Novel QTL for Stripe Rust Resistance on Chromosomes 4A and 6B in Soft White Winter Wheat Cultivars
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Field Experiments
2.3. Statistical Analysis
2.4. Marker Analysis
Linkage Map Construction and QTL Analysis
2.5. QTL Validation
3. Results
3.1. Field Disease Evaluations
3.2. Linkage Map Construction
3.3. QTL Identification
| QTL | Trait | Year | Location | cM Peak | Confidence Interval | Nearest Marker | LOD | R2 | Additive Effect | Parent |
|---|---|---|---|---|---|---|---|---|---|---|
| QYrel.wak-4A | IT | 2012 | PU | 98.9 | 94.2–103.3 | Xiwa6885 | 6.09 | 0.14 | 0.64 | Eltan |
| CF | 97 | 90.8–112.2 | Xiwa6885 | 8.73 | 0.17 | 1.12 | ||||
| CF | 99.8 | 96.6–101.9 | Xiwa6885 | 3.01 | 0.05 | 0.96 | ||||
| IT | 2013 | CF | 90.8 | 81–101.2 | Xiwa6193/Xiwa2606 | 3.51 | 0.09 | 0.65 | ||
| DS | 2013 | CF | 83 | 78.8–101.7 | Xiwa6193/Xiwa2606 | 5.19 | 0.16 | 10.1 | ||
| IT | 2014 | PU | 94 | 78.6–101.7 | Xiwa6193/Xiwa2606 | 3.94 | 0.09 | 0.55 | ||
| QYrfi.wak-6B | IT | 2012 | PU | 40.7 | 30.9–90 | Xiwa6599 | 7.21 | 0.31 | 1.41 | Finch |
| 2013 | CF | 46.2 | 32.8–68.6 | Xiwa6599 | 5.34 | 0.15 | 0.91 | |||
| 2014 | PU | 46.4 | 33.4–68.6 | Xiwa6599 | 4.46 | 0.12 | 0.47 | |||
| DS | 2012 | PU | 45.2 | 32.9–78.1 | Xiwa6599 | 5.00 | 0.15 | 5.62 | ||
| 2013 | CF | 46.2 | 33.4–63.7 | Xiwa6599 | 3.94 | 0.10 | 7.44 |
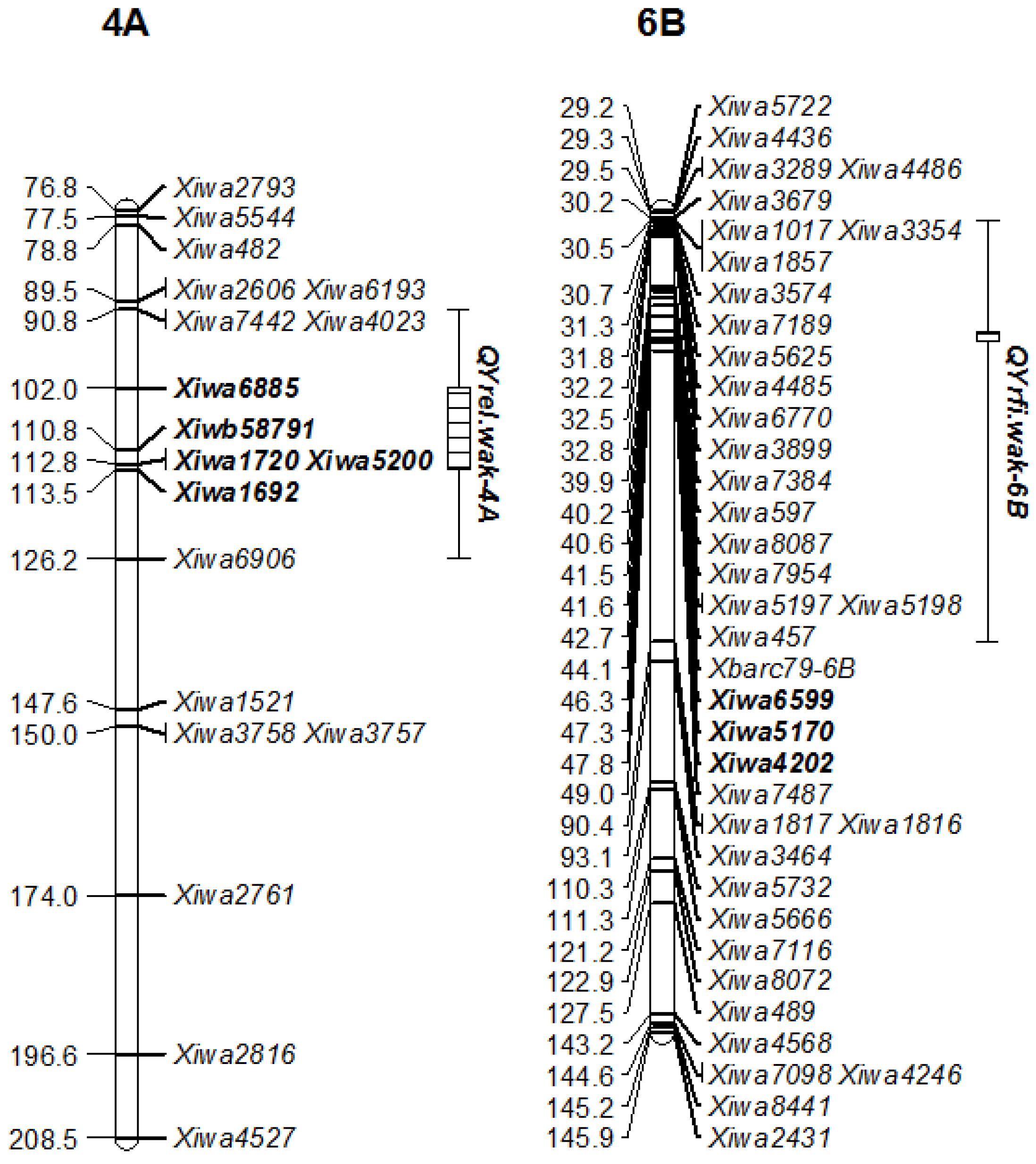

3.4. QTL Comparisons
| Variety | QYrel.wak-4A Markers | QYrfi.wak-6B Markers | ||||
|---|---|---|---|---|---|---|
| Xiwa5200 | Xiwa1720 | Xiwa6885 | Xiwa1692 | Xiwa5170 | Xiwa6599 | |
| Finch | C | A | C | C | G | T |
| Eltan | T | G | T | T | A | C |
| Stephen | C | A | C | C | G | C |
| Avocet | C | A | T | C | - | - |
| Heines VII | C | A | T | T | - | - |
| Nord Deprez | C | A | C | T | - | - |
| Minister | C | A | T | C | - | - |
| AUS91456 | T | G | T | C | - | - |
| Luke | T | A/G | C/T | C | - | - |
| Recital | - | - | - | - | G | C |
| Janz | - | - | - | - | G | C |
| Pingyuan 50 | - | - | - | - | G | C |
| Pavon 76 | - | - | - | - | A/G | C/T |
| Chromosome | Marker | Finch | Eltan | Variety | Repeat Length bp | Source |
|---|---|---|---|---|---|---|
| 6B | Xgwm58 | - | - | Pavon76 | 133 | Williams et al. [38] |
| 6B | Xbarc136 | - | - | Stephens | 280 | Santra et al. [36] |
| 6B | Xgwm193 | - | - | Renan | -- a | Dedryver et al. [39] |
| 6B | Xgdm113 | - | - | Stephens | 163 | Santra et al. [36] |
| 6B | Xbarc101 | - | - | Stephens | 185 | Santra et al. [36] |
| 6B | Xgwm626 | + | - | Pavon 76 | 101 | Williams et al. [38] |
| 6B | Xgwm132 | + | + | Stephens | 118 | Santra et al. [36] |
| 6B | Xgwm136 | - | - | Pingyuan 50 | 275 | Lan et al. [40] |
