Abstract
Although mulching is a widely used agronomic practice, its effects on the rhizosphere remain poorly understood. Here, we employed amplicon and transcriptomic sequencing to investigate variations in a grapevine rhizosphere system under mulch treatment (rice straw + felt + plastic film). Analyzing 16S and intergenic spacer (ITS) rRNA sequences indicated that the Shannon and Simpson indices of the bacterial and fungal communities increased markedly under mulch treatment. The bacterial and fungal compositions varied significantly between the control and mulch treatments. Mulching enriched for potentially beneficial microbes that confer disease resistance to plants or participate in nitrogen metabolism (Kaistobacter, Ammoniphilus, Lysobacter, Ammoniphilus, Alicyclobacillus, Aquicella, Nitrospira, Chaetomium, and Microascus), whereas more potentially pathogenic microbes (Fusarium and Gibberella) were detected in the control. Moreover, certain bacteria and fungi exhibited different correlations with the root transcriptome functions of the MEBlue module. The complexity of the bacterial and fungal co-occurrence networks increased with higher node numbers, positive and negative links after mulching. Following mulching, the rhizosphere showed elevated pH, organic matter, and catalase activities, and decreased sucrase and cellulase and β-glucosidase activities. Our results provide comprehensive data showing how a grapevine rhizosphere system responded to mulching treatment and shed important insight into mulching practices for fruit trees.
1. Introduction
Mulching is a conventional management technique that is widely used in agricultural production. Mulch forms a barrier that covers soil with organic or inorganic materials and plays positive roles in suppressing weed growth, maintaining the soil-water content, controlling the soil temperature, improving soil function, promoting plant growth, and producing high-quality products [1,2,3]. Li et al., 2021 found that straw mulching affected the root growth of faba beans by changing the soil-nutrient supply and soil moisture [4]. Shirzadi et al., 2020 showed that the use of plastic mulch increased the onion yield by 29% [5]. Li et al., 2022 reported that plastic-film mulch changed the root-related microbiomes of maize plants and increased root growth [6]. However, little is known regarding the relationship between soil conditions and plant growth under a combination of organic and inorganic mulch treatment.
The rhizosphere functions as the key zone for interactions between plants and soils; a dynamic zone that integrates energy exchange and nutrient cycling among plants, soil, and microbial communities [7,8]. Rhizosphere microorganisms play pivotal roles in plant growth, plant resistance to abiotic and biotic stresses, and plant yields and qualities [9,10,11]. Rhizosphere microbes influence many aspects of plant growth, including the circadian clock and nutrient levels [12,13]. The results of various studies have shown that the relative abundances of rhizosphere microbes change under mulch treatment. Juhos et al., 2023 found that straw mulch could improve microbiological parameters when compared to wool mulch [14]. Recent findings have shown that organic mulch helps regulate soil-microbial communities in tea and litchi [15,16]. In addition, plastic mulch films (biodegradable mulch and polyethylene mulch) can increase microbial abundances in the rhizosphere [17].
Previous reports have also shown that root growth and development can be influenced by mulch treatment. Film mulching confined maize root growth under dry conditions and inhibited excessive root growth under wet conditions [18]. Film mulching can also significantly increase the length, diameter, and weight of root radishes [19]. Similar to film mulching, wheat-straw mulching promotes plant root growth [20].
Grapevines are economically important fruit trees that are cultivated worldwide. Grapevines are also important commercial fruit trees in China. The grapevine cultivation area of China is 582,728 ha and the total fruit yield was 19.3 t in 2021 (https://www.fao.org/faostat/zh/#data/QCL, accessed on 12 June 2023). Mulching is widely used in vineyards [21,22]. Currently, mulching is considered a useful tool for adapting to climate change in the grapevine industry [23]. However, few studies have focused on the responses of different grapevine rhizosphere systems to mulch treatments. In this study, we aimed to (1) clarify the variations in soil microbial compositions and functions that occur under mulch treatment and (2) analyze the relationship between the root system and soil microbes under mulching conditions.
