Development and Integration of an SSR-Based Molecular Identity Database into Sugarcane Breeding Program
Abstract
:Conflicts of Interest
References
- Pan, Y.-B. Biotechnology: Impact on sugarcane agriculture and industry. Sugar Tech 2012, 14, 1–2. [Google Scholar] [CrossRef]
- Wu, J.; Huang, Y.; Lin, Y.; Fu, C.; Liu, S.; Deng, Z.; Li, Q.; Huang, Z.; Chen, R.; Zhang, M. Unexpected Inheritance Pattern of Erianthus arundinaceus Chromosomes in the Intergeneric Progeny between Saccharum spp. and Erianthus arundinaceus. PLoS ONE 2014, 9, e110390. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.-B. Databasing molecular identities of sugarcane (Saccharum spp.) clones constructed with microsatellite (SSR) DNA markers. Am. J. Plant Sci. 2010, 1, 87–94. [Google Scholar] [CrossRef]
- Pan, Y.-B.; Scheffler, B.S.; Richard, E.P., Jr. High throughput genotyping of commercial sugarcane clones with microsatellite (SSR) DNA markers. Sugar Tech 2007, 9, 176–181. [Google Scholar]
- Bischoff, K.P.; Gravois, K.A. The development of new sugarcane varieties at the LSU Ag center. J. Am. Sugar Cane Technol. 2004, 24, 142–164. [Google Scholar]
- Cordeiro, G.M.; Taylor, G.O.; Henry, R.J. Characterisation of microsatellite markers from sugarcane (Saccharum sp.), a highly polyploid species. Plant Sci. 2000, 155, 161–168. [Google Scholar] [CrossRef]
- Pan, Y.-B. Highly polymorphic microsatellite DNA markers for sugarcane germplasm evaluation and variety identity testing. Sugar Tech 2006, 8, 246–256. [Google Scholar] [CrossRef]
- Tew, T.L.; White, W.H.; Legendre, B.L.; Grisham, M.P.; Dufrene, E.O.; Garrison, D.D.; Veremis, J.C.; Pan, Y.-B.; Richard, E.P., Jr.; Miller, J.D. Registration of ‘HoCP 96–540’ Sugarcane. Crop Sci. 2004, 45, 785–786. [Google Scholar]
- Tew, T.L.; Burner, D.M.; Legendre, B.L.; White, W.H.; Grisham, M.P.; Dufrene, E.O.; Garrison, D.D.; Veremis, J.C.; Pan, Y.-B.; Richard, E.P., Jr. Registration of ‘Ho 95–988’ Sugarcane. Crop Sci. 2005, 45, 1660–1661. [Google Scholar] [CrossRef]
- Tew, T.L.; Dufrene, E.O., Jr.; Garrison, D.D.; White, W.H.; Grisham, M.P.; Pan, Y.-B.; Richard, E.P., Jr.; Legendre, B.L.; Miller, J.D. Registration of ‘HoCP 00–950’ sugarcane. J. Plant Regist. 2009, 3, 42–50. [Google Scholar] [CrossRef]
- Tew, T.L.; Dufrene, E.O., Jr.; Cobill, R.M.; Garrison, D.D.; White, W.H.; Grisham, M.P.; Pan, Y.-B.; Legendre, B.L.; Richard, E.P., Jr.; Miller, J.D. Registration of ‘HoCP 91–552’ sugarcane. J. Plant Regist. 2011, 5, 181–190. [Google Scholar] [CrossRef]