| 6B | Xgwm361 | - | - | Pingyuan 50 | 123 | Lan et al. [40] |
| 6B | Xuhw89 | - | - | T. turgidum spp. dicoccodes | 124 | Uauy et al. [37] Ramburan et al. [41] |
| 4A | Xgwm160 | - | - | Avocet/IDO444 (Kariega) | 209 | Chen et al. [42] |
| 4A | Xbarc70 | + | - | Stephens | 247 | Vazquez et al. [43] |
| Variety | Gene/QTL | Resistance Type a | Location (cM) | Source |
|---|---|---|---|---|
| Heines VII | YrHVII | ASR | Location unknown | Chen et al. [44] |
| Minister | YrMin | ASR | Location unknown | Chen et al. [44] |
| Nord Deprez | YrND | ASR | Location unknown | Chen et al. [44] |
| Stephens | Qyrst.orr-4AL | APR | 44.8 | Vazquez et al. [43] |
| AUS91456 | Yr51 | ASR | 104.6 | Randhawa et al. [32] |
| IDO444 | Qyrid.ui-4A | APR | Long arm | Chen et al. [42] |
| Lalbahadur | Yr60 | ASR | Distal end of 4AL | Herrera-Foessel et al. [33] |
| Kariega | QYr.sgi-4A1 | Early field resistance only | Long arm | Ramburan et al. [41] |
| Avocet S | QYr.sgi-4A2 | Identified in a growth chamber experiment | Long arm | Ramburan et al. [41] |
| Variety | Yr60 SSR Markers (bp) | QYrel.wak-4A KASP | |||
|---|---|---|---|---|---|
| Xwmc776 | Xwmc219 | Xwmc313 | Xiwa5200 a | Xiwa1692 | |
| Chinese | 226 | 222 | 215 | + b | + |
| Spring | |||||
| 4AS 0-0.2 c | 226 | 222 | 215 | + | + |
| 4AS 0-0.63 | 226 | 222 | 215 | + | + |
| 4AS 0-0.76 | 226 | 222 | 215 | + | + |
| 4AS 0-1.0 | 226 | 222 | 215 | + | + |
| 4AL 0-0.43 | - | - | - | + | + |
| 4AL 0-0.66 | - | - | - | + | + |
| 4AL 0-0.73 | - | - | - | + | + |
| 4AL 0-0.79 | - | - | - | + | + |
| 4AL 0-0.82 | - | - | - | + | + |
| 4AL 0-0.85 | - | - | - | + | + |
| 4AL 0-1.0 | 226 | 222 | 215 | + | + |
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chen, X.M. Epidemiology and control of stripe rust [Puccinia striiformis f. sp. tritici] on wheat. Can. J. Plant Path. 2005, 27, 314–337. [Google Scholar] [CrossRef]
- Wellings, C.R. Global status of stripe rust: A review of historical and current threats. Euphytica 2011, 179, 129–141. [Google Scholar] [CrossRef]
- Chen, X.M. Challenges and solutions for stripe rust control in the United States. Aust. J. Agric. Res. 2007, 58, 648–655. [Google Scholar] [CrossRef]
- Line, R.F. Stripe rust of wheat and barley in North America: A retrospective historical review. Annu. Rev. Phytopathol. 2002, 40, 75–118. [Google Scholar] [CrossRef] [PubMed]
- Qayoum, A.; Line, R.F. High-temperature, adult-plant resistance to stripe rust of wheat. Phytopathology 1985, 75, 1121–1125. [Google Scholar] [CrossRef]
- Chen, X.M. Review Article: High-temperature adult-plant resistance, key for sustainable control of stripe rust. Am. J. Plant Sci. 2013, 4, 608–627. [Google Scholar] [CrossRef]