2. Materials and Methods
2.1. Study Site and Experimental Design
A field study was conducted in a vineyard in a plastic-covered greenhouse at the Jiangsu Academy of Agricultural Sciences, Nanjing, Jiangsu Province, China (32°02′ N, 118°52′ E). In this study, conventional tillage with no mulching was used as the control group, and rice straw (5 cm thick) + felt (200 g∙m2) + plastic film (0.08 mm) was used as the mulch treatment (mulch group). Eight-year-old ’Zijinhongxia’ (Vitis vinifera L.) grapevines were used as materials. Three replicates were performed for each treatment group. For each replicate, we included three grapevines. The grapevines were separated by 1.8 m when they were planted. The grapevines were spur-pruned in December 2021. This study was conducted in February 2022. An intelligent temperature recorder L92-1 (Hangzhou Loggertech Co., Ltd., Hangzhou, China) was used to monitor the soil temperature every 10 min at a depth of 10 cm, near the root of each grapevine. Soil properties were: available N = 0.36 g/kg, available P = 0.63 g/kg, available K = 0.55 g/kg, total N = 2.33 g/kg, total P = 2.58 g/kg, total K = 17.42 g/kg, pH = 4.76, and organic matter = 34.39 g/kg. The soil was irrigated 3 days before treatment. No irrigation was performed during treatment.
2.2. Plant and Soil Sampling
Root tissue and rhizosphere soil samples were obtained when the grapevines grew to 4–5 leaves. The surface soil was removed, and only the soil adhering to the roots (0–4 mm) at a depth of 0–10 cm was collected by gentle brushing and used as the grapevine rhizosphere soil [24]. The rhizosphere soil was then pooled and sieved through a 40-mesh sieve, after which the fine roots were removed and homogeneously mixed. The root tissues were cleaned with double-distilled H2O, wiped with sterile filter paper, and quickly frozen in liquid nitrogen. The soil (approximate 100 g) and plant samples (approximate 5 g) were immediately transported to our laboratory. Root tissues were stored at −80 °C for subsequent RNA extraction. A portion of each soil sample was stored in an airtight cooler to determine soil-pH values, organic matter contents, and soil-enzyme activities (i.e., catalase (CAT), sucrase, and cellulase activities). Another portion of each soil sample (approximate 20 g) was stored at −80 °C (for determining β-glucosidase activities and extracting total DNA).
2.3. 16S rRNA and Intergenic Spacer (ITS) rRNA Amplicon Sequencing and Data Processing
Total soil DNA was extracted from rhizosphere soil sample using the E.Z.N.A.® Soil DNA Kit (Omega Bio-tek, Norcross, GA, USA). The DNA quality was assessed using a Fragment Analyzer 5400 (Agilent Technologies, Selangor, Malaysia), and the DNA quantity was measured using a Nanodrop ND-2000 instrument (Nanodrop Technologies, Wilmington, DE, USA). The V3–V4 region of 16S rRNA was amplified using the primers, 341F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGG TWTCTAAT-3′). The ITS rRNA region of ITS1-1F was amplified using the primers, ITS1-1F-F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS1-1F-R (5′-GCTGCGTTCTTCATCGATGC-3′). Polymerase chain reaction (PCR) analysis was performed as described by Wang et al. [24]. PCR products were extracted from 2% agarose gels and further purified using an AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) and quantified using QuantiFluor™-ST (Promega, Madison, WI, USA). The purified amplicons were pooled in equimolar concentrations and paired-end sequenced (2 × 300 base pairs [bp]) on an Illumina NovaSeq6000 instrument (Illumina, San Diego, CA, USA), according to standard protocols. The resulting raw sequences were filtered, trimmed, denoised, and merged, and chimeric sequences were identified and removed using the DADA2 plugin of QIIME2 to identify amplicon sequence variants (ASVs) [25]. A taxonomy table was generated by aligning the ASV sequences with Greengenes [26].
2.4. Root RNA Extraction and High-Through Sequencing
Total RNA was extracted from the root tissue using an RNAprep Pure Plant Kit (TIANGEN, Beijing, China). RNA concentrations and purities were measured using a Nanodrop ND-2000 instrument (Nanodrop Technologies, Wilmington, DE, USA). We used 1 μg RNA per sample as input material for the RNA sample preparations. Sequencing libraries were constructed using the NEBNext®UltraTM RNA Library Prep Kit from Illumina® (New England Biolabs, Ipswich, MA, USA). PCR products were purified (AMPure XP system), and the libraries were quantified using an Agilent Bioanalyzer 2100 system. The library preparations were sequenced on the Illumina platform with 150 bp paired-ends.
