, S.; Otten, W.; Dupuy, L.; White, P.; Valentine, T. Challenges and opportunities for quantifying roots and rhizosphere interactions through imaging and image analysis. Plant Cell Environ. 2015, 38, 1213–1232. [Google Scholar] [CrossRef] [PubMed]
- Roose, T.; Keyse, S.; Daly, K.; Carminati, A.; Otten, W.; Vetterlein, D.; Peth, S. Challenges in imaging and predictive modeling of rhizosphere processes. Plant Soil 2016, 407, 9–38. [Google Scholar] [CrossRef]
- Lafond, J.; Han, L.; Dutilleul, P. Concepts and analyses in the CT scannning of root systems and leaf canopies: A timely summary. Front. Plant Sci. 2015, 6, 1111. [Google Scholar] [CrossRef] [PubMed]
- Wachsman, G.; Sparks, E.; Benfey, P. Genes and networks regulating root anatomy and architecture. New Phytol. 2015, 208, 26–38. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J. Root phenes for enhanced soil exploration and phosphorus acquisition: Tools for future crops. Plant Physiol. 2011, 156, 1041–1049. [Google Scholar] [CrossRef] [PubMed]
- Gumayao, R.; Chin, J.; Pariasca-Tanaka, J.; Pesaresi, P.; Catausan, S.; Dalid, C.; Slamet-Loedin, I.; Tecson-Mendoza, E.; Wissuwa, M.; Heuer, S. The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature 2013, 488, 535–539. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Naugler Klassen, T.; Keyes, S.; Daly, M.; Jones, D.; Mavrogordato, M.; Sinclair, I.; Roose, T. Imaging the interaction of roots and phosphate fertiliser granules using 4D X-ray tomography. Plant Soil 2015, 401, 125–134. [Google Scholar] [CrossRef]
- Rich, S.; Watt, M. Soil conditions and cereal root system architecture: Review and considerations for linking Darwin and Weaver. J. Exp. Bot. 2013, 64, 1193–1208. [Google Scholar] [CrossRef] [PubMed]
- Koevoets, I.; Venema, J.; Elzenga, J.; Testerink, C. Roots Withstanding their Environment: Exploiting Root System Architecture Responses to Abiotic Stress to Improve Crop Tolerance. Front. Plant Sci. 2016, 7, 1335. [Google Scholar] [CrossRef] [PubMed]
- McMichael, B.; Burke, J. Soil temperature and root growth. HortScience 1998, 33, 947–951. [Google Scholar]
- Rey, T.; Schornack, S. Interactions of beneficial and detrimental root-colonizing filamentous microbes with plant hosts. Genome Biol. 2013, 14, 121. [Google Scholar] [CrossRef] [PubMed]
- Hacquard, S.; Kracher, B.; Hiruma, K.; Münch, P.; Garrido-Oter, R.; Thon, M.; Weimann, A.; Damm, U.; Dallery, J.F.; Hainaut, M.; et al. Survival trade-offs in plant roots during colonization by closely related beneficial and pathogenic fungi. Nat. Commun. 2016, 7, 11362. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Dutilleul, P.; Prasher, S.O.; Beaulieu, C.; Smith, D.L. Assessment of common scab-inducing pathogen effects on potato underground organs via computed tomography scanning. Phytopathology 2008, 98, 1118–1125. [Google Scholar] [CrossRef] [PubMed]
- Sturrock, C.J.; Woodhall, J.; Brown, M.; Walker, C.; Mooney, S.J.; Ray, R.V. Effects of damping-off caused by Rhizoctonia solani anastomosis group 2-1 on roots of wheat and oil seed rape quantified using X-ray Computed Tomography and real-time PCR. Front. Plant Sci. 2015, 6, 461. [Google Scholar] [CrossRef] [PubMed]
- McNear, D., Jr. The Rhizosphere—Roots, Soil and Everything In Between. Nat. Educ. Knowl. 2013, 4, 1. [Google Scholar]
- Philippot, L.; Raaijmakers, J.; Lemanceau, P.; van der Putten, W. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Stringlis, I.; Proietti, S.; Hickman, R.; Van Verk, M.; Zamioudis, C.; Pieterse, C. Root transcriptional dynamics induced by beneficial rhizobacteria and microbial immune elicitors reveal signatures of adaptation to mutualists. Plant J. 2018, 93, 166–180. [Google Scholar] [CrossRef] [PubMed]