- Gerechter-Amitai, Z.K.; van Silfout, C.H.; Grama, A.; Kleitman, F. Yr15 a new gene for resistance to Puccinia strifformis in Triticum dicoccoides sel. G-25. Euphytica 1989, 43, 187–190. [Google Scholar] [CrossRef]
- Wan, A.; Chen, X.M. Virulence characterization of Puccinia striiformis f. sp. tritici using a new set of Yr single-gene line differentials in the United States in 2010. Plant Dis. 2014, 98, 1534–1542. [Google Scholar]
- Campbell, M.A.; Fitzgerald, H.A.; Ronald, P.C. Engineering pathogen resistance in crop plants. Transgen. Res. 2002, 11, 599–613. [Google Scholar] [CrossRef]
- Roelfs, A.P.; Singh, R.P.; Saar, E.E. Rust Diseases of Wheat: Concepts and Methods of Disease Management; CIMMYT: Mexico, D.F., Mexico, 1992; pp. 1–81. [Google Scholar]
- McIntosh, R.A.; Wellings, R.A.; Park, R.F. Wheat Rusts: An Atlas of Resistance Genes; CSIRO Publications: East Melbourne, VIC, Australia, 1995; pp. 20–26. [Google Scholar]
- McIntosh, R.A.; Yamazaki, Y.; Dubcovsky, J.; Rogers, J.; Morris, C.; Appels, R.; Xia, X.C. Catalogue of Gene Symbols for Wheat. In Proceedings of the 12th International Wheat Genetics Symposium, Yokohama, Japan, 8 September 2013; Springer: Tokyo, Japan, 2015. [Google Scholar]
- Distelfeld, A.; Uauy, C.; Fahima, T.; Dubcovsky, J. Physical map of the wheat high-grain protein content gene Gpc-B1 and development of a high-throughput molecular marker. New Phytol. 2006, 169, 753–763. [Google Scholar] [CrossRef] [PubMed]
- Murphy, L.R.; Santra, D.; Kidwell, K.; Yan, G.Y.; Chen, X.M.; Campbell, K.G. Linkage maps of wheat stripe rust resistance genes Yr5 and Yr15 for use in marker-assisted selection. Crop Sci. 2009, 49, 1786–1790. [Google Scholar] [CrossRef]
- Collard, B.C.Y.; Mackill, D.J. Marker-assisted selection: An approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. B 2008, 303, 557–572. [Google Scholar] [CrossRef] [PubMed]
- Campbell, K.G.; Allan, R.E.; Anderson, J.; Pritchett, J.A.; Little, L.M.; Morris, C.F.; Line, R.F.; Chen, X.; Walker-Simmons, M.K.; Carter, B.P.; Burns, J.W.; Jones, S.S.; Reisenauer, P.E. Registration of “Finch” Wheat. Crop Sci. 2005, 45, 1656–1657. [Google Scholar] [CrossRef]
- Peterson, C.J.; Allan, R.E. Registration of “Eltan” Wheat. Crop Sci. 1991, 31, 1704. [Google Scholar] [CrossRef]
- McNeal, F.H.; Konzak, C.F.; Smith, E.P.; Tate, W.S.; Russell, T.S. A Uniform System for Recording and Processing Cereal Research Data; USDA-ARS: Washington, DC, USA, 1971; pp. 34–121. [Google Scholar]