2.5. Root Transcriptomic Data Analysis
The original data were filtered and reads with adapters were removed using Fastp software (v.0.19.3). The clean reads were mapped to a reference genome (Vitis vinifera, wine grape, BioProject: PRJEA18785). Gene-expression levels were estimated by calculating fragments per kilobase of transcript per million fragments mapped reads values using Feature Counts software (v1.6.2) and StringTie software (v1.3.4d). Differentially expressed genes (DEGs) between the two treatments were analyzed using DESeq2 software (v1.22.1) and edgeR software (v3.24.3). P values were corrected using the Benjamini–Hochberg method. Corrected p values and |log2 fold-change [FC]| values were used as thresholds for significantly different expressions. Enrichment analysis was performed using a hypergeometric test. Hypergeometric-distribution testing was performed for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway-enrichment analysis. Gene Ontology (GO) terms were used for gene-set-enrichment analysis.
2.6. Statistical Analysis
Variance analysis of soil-physicochemical properties and enzymes was performed using a t test. Alpha diversity indices (including the Chao1, observed features, Shannon, and Simpson indices) were calculated to estimate microbial diversity. Alpha-diversity variance analysis was performed using the Kruskal–Wallis test. Beta-diversity distance measurements were performed based on Bray–Curtis analysis to analyze structural variations in microbial communities across samples through principal-coordinate analysis (PCoA). Co-occurrence analyses were conducted to determine the network characteristics of the rhizosphere microbial communities, using the following criteria: Spearman correlation > 0.5 and p < 0.05.
A correlation heatmap was constructed to determine the relationships between the root transcriptome and microbial communities at the genus level (Spearman correlation > 0.6 and p < 0.05).
3. Results
3.1. Rhizosphere Soil Physicochemical Properties and Enzyme
The pH values (4.78 and 5.61 in the control and mulch groups, respectively; t test, p < 0.0001) and organic-matter contents (35.24 g/kg and 38.66 g/kg in the control and mulch groups, respectively; t test, p < 0.001) were significantly higher after treatment with mulch than in the control group (Figure 1). The catalase activity (1.04 mg/g and 1.27 mg/g 20 min in the control and mulch groups, respectively; t test, p < 0.01) also significantly increased. However, the activities of cellulase (3.10 U/g and 1.55 U/g in the control and mulch groups, respectively; t test, p < 0.01), sucrase (20.24 U/g and 16.10 U/g in the control and mulch groups, respectively; t test, p < 0.01), and β-glucosidase (191.92 U/g and 151.00 U/g in and the control and mulch groups, respectively; t test, p < 0.01) decreased significantly. In addition, the soil temperature difference was lower in the mulch group than in the control group (Table S1).
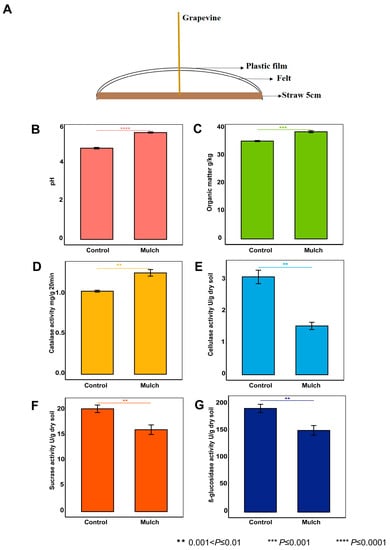
Figure 1.
Study design (A) for investigating the effects of mulching treatment on the physicochemical properties (B–G) of rhizosphere soil.
3.2. Amplicon-Sequencing Results
3.3. Composition and Function of Rhizosphere Soil Bacteria after Mulch Treatment
In this study, we performed 16S rRNA-amplicon sequencing to detect the responses of rhizosphere bacteria to mulching. We identified 2519 and 3511 ASVs in the control and mulch groups, respectively (Figure S1). The taxonomic affiliations showed that the rhizosphere contained two kingdoms, 34 phyla, 103 classes, 146 orders, 186 families, 235 genera, and 107 species (Figure S1).