- Piñeros, M.; Larson, B.; Shaff, J.; Schneider, D.; Falcão, A.; Yuan, L.; Clark, R.; Craft, E.; Davis, T.; Pradier, P.; et al. Evolving technologies for growing, imaging and analyzing 3D root system architecture of crop plants. J. Integr. Plant Biol. 2016, 58, 230–241. [Google Scholar] [CrossRef] [PubMed]
- Balduzzi, M.; Binder, B.; Bucksch, A.; Chang, C.; Hong, L.; Iyer-Pascuzzi, A.; Pradal, C.; Sparks, E. Reshaping Plant Biology: Qualitative and Quantitative Descriptors for Plant Morphology. Front. Plant Sci. 2017, 3, 117. [Google Scholar] [CrossRef] [PubMed]
- Lobet, G.; Koevoets, I.; Noll, M.; Meyer, P.; Tocquin, P.; Pages, L.; Perilleux, C. Using a Structural Root System Model to Evaluate and Improve the Accuracy of Root Image Analysis Pipelines. Front. Plant Sci. 2017, 8, 447. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, J.; Kirchgessner, N.; Colombi, T.; Walter, A. Rapid phenotyping of crop root systems in undisturbed field soils using X-ray computed tomography. Plant Methods 2015, 11, 41. [Google Scholar] [CrossRef] [PubMed]
- Metzner, R.; Eggert, A.; Dusschoten, D.; Pflugfelder, D.; Gerth, S.; Schurr, U.; Uhlmann, N.; Jahnke, S. Direct comparison of MRI and X-ray CT technologies for 3D imaging of root systems in soil: potential and challenges for root trait quantification. Plant Methods 2015, 11, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, N.; Aggarwal, L. Automated medical image segmentation techniques. J. Med. Phys. 2010, 35, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.; Brown, K. New roots for agriculture: exploiting the root phenome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012, 367, 1598–1604. [Google Scholar] [CrossRef] [PubMed]
- Ginat, D.; Gupta, R. Advanced in Computed Tomography Imaging Technology. Annu. Rev. Biomed. Eng. 2014, 16, 431–453. [Google Scholar] [CrossRef] [PubMed]
- Larobina, M.; Murino, L. Medical Image File Formats. J. Digit. Imaging 2014, 27, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Bryant, J.; Drage, N.; Richmond, S. CT Number Definition. Radiat. Phys. Chem. 2012, 81, 358–361. [Google Scholar] [CrossRef]
- Kalender, W. Computed Tomography; Publicis MCD Verlag: Oslo, Norway, 2000. [Google Scholar]
- Tjong, W.; Kazakia, G.J.; Burghardt, A.J.; Majumdar, S. The effect of voxel size on high-resolution peripheral computed tomography measurements of trabecular and cortical bone microstructure. Med. Phys. 2012, 39, 1893–1903. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, B.A. Effect of micro-computed tomography voxel size and segmentation method on trabecular bone microstructure measures in mice. Bone Rep. 2016, 5, 136–140. [Google Scholar] [CrossRef] [PubMed]
- Downie, H.; Holden, N.; Otten, W.; Spiers, A.J.; Valentine, T.A.; Dupuy, L.X. Transparent soil for imaging the rhizosphere. PLoS ONE 2012, 7, e44276. [Google Scholar] [CrossRef] [PubMed]
- Mooney, S.; Pridmore, T.; Helliwell, J.; Bennett, M. Developing X-ray Computed Tomography to non-invasively image 3-D root systems architecture in soil. Plant Soil, 2012, 352, 1–22. [Google Scholar] [CrossRef]
- Gonzalez, R.; Woods, R. Digital Image Processing, 4th ed.; Pearson: London, UK, 2017. [Google Scholar]
- Gonzalez, R.; Woods, R.; Eddins, S. Digital Image Processing Using MATLAB, 2nd Edition; McGraw Hill India: New York, NY, USA, 2010. [Google Scholar]
- Solomon, C.; Breckon, T. Fundamentals of Digital Image Processing; Wiley-Blackwell: Hoboken, NJ, USA, 2011. [Google Scholar]