- Line, R.F.; Qayoum, A. Virulence, Aggressiveness, Evolution, and Distribution of Races of Puccinia Striiformis (the Cause of Stripe Rust of Wheat) in North America, 1968–1987. U.S. Department of Agriculture Technical Bulletin No. 1788; National Technical Information Service: Springfield, VA, USA, February 1992; p. 44. [Google Scholar]
- Peterson, R.F.; Campbell, A.B.; Hannah, A.E. A diagrammatic scale for estimating rust intensity on leaves and stems of cereals. Can. J. Res. 1948, 26, 496–500. [Google Scholar] [CrossRef]
- Holland, J.B.; Nyquist, W.E.; Cervantes-Martinez, C.T. Estimating and interpreting heritability for plant breeding: An update. In Plant Breeding Reviews; Janick, J., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2003; Volume 22. [Google Scholar]
- Rӧder, M.S.; Korzun, V.; Wendehake, K.; Plaschke, J.; Tixier, M.; Leroy, P.; Ganal, W. (1998) A microsatellite map of wheat. Genetics 1998, 149, 2007–2023. [Google Scholar]
- Oetting, W.S.; Lee, H.K.; Flanders, D.J.; Wiesner, T.A.; Sellers, T.A.; King, R.A. Linkage analysis with multiplexed short tandem repeat polymorphisms using infrared fluorescence and M13 tailed primers. Genomics 1995, 30, 450–458. [Google Scholar] [CrossRef] [PubMed]
- Cavanagh, C.; Chao, S.; Wang, S.; Huang, B.E.; Stephen, S.; Kiani, S.; Forrest, K.; Saintenac, C.; Brown-Guedira, G.; Akhunova, A.; et al. Genome-wide comparative diversity multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc. Natl. Acad. Sci. USA 2013, 110, 8057–8062. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wong, D.; Forrest, K.; Allen, A.; Chao, S.; Huang, B.; Maccafern, M.; Salvi, S.; Milner, S.; Cattivelli, L.; et al. Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol. J. 2014, 12, 787–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Ooijen, J.W. JoinMap 4, Software for Calculation of Genetic Linkage Maps in Experimental Populations; Kyazma B.V.: Wageningen, The Netherlands, 2006. [Google Scholar]
- Basten, J.C.; Weir, B.S.; Zeng, Z.B. Windows QTL Cartographer 2.5; North Carolina State University, Department of Statistics: Raleigh, NC, USA, 2012. [Google Scholar]
- Kosambi, D. The estimation of map distances from recombination values. Ann. Eugen. 1943, 12, 172–175. [Google Scholar] [CrossRef]
- Chaky, J.M. Advanced Backcross QTL Analysis in a Mating Between Glycine max and Glycine soja. Master’s Thesis, University of Nebraska, Lincoln, NE, USA, 2003. [Google Scholar]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Rosewarne, G.; Herrera-Foessel, S.A.; Singh, R.P.; Huerto-Espino, J.; Lan, C.X.; He, Z.H. Quantitative trait loci for stripe rust resistance in wheat. Theor. Appl. Genet. 2013, 126, 2427–2449. [Google Scholar] [CrossRef] [PubMed]