We detected 880 enriched ASVs (34.93% in the control group, 25.06% in the mulch group) that overlapped between the two treatments (Figure 2A). As shown in Figure 2B, the most abundant phyla were Proteobacteria (average: 70.17%, control group; 59.47%, mulch group), Actinobacteria (average: 12.43%, control group; 12.37%, mulch group), Gemmatimonadetes (average: 4.99%, control group; 7.91%, mulch group), Acidobacteria (average: 4.22%, control group; 5.34%, mulch group), Bacteroidetes (average: 1.79%, control group; 4.16%, mulch group), Firmicutes (average: 1.37%, control group; 3.02%, mulch group), TM7 (average: 1.28%, control group; 2.76%, mulch group), Chloroflexi (average: 1.67%, control group; 1.83%, mulch group), Crenarchaeota (average: 0.31%, control group; 0.95%, mulch group), and Verrucomicrobia (average: 0.41%, control group; 0.70%, mulch group). At the genus level, significant differences were observed in the relative abundances of 19 genera (Figure 2E). Among these, Kaistobacter was the most abundant in the mulch group and Rhodoplanes was the most abundant in the control group. The relative abundances of Rathayibacter, Nocardioides, B-42, Uliginosibacterium, Alkanibacter, Luteimonas, Dermacoccus, and Peredibacter were significantly higher in the control group than in the mulch group, whereas the relative abundances of Oceanobacillus, Wandonia, Alicyclobacillus, Ammoniphilus, Reyranella, Nitrospira, Oryzihumus, Aquicella, and Lysobacter were lower in the control group than in the mulch group (Figure 2E).
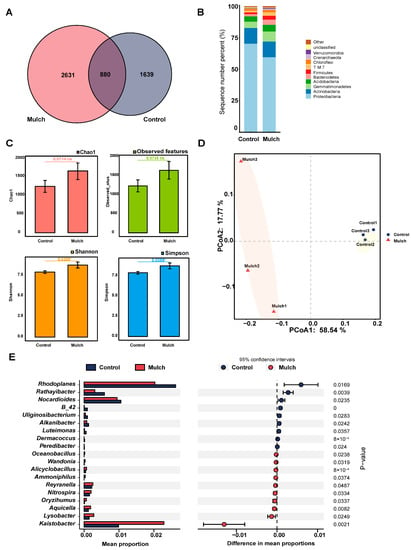
Figure 2.
Rhizosphere bacteria in the control and mulch groups. (A) Amplicon sequence variants (ASVs). (B) Distribution of ASVs at the phylum level. (C) Bacterial alpha-diversities following different treatments. * p < 0.05, between different treatments. (D) Principal-component analysis (PCoA) of bacteria based on Bray–Curtis distances. (E) Different frequencies of bacteria at the genus level under different treatments.
Next, bacterial alpha diversity was estimated using the Chao1, observed features, Shannon, and Simpson indices. The Chao1 and observed feature indices (t test, p > 0.05) showed no significant differences between the control and mulch treatments (Figure 2C), whereas the Shannon index (t test, p < 0.05) and Simpson index (t test, p < 0.05) were higher in the mulch group than in the control group.
However, PCoA analysis of the beta diversity, based on Bray–Curtis analysis, showed that the control samples were clearly separated from the mulch samples (Figure 2D).
3.4. Composition and Function of RhizosphereSoil Fungi after Mulch Treatment
The responses of the rhizosphere fungi were detected using ITS-amplicon sequencing. We found 488 and 620 ASVs in the control and mulch-treated groups, respectively (Figure S2). The taxonomic affiliations showed that the rhizosphere contained 6 kingdoms, 10 phyla, 23 classes, 48 orders, 84 families, 137 genera, and 165 species (Figure S2).
There were 156 enriched ASVs (average: 25.16%, control group; 31.97%, mulch group) that overlapped between the two treatments (Figure 3A). The relative abundances of the major phyla are shown in Figure 2B. The most abundant phyla were Ascomycota (average: 82.76%, control group; 71.83%, mulch group), Mortierellomycota (average: 13.97%, control group; 11.17%, mulch group), Basidiomycota (average: 0.65%, control group; 1.15%, mulch group), Zoopagomycota (average: 0.44%, control group; 0.63%, mulch group), Chytridiomycota (average: 0.18%, control group; 0.71%, mulch group), Rozellomycota (average: 0.16%, control group; 2.68%, mulch group), Aphelidiomycota (average: 0.12%, control group; 0.43%, mulch group), Mucoromycota (average: 0.08%, control group; 0.08%, mulch group), and Cercozoa (average: 0.05%, group; 0.04%, mulch group). At the genus level, a significant difference was observed in the relative abundances of 13 genera (Figure 3E). The relative abundances of Trichocladium, Fusarium, Gibberella, Monilia, and Stephanonectria were higher in the control group than in the mulch group. The relative abundances of Bionectria, Lophiostoma, Microascus, and Chaetomium were higher in the mulch group than in the control group (Figure 3E).