- Wang, H.; Mossa, R.; Chen, X.; Stanley, R.J.; William, V.; Stoecker, W.V.; Celebi, M.E.; Malters, J.M.; Grichnik, J.M.; Marghoob, A.A.; et al. Modified watershed technique and post-processing for segmentation of skin Lesions in dermoscopy images. Comput. Med. Imaging Graph. 2011, 35, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Lai, M. Deep Learning for Medical Image Segmentation. arXiv, 2015; arXiv:1505.02000v1. [Google Scholar]
- Zelditch, M.; Swiderski, D.; Sheets, H.; Fink, W. Geometric Morphometrics For Biologists: A Primer; Elsevier Academic Press: New York, NY, USA, 2004. [Google Scholar]
- Rogers, E.; Monaenkova, D.; Mijar, M.; Nori, A.; Goldman, D.; Benfey, P. X-Ray computed tomography reveals the response of root system architecture to soil texture. Plant Physiol. 2016, 171, 2028–2040. [Google Scholar] [CrossRef] [PubMed]
- Morris, E.; Griffiths, M.; Golebiowska, A.; Mairhofer, S.; Burr-Hersey, J.; Goh, T.; von Wangenheim, D.; Atkinson, B.; Sturrock, C.; Lynch, J.; et al. Shaping 3D Root System Architecture. Curr. Biol. 2017, 27, R919–R930. [Google Scholar] [CrossRef] [PubMed]
- Blunk, S.; Malik, A.H.; de Heer, M.I.; Ekblad, T.; Bussell, J.; Sparkes, D.; Fredlund, K.; Sturrock, C.J.; Mooney, S.J. Quantification of seed-soil contact of sugar beet (Beta vulgaris) using X-ray Computed Tomography. Plant Methods 2017, 13, 71. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.; Han, L.; Dutilleul, P.; Smith, D.L. Computed tomography scanning can monitor the effects of soil medium on root system development: An example of salt stress in corn. Front. Plant Sci. 2015, 6, 256. [Google Scholar] [CrossRef] [PubMed]
- Mairhofer, S.; Zappala, S.; Tracy, S.; Sturrock, C.; Bennett, M.J.; Mooney, S.J.; Pridmore, T.P. Recovering complete plant root system architectures from soil via X-ray μ-Computed Tomography. Plant Methods 2013, 9, 8. [Google Scholar] [CrossRef] [PubMed]
- Mairhofer, S.; Zappala, S.; Tracy, S.R.; Sturrock, C.; Bennett, M.; Mooney, S.J.; Pridmore, T. RooTrak: Automated recovery of three-dimensional plant root architecture in soil from x-ray microcomputed tomography images using visual tracking. Plant Physiol. 2012, 158, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Lesage, D.; Angelini, E.; Bloch, I.; Funka-Lea, G. A review of 3D vessel lumen segmentation techniques: Models, features, and extraction schemes. Med. Image Anal. 2009, 13, 819–845. [Google Scholar] [CrossRef] [PubMed]
- Frangi, A.; Niessen, Q.; Vincken, K.; Viergever, M. Multiscale vessel enhancement filtering. In MICCAI’98. MICCAI 1998; Lecture Notes in Computer Science; Wells, W., Colchester, A., Delp, S., Eds.; Springer: Berlin/Heidelberg, Germany, 1998; Volume 1496, pp. 130–137. [Google Scholar]
- Lo, P.; Sporring, J.; Ashraf, H.; Pedersen, J.; de Bruijne, M. Vessel-guided airway tree segmentation: A voxel classification approach. Med. Image Anal. 2010, 14, 527–538. [Google Scholar] [CrossRef] [PubMed]
- Schulz, H.; Postma, J.; van Dusschoten, D.; Scharr, H.; Behnke, S. Plant root system analysis from MRI Images. In Computer Vision, Imaging and Computer Graphics; Theory and Application; Csurka, G., Kraus, M., Laramee, R., Richard, P., Braz, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; Volume 359, pp. 411–425. [Google Scholar]
- Stingaciu, L.; Schulz, H.; Pohlmeier, A.; Behnke, S.; Zilken, H.; Javaux, M.; Vereecken, H. In Situ Root System Architecture Extraction from Magnetic Resonance Imaging for Water Uptake Modeling. Vadose Zone J. 2013, 12. [Google Scholar] [CrossRef]
- Gibbs, J.; Pound, M.; French, A.; Pridmore, T. Active camera placement for 3D reconstruction of plant shoots. In Proceedings of the 2nd Asia-Pacific Plant Phenotyping Conference, Nanjing, China, 23–25 March 2018; Available online: http://www.appp-con.org/ (accessed on 10 April 2018).