- White, W.H.; Cobill, R.M.; Tew, T.L.; Burner, D.M.; Grisham, M.P.; Dufrene, E.O., Jr.; Pan, Y.-B.; Richard, E.P., Jr.; Legendre, B.L. Registration of ‘Ho 00–961’ Sugarcane. J. Plant Regist. 2011, 5, 332–338. [Google Scholar] [CrossRef]
- Hale, A.L.; Dufrene, E.O.; Tew, T.L.; Pan, Y.-B.; Viator, R.P.; White, P.M.; Veremis, J.C.; White, W.H.; Cobill, R.; Richard, E.P.; et al. Registration of ‘Ho 02–113’ Sugarcane. J. Plant Regist. 2011, 7, 51–57. [Google Scholar] [CrossRef]
- Pan, Y.-B.; Miller, J.D.; Schnell, R.J.; Richard, E.P.; Wei, Q. Application of microsatellite and RAPD fingerprints in the Florida sugarcane variety program. Sugar Cane Int. 2003, 189, 19–28. [Google Scholar]
- Chandra, A.; Grisham, M.P.; Pan, Y.-B. Allelic divergence and cultivar-specific SSR alleles revealed through fluorescence-labeled SSR markers based capillary electrophoregrams in highly polyploid species sugarcane. Genome 2014, 57, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.-B.; Tew, T.L.; Schnell, R.J.; Viator, R.P.; Richard, E.P., Jr.; Grisham, M.P.; White, W.H. Microsatellite DNA marker-assisted selection of Saccharum spontaneum cytoplasm-derived germplasm. Sugar Tech 2006, 8, 23–29. [Google Scholar] [CrossRef]
- Pan, Y.-B.; Cordeiro, G.M.; Richard, E.P., Jr.; Henry, R.J. Molecular genotyping of sugarcane clones with microsatellite DNA markers. Maydica 2003, 48, 319–329. [Google Scholar]
- Tew, T.L.; Pan, Y.-B. Microsatellite (simple sequence repeat) marker-based paternity analysis of a seven-parent sugarcane polycross. Crop Sci. 2010, 50, 1401–1408. [Google Scholar] [CrossRef]
- Pan, Y.-B.; Liu, P.; Que, Y. Independently segregating simple sequence repeats (SSR) alleles in polyploid sugarcane. Sugar Tech 2015, 17, 235–242. [Google Scholar] [CrossRef]
- Lu, X.; Zhou, Z.; Pan, Y.-B.; Chen, C.Y.; Zhu, J.; Chen, P.H.; Li, Y.-R.; Cai, Q.; Chen, R.K. Segregation analysis of microsatellite (SSR) markers in sugarcane polyploids. Genet. Mol. Res. 2015, 14, 18384–18395. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Chandra, A.; Que, Y.; Chen, P.-H.; Grisham, M.P.; White, W.H.; Dalley, C.D.; Tew, T.L.; Pan, Y.-B. Identification of QTLs controlling sucrose content based on an enriched genetic linkage map of sugarcane (Saccharum spp. hybrids) cultivar ‘LCP 85–384’. Euphytica 2016, 207, 527–549. [Google Scholar] [CrossRef]
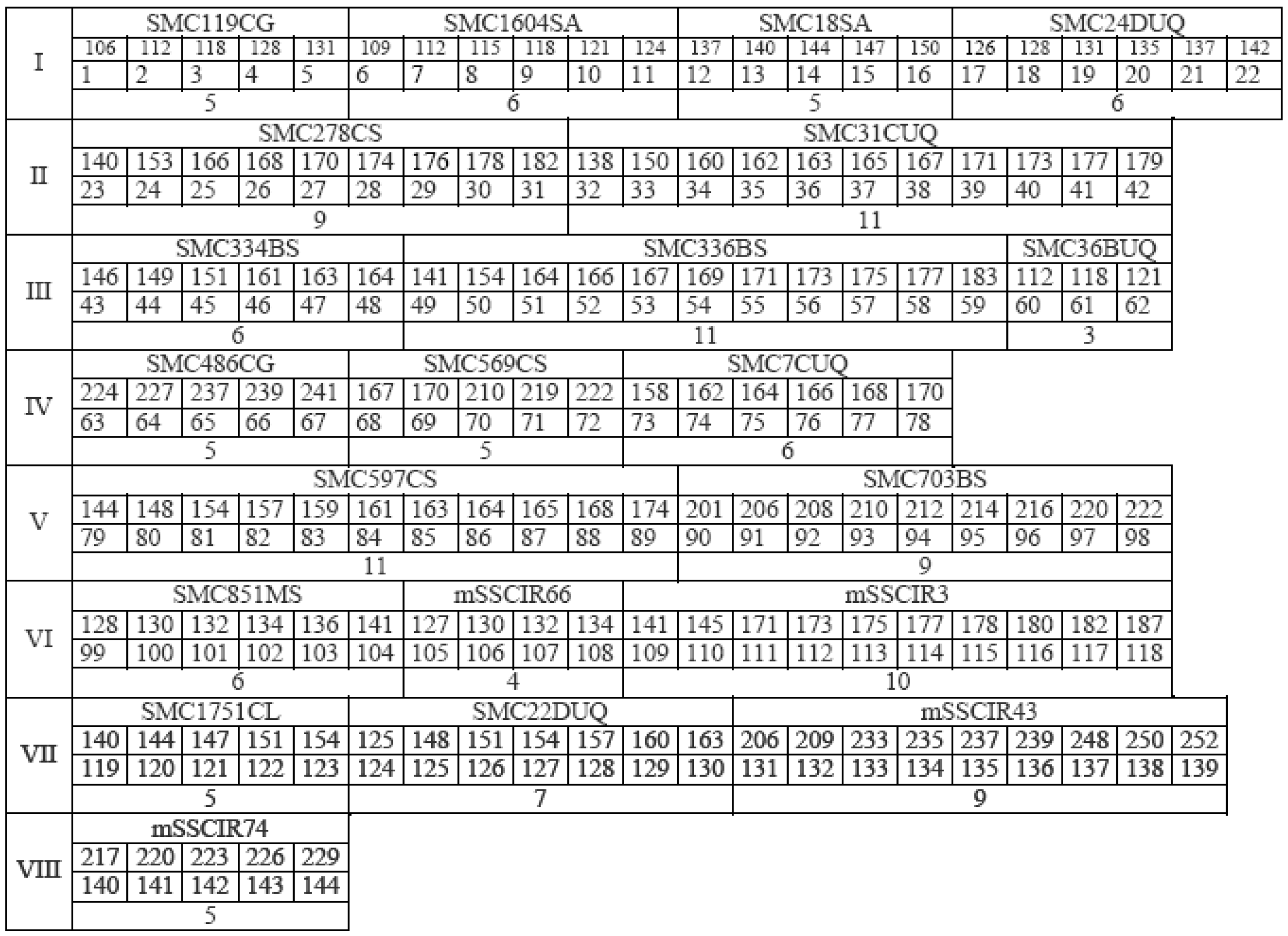