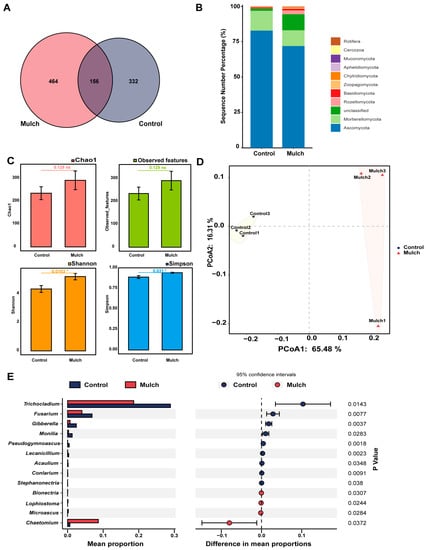
Figure 3.
Rhizosphere fungi in the control and mulch groups. (A) Amplicon sequence variants (ASVs). (B) Distribution of ASVs at the phylum level. (C) Fungi alpha-diversities after different treatments. * p < 0.05, between different treatments. (D) PCoA of fungi based on Bray–Curtis distances. (E) Different frequencies of fungi at the genus level under different treatments.
The fungal alpha diversity was also evaluated. Chao1 index analysis (t test, p > 0.05) and observed features index analysis (t test, p > 0.05) revealed no significant differences between the control and mulch groups, although the Shannon index (t test, p < 0.05) and Simpson index (t test, p < 0.05) were significantly higher in the mulch group than in the control group (Figure 3C). However, the beta-diversity results showed that the control and mulch-treated groups clustered together (Figure 3D).
3.5. Microbial Co-Occurrence Networks in Rhizosphere Soil
Bacterial co-occurrence-network analyses revealed 98 and 1420 nodes and edges in the control group, respectively, 100 and 1464 nodes and edges in the mulch group, respectively (Figure 4). The average node degree was 28.98 in the control group and 29.28 in the mulch group. The density was higher in the control group than in the mulch group (0.299 vs. 0.296). The closeness was higher in the mulching treatments. Fungal co-occurrence-network analyses that the edge number, density, average node degree, and closeness were 926, 0.199, 19.09, and 0.044 in the control group, respectively, whereas those parameters were 1014, 0.179, 18.95, and 0.086, respectively, in the mulch group (Figure 4). Both the bacterial and fungal co-occurrence networks had greater positive and negative links in the mulch group than in the control group.
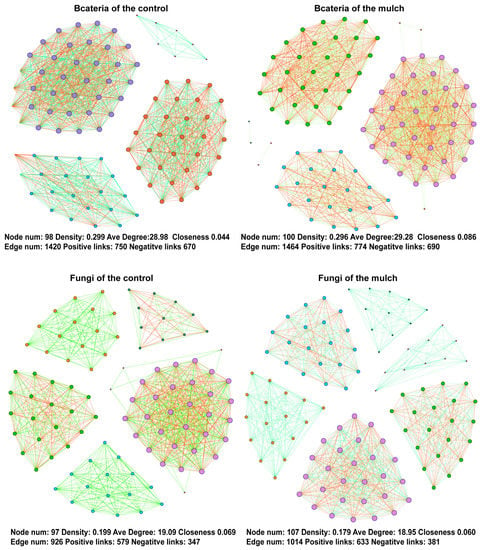
Figure 4.
Co-occurrence networks for the bacterial and fungal rhizospheres in the control and mulch groups.