- Pace, J.; Lee, N.; Naik, H.; Ganapathysubramanian, B.; Lübberstedt, T. Analysis of Maize (Zea mays L.) Seedling Roots with the High-Throughput Image Analysis Tool ARIA (Automatic Root Image Analysis). PLoS ONE 2014, 9, e108255. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Helmann, T.; Lo, P.; Sumkauskalte, M.; Puderbach, M.; de Bruijne, M.; Melnzer, H.; Wegner, I. Statistical tracking of tree-like tubular structures with efficient branching detection in 3D medical image data. Phys. Med. Biol. 2012, 57, 5325–5342. [Google Scholar] [CrossRef] [PubMed]
- Schaap, M.; Manniesing, R.; Smal, I.; van Walsum, T.; van der Lugt, A.; Niessen, W. Bayesian tracking of tubular structures and its application to carotid arteries in CTA. Medical Image Computing and Computer Assisted Intervention - MICCAI 2007, 10, 562–570. [Google Scholar] [PubMed]
- Lesage, D.; Angelini, E.; Bloch, I.; Funka-Lea, G. Medial-based Bayesian tracking for vascular segmentation: Application to coronary arteries in 3d CT angiography. In Proceedings of the IEEE International Symposium on Biomedical Imaging. IEEE, Paris, France, 14–17 May 2008; Volume 1496, pp. 268–271. [Google Scholar]
- Carroni, M.; Saibil, H. Cryo electron microscopy to determine the structure of macromolecular complexes. Methods 2016, 95, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Punjabi, A.; Rubinstein, J.; Fleet, D.; Brusker, M. cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 2017, 14, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Dorkenwald, S.; Schubert, P.; Killinger, M.; Urban, G.; Mikula, S.; Svara, F. Automated synaptic connectivity inference for volume electron microscopy. Nat. Methods 2017, 14, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Brieman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Marcus, G. Deep Learning: A Critical Appraisal. arXiv, 2018; arXiv:1505.02000v1. [Google Scholar]
- Tomè, D.; Monti, F.; Baroffio, L.; Bondi, L.; Tagliasacchi, M.; Tubaro, S. Deep convolutional neural networks for pedestrian detection. arXiv, 2015; arXiv:1510.03608v5. [Google Scholar]
- Elhaj, F.; Salim, N.; Harris, A.; Swee, T.; Ahmed, T. Arrhythmia recognition and classification using combined linear and nonlinear features of ECG signals. Comput. Methods Programs Biomed. 2016, 127, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Bazarghan, M.; Jaberi, Y.; Amandi, R.; Abedi, M. Automatic ECG Beat Arrhythmia Detection. arXiv, 2012; arXiv:1209.0167v3. [Google Scholar]
- Felzenszwal, P.; Huttenlocher, D. Efficient Graph-Based Image Segmentation. Int. J. Comput. Vis. 2004, 59, 167–181. [Google Scholar] [CrossRef]
- Iassonov, P.; Gebrenegus, T.; Tuller, M. Segmentation of X-ray computed tomography images of porous materials: A crucial step for characterization and quantitative analysis of pore structures. Water Resour. Res. 2009, 45, W09415. [Google Scholar] [CrossRef]
- Petersen, J.; Nielsen, M.; Lo, P.; Saghir, Z.; Dirksen, A.; de Brujne, M. Optimal graph based segmentation using flow lines with application to airway wall segmentation. Inf. Process. Med. Imaging 2011, 22, 46–60. [Google Scholar]
- Janusch, I.; Kropatsch, G.W.B. Topological Image Analysis and (Normalised) Representations for Plant Phenotyping. In Proceedings of the 16th International Symposium on Symbolic and Numeric Algorithms for Scientific Computing, Timisoara, Romania, 22–25 September 2014. [Google Scholar]
- Wohlhart, P.; Lepetit, V. Novel Concepts for Recognition and Representation of Structure in Spatio-Temporal Classes of Images. In Proceedings of the 20th Computer Vision Winter Workshop, Seggau, Austria, 9–11 February 2015. [Google Scholar]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Girish, V.; Vijayalakshmi, A. Affordable image analysis using NIH Image/ImageJ. Indian J. Cancer 2004, 41, 47. [Google Scholar] [PubMed]
- Flavel, R.; Guppy, C.; Tighe, M.; Watt, M.; McNeill, A.; Young, I. Non-destructive quantification of cereal roots in soil using high-resolution X-ray tomography. J. Exp. Bot. 2012, 63, 2503–2511. [Google Scholar] [CrossRef] [PubMed]
- Flavel, R.; Guppy, C.; Rabbi, S.; Young, I. An image processing and analysis tool for identifying and analysing complex plant root systems in 3D soil using non-destructive analysis: Root1. PLoS ONE 2017, 12, e0176433. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Schneider, H.; Burridge, J.; Ascanio, A.; Wojciechowski, T.; Topp, C.; Lynch, J.; Weitz, J.; Bucksch, A. Digital imaging of root traits (DIRT): A high-throughput computing and collaboration platform for field-based root phenomics. Plant Methods 2015, 11, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Z.; Valdes, C.; Clarke, J. Existing and Potential Statistical and Computational Approaches for the Analysis of 3D CT Images of Plant Roots. Agronomy 2018, 8, 71. https://doi.org/10.3390/agronomy8050071
Xu Z, Valdes C, Clarke J. Existing and Potential Statistical and Computational Approaches for the Analysis of 3D CT Images of Plant Roots. Agronomy. 2018; 8(5):71. https://doi.org/10.3390/agronomy8050071
Chicago/Turabian StyleXu, Zheng, Camilo Valdes, and Jennifer Clarke. 2018. "Existing and Potential Statistical and Computational Approaches for the Analysis of 3D CT Images of Plant Roots" Agronomy 8, no. 5: 71. https://doi.org/10.3390/agronomy8050071