| SSR Name | Repeat Motif | Forward Primer Sequence (5′ to 3′) Reverse Primer Sequence (5′ to 3′) | Annealing Temp (°C) |
|---|---|---|---|
| SMC119CG | (TTG)12 | TTCATCTCTAGCCTACCCCAA | 58 |
| AGCAGCCATTTACCCAGGA | |||
| SMC1604SA | (TGC)7 | AGGGAAAAGGTAGCCTTGG | 58 |
| TTCCAACAGACTTGGGTGG | |||
| SMC18SA | (CGA)10 | ATTCGGCTCGACCTCGGGAT | 62 |
| AGTCGAAAGGTATAATAGTGTTAC | |||
| SMC24DUQ | (TG)13 | CGCAACGACATATACACTTCGG | 64 |
| CGACATCACGGAGCAATCAGT | |||
| SMC278CS | (TG)19 (AG)25 | TTCTAGTGCCAATCCATCTCAGA | 64 |
| CATGCCAACTTCCAAACAGACT | |||
| SMC31CUQ | (TC)10 (AC)22 | CATGCCAACTTCCAATACAGACT | 62 |
| AGTGCCAATCCATCTCAGAGA | |||
| SMC334BS | (TG)36 | CAATTCTGACCGTGCAAAGAT | 60 |
| CGATGAGCTTGATTGCGAATG | |||
| SMC336BS | (TG)23(AG)19 | ATTCTAGTGCCAATCCATCTCA | 62 |
| CATGCCAACTTCCAAACAGAC | |||
| SMC36BUQ | (TTG)7 | GGGTTTCATCTC TAGCCTACC | 64 |
| TCAGTAGCAGAGTCAGACGCTT | |||
| SMC486CG | (CA)34 | GAAATTGCCTCCCAGGATTA | 58 |
| CCAACTTGAGAATTGAGATTCG | |||
| SMC569CS | (TG)37 | GCGATGGTTCCTATGCAACTT | 62 |
| TTCGTGGCTGAGATTCACACTA | |||
| SMC7CUQ | (CA)10 (C)4 | GCCAAAGCAAGGGTCACTAGA | 60 |
| AGCTCTATCAGTTGAAACCGA | |||
| SMC597CS | (AG)31 | GCACACCACTCGAATAACGGAT | 64 |
| AGTATATCGTCCCTGGCATTCA | |||
| SMC703BS | (CA)12 | GCCTTTCTCCAAACCAATTAGT | 62 |
| GTTGTTTATGGAATGGTGAGGA | |||
| SMC851MS | (AG)29 | ACTAAAATGGCAAGGGTGGT | 58 |
| CGTGAGCCCACATATCATGC | |||
| mSSCIR66 | (GT)43GC (GT)6 | AGGTGATTTAGCAGCATA | 48 |
| CACAAATAAACCCAATGA | |||
| mSSCIR3 | (GT)28 | ATAGCTCCCACACCAAATGC | 60 |
| GGACTACTCCACAATGATGC | |||
| SMC1751CL | (TGC)7 | GCCATGCCCATGCTAAAGAT | 60 |
| ACGTTGGTCCCGGAACCG | |||
| SMC22DUQ | (CAG)5C (AGG)5 | CCATTCGACGAAAGCGTCCT | 62 |
| CAAGCGTTGTGCTGCCGAGT | |||
| mSSCIR43 | (GT)3(AT)2(GT)29 | ATTCAACGATTT TCACGAG | 52 |
| AACCTAGCAATTTACAAGAG | |||
| mSSCIR74 | (CGC)9 | GCGCAAGCCACACTGAGA | 54 |
| ACGCAACGCAAAACAACG |
© 2016 by the author; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, Y.-B. Development and Integration of an SSR-Based Molecular Identity Database into Sugarcane Breeding Program. Agronomy 2016, 6, 28. https://doi.org/10.3390/agronomy6020028
Pan Y-B. Development and Integration of an SSR-Based Molecular Identity Database into Sugarcane Breeding Program. Agronomy. 2016; 6(2):28. https://doi.org/10.3390/agronomy6020028
Chicago/Turabian StylePan, Yong-Bao. 2016. "Development and Integration of an SSR-Based Molecular Identity Database into Sugarcane Breeding Program" Agronomy 6, no. 2: 28. https://doi.org/10.3390/agronomy6020028