3.6. Transcriptome Analysis of Root Tissues after Mulch Treatment
We obtained 270,193,408 reads from six transcriptome libraries after quality-control analysis; the Q20 rates were all over 97%, and the Q30 rates were all over 93% (Table S4). The root DEGs in 4–5 leaves were screened based on a log2 FC value of ≥1 and an adjusted p value of <0.05. We identified 1236 DEGs, of which 494 and 742 were downregulated or upregulated, respectively (Figure 5A,B). The DEGs were further studied by KEGG pathway-enrichment analysis, which highlighted several significantly enriched KEGG terms, including stilbenoid, diarylheptanoid, and gingerol biosynthesis; phenylpropanoid biosynthesis; plant-pathogen interaction; biosynthesis of amino acids; phenylalanine metabolism; and flavonoid biosynthesis (Figure 5C).
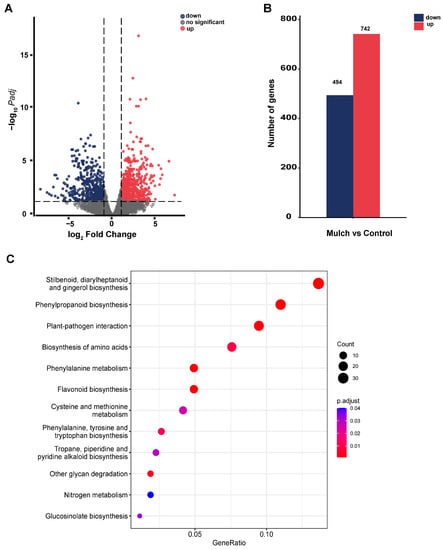
Figure 5.
The number and functions of root-tissue differentially expressed genes (DEGs). (A) Volcano-plot analysis of DEGs in the control and mulch groups. (B) Number of DEGs. (C) Kyoto Encyclopedia of Genes and Genomes (KEGG)-based pathway enrichment associated with DEGs in the control and mulch groups.
Weighted correlation network analysis (WGCNA) was conducted to explore key gene functions in the control and mulch groups (Figure S3). Eight modules were identified, and the enriched functions of each module were analyzed. In the MEblack module, the most enriched pathways included DNA replication, flavonoid biosynthesis, SNARE interactions in vesicular transport and circadian rhythm plant (Figure 6A); in the MEgreen module, the most enriched pathways were enriched in ribosome, biosynthesis of amino acids, protein processing in endoplasmic reticulum, and some metabolisms (Figure 6B); in MEblue, the top pathways were plant-pathogen interaction, plant hormone signal transduction, and phenylpropanoid biosynthesis (Figure 6C); in MEyellow, the top pathways were oxidative phosphorylation, terpenoid backbone biosynthesis, and peroxisome (Figure 6D); in MEturquoise, the most enriched pathways were biosynthesis of cofactors, biosynthesis of amino acids, and endocytosis (Figure 6E); in MEred, the phagosome, DNA replication, and mismatch repair pathway terms were mainly enriched (Figure 6F); in MEbrown, spliceosome, mRNA surveillance pathway and nucleocytoplasmic transport were the top pathway terms (Figure 6G). No pathways were enriched in MEpink.
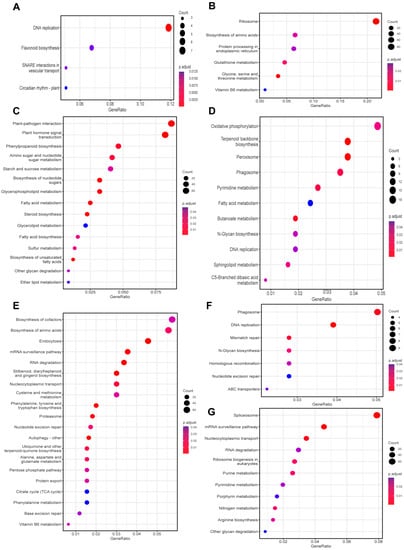
Figure 6.
KEGG-based pathway-enrichment analysis of different modules based on weighted correlation-network analysis (WGCNA). (A) MEblack; (B) MEGreen; (C) MEblue; (D) MEyellow; (E) METurquoise; (F) MERed; (G) MEbrown.