- Randhawa, M.; Bansal, U.; Valárik, M.; Klocová, B.; Doležel, J.; Bariana, H. Molecular mapping of stripe rust resistance gene Yr51 in chromosome 4AL of wheat. Theor. Appl. Genet. 2014, 127, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Foessel, S.A.; Singh, R.P.; Lan, C.X.; Huerta-Espina, J.; Calvo-Salazar, V.; Bansal, U.K.; Bariana, H.S.; Lagudah, E.S. Yr60, a gene conferring moderate resistance to stripe rust in wheat. Plant Dis. 2015, 99, 508–511. [Google Scholar] [CrossRef]
- Endo, T.R.; Gill, B.S. The deletion stocks of common wheat. J. Hered. 1996, 87, 295–307. [Google Scholar] [CrossRef]
- Annual Stripe Rust Race Data Reports. Available online: http://striperust.wsu.edu/races/stripe-rust-race-data.html (accessed on 26 October 2015).
- Santra, D.K.; Chen, X.M.; Santra, M.; Campbell, K.G.; Kidwell, K.K. Identification and mapping QTL for high-temperature adult-plant resistance to stripe rust in winter wheat (Triticum aestivum L.) cultivar “Stephens”. Theor. Appl. Genet. 2008, 117, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Uauy, C.; Brevis, J.; Chen, X.; Khan, I.; Jackson, L.; Chicaiza, O.; Distelfeld, A.; Fahima, T.; Dubcovsky, J. High-temperature adult-plant (HTAP) stripe rust resistance gene Yr36 from Triticum turgidum ssp. dicoccoides is closely linked to the grain protein content gene Gpc-B1. Theor. Appl. Genet. 2005, 112, 97–105. [Google Scholar] [PubMed]
- Williams, H.M.; Singh, R.P.; Huerta-Espino, J.; Palacios, G.; Suenaga, K. Characterization of genetic loci conferring adult plant resistance to leaf rust and stripe rust in spring wheat. Genome 2006, 49, 977–990. [Google Scholar] [CrossRef] [PubMed]
- Dedryver, F.; Paillard, S.; Mallard, S.; Robert, O.; Trottet, M.; Nègre, S.; Verplancke, G.; Jahier, J. Characterization of genetic components involved in durable resistance to stripe rust in the bread wheat “Renan”. Phytopathology 2009, 99, 968–973. [Google Scholar] [CrossRef] [PubMed]
- Lan, C.; Liang, S.; Zhou, X.; Zhou, G.; Lu, Q.; Xia, X.; He, Z. Identification of genomic regions controlling adult-plant stripe rust resistance in Chinese landrace Pingyuan 50 through bulked segregant analysis. Phytopathology 2010, 100, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Ramburan, V.P.; Pretorius, Z.A.; Louw, J.H.; Boyd, L.A.; Smith, P.H.; Boshoff, W.H.P.; Prins, R. A genetic analysis of adult plant resistance to stripe rust in the wheat cultivar Kariega. Theor. Appl. Genet. 2004, 108, 1426–1433. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chu, C.; Souza, E.; Guttieri, M.; Chen, X.; Xu, S.; Hole, D.; Zemetra, R. Genome-wide identification of QTL conferring high-temperature adult-plant (HTAP) resistance to stripe rust (Puccinia striiformis f. sp. tritici) in wheat. Mol. Breed. 2011, 29, 791–800. [Google Scholar] [CrossRef]