3.7. Variations in Root Transcriptome Relationships with Bacterial and Fungal Communities
The Spearman correlation heatmap showed that five phyla (Acidobacteria, Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria) correlated significantly with the seven root transcriptome modules (Figure 7), i.e., not including the MEpink module. At the genus level, Edaphobacter (Acidobacteria) negatively correlated with MEblack. In the Actinobacteria phylum, eight genera showed significant correlations with root transcriptome modules. Among them, Salinibacterium, Oryzihumus, Sphaerisporangium and Dermacoccus exhibited inverse correlations with the MEturquoise and MEblue modules. Pseudonocardia and Mycobacterium were significantly correlated only with the MEturquoise, Nocatdioides was clearly correlated with MEblue, and Geodermatophilus specifically correlated with MEred. In Bacteroidetes, Wandonia showed inverse correlations with MEturquoise and MEblue, Pedobacter also showed the opposite trend for MEbrown and MEyellow. In Firmicutes, Alicyclobacillus and Paenibacillus significantly positively correlated with MEblue. Faecalibacterium and Sporosarcina showed the same positive correlation trend in MEyellow and MEbrown, and Oceanobacillus correlated negatively with MEturquoise and MEbrown. Thirty genera showed significant correlations with the seven root transcriptome modules (Figure 7). In the fungal communities, four phyla correlated significantly with the root transcriptome module (Figure 8). Among these, Rhizophydium (Chytridiomycota) correlated specifically with MEred, and Thaumatomonas (Cercozoa) correlated significantly with MEbrown and MEyellow. Saitozyma and Oliveonia (Basidiomycota) correlated with MEturquoise. Ascomycota, including 26 genera, correlated significantly with the seven root transcriptome modules (Figure 8).
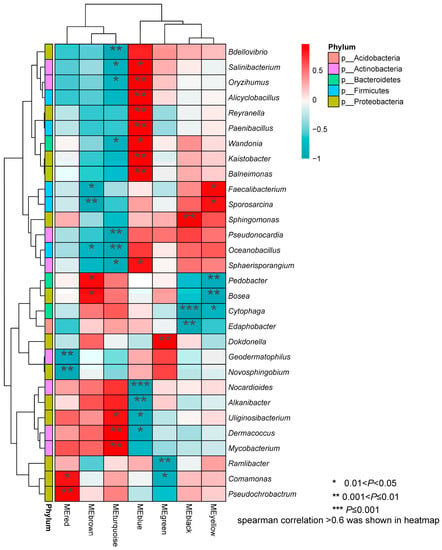
Figure 7.
Correlation heat map for bacterial genera with different KEGG enrichment modules.
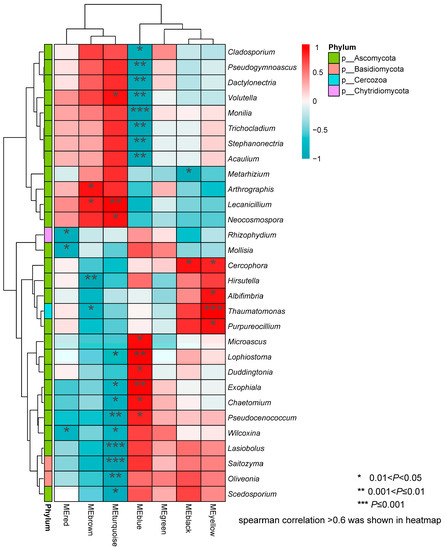
Figure 8.
Correlation heat map for fungal genera with different KEGG enrichment modules.
Redundancy analysis (RDA) plots were conducted to determine the soil microbial enrichment level in relation to root transcriptome. Bacterium enrichment levels MEpink (p = 0.313), MEblue (p = 0.092), MEblack (p = 0.333), MEyellow (p = 0.324), MEgreen (p = 0.510), MEred (p = 0.194), MEbrown (p = 0.219) and MEturquoise (p = 0.432) had no significant effect on the bacterial genera (Figure 9A). Relating to fungi, only MEblack (p = 0.003) had a strong effect on fungal genera. Compared with other fungi, Scedosporium, Agaricus, Candida, and Lasiobolus were influenced by MEblack to a greater extent (Figure 9B).
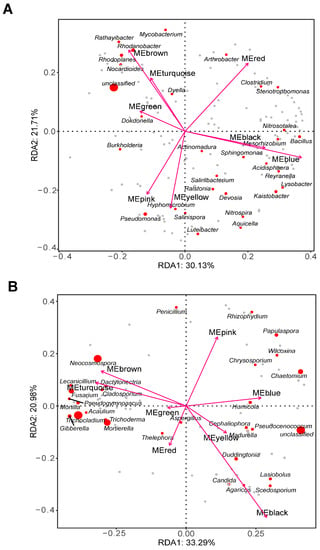
Figure 9.