- Vazquez, M.D.; Peterson, C.J.; Riera-Lizarazu, O.; Chen, X.; Heesacker, A.; Ammar, K.; Crossa, J.; Mundt, C.C. Genetic analysis of adult plant, quantitative resistance to stripe rust in wheat cultivar “Stephens” in multi-environment trials. Theor. Appl. Genet. 2012, 124, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.M.; Line, R.F.; Jones, S.S. Chromosomal locations of genes for resistance to Puccinia striiformis in winter wheat cultivars Heines VII, Clement, Moro, Tres and Daws. Phytopathology 1995, 85, 1362–1367. [Google Scholar] [CrossRef]
- IWGSC. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 2014, 345, 286. [Google Scholar]
- Neumann, K.; Kobiljski, B.; Dencic, S.; Varshney, R.K.; Börner, A. Genome-wide association mapping: A case study in bread wheat (Triticum aestivum L.). Mol. Breed. 2011, 27, 37–58. [Google Scholar] [CrossRef]
- Zegeye, H.; Rasheed, A.; Makdis, F.; Badebo, A.; Ogbonnaya, F.C. Genome-wide association mapping for seedling and adult plant resistance to stripe rust in synthetic hexaploid wheat. PLoS ONE 2014, 9, e105593. [Google Scholar] [CrossRef] [PubMed]
- Poland, J.A.; Brown, P.J.; Sorrells, M.E.; Jannick, J-L. Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS ONE 2012, 7, e32253. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.M.; (USDA-ARS Research Plant Pathologist, Pullman, WA, USA). Personal communication, 2015.
- Carter, A.H.; Chen, X.M.; Garland-Campbell, K.; Kidwell, K.K. Identifying QTL for high temperature adult-plant resistance to stripe rust (Puccinia striiformis f. sp. tritici) in the spring wheat (Triticum aestivum L.) cultivar “Louise”. Theor. Appl. Genet. 2009, 119, 1119–1128. [Google Scholar] [PubMed]
- Fu, D.; Uauy, C.; Distelfeld, A.; Blechl, A.; Epstein, L.; Chen, X.; Sela, H.; Fahima, T.; Dubcovsky, J. A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science 2009, 323, 1357–1360. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klarquist, E.F.; Chen, X.M.; Carter, A.H. Novel QTL for Stripe Rust Resistance on Chromosomes 4A and 6B in Soft White Winter Wheat Cultivars. Agronomy 2016, 6, 4. https://doi.org/10.3390/agronomy6010004
Klarquist EF, Chen XM, Carter AH. Novel QTL for Stripe Rust Resistance on Chromosomes 4A and 6B in Soft White Winter Wheat Cultivars. Agronomy. 2016; 6(1):4. https://doi.org/10.3390/agronomy6010004
Chicago/Turabian StyleKlarquist, Emily F., Xianming M. Chen, and Arron H. Carter. 2016. "Novel QTL for Stripe Rust Resistance on Chromosomes 4A and 6B in Soft White Winter Wheat Cultivars" Agronomy 6, no. 1: 4. https://doi.org/10.3390/agronomy6010004