Redundancy analysis (RDA) plots of root transcriptome KEGG enrichment module related to soil bacterial genera (A) and fungal genera (B). Top 30 genera were presented.
3.8. Grapevine Growth Status and Phenological Date
As shown in Figure 10A, at the same date, the growth and development of grapevine was more advanced in the mulch group than that in the control. Figure 10B shows that the start dates of bud burst, flowering, and berry ripening were all earlier (5–7 d) in the mulch group than that in the control group.
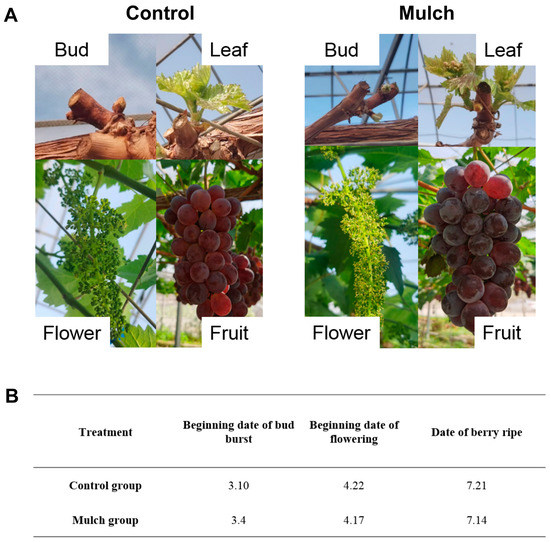
Figure 10.
Grapevine growth status at the same date in the control and mulch group (A), and phenological date (B).
4. Discussion
Soil temperature is a key factor in plant growth [27,28]. Mulching reduces the maximum soil temperature in potato “Kufri Jyoti” [29]. Bio-based films and biodegradable paper can also decrease soil temperature in tomato “Fentailang” [30]. As previously reported, the maximum soil temperature decreases after straw mulching, whereas the minimum temperature increases [31]. In this study, both the maximum and minimum soil temperatures decreased after mulch treatment, but the temperature difference was 1.7 °C lower in the mulch group than in the control group (Table S1). The results showed that mulch treatment had a better heat-preservation effect than control treatment. The organic matter content and pH were found to be key factors affecting the soil quality. Previous data showed that mulching increased the soil-organic matter content and Ph in maize “Sygenta-8611”, in wheat, and Gold Milenium apples on M.9 rootstock [32,33,34], in agreement with our current findings (Figure 1). The results showed that the soil quality improved to some degree.
In addition, soil-enzyme activity responded to mulching treatments, and CAT, sucrose, and cellulase were more sensitive to mulching treatments than to other soil properties [35]. Beta-glucosidase activity is one of the most sensitive and reliable indices for assessing soil quality [36]. This study determined the activities of four soil enzymes. Zhang et al., 2021 found that soil CAT activity increased under continuous straw mulch [37]. Shi et al., 2022 showed that plastic films promoted CAT activity in potato fields [38]. CAT activities showed a similar trend after mulch treatment in this study. Some findings showed that mulching increased β-glucosidase and sucrase activities [32,38]; however, we observed a different trend. Soil-enzymatic activity has been correlated with plant growth and development. For instance, Wang et al., (2014) indicated that soil urease, phosphatase, and-glucosidase activities were associated with different trends in vegetative and reproductive growth after mulch treatment [39]. In general, the grapevine soil-enzyme activities responded differently to mulching treatments.
Soil microbiomes play key roles in driving soil functions. For bacteria, the dominant phyla detected in this study were most prevalent in the plant rhizosphere. The results of previous studies showed that Proteobacteria, Actinobacteria, Crenarchaeota, Verrucomicrobia, Gemmatimonadetes, Acidobacteria, Firmicutes, and Chloroflexi were the major bacterial phyla in grapevines [10,40]. TM7 has also been detected in maize rhizosphere soils [41]. Compared to the control group, Kaistobacter, Lysobacter, Aquicella, Nitrospira, and Ammoniphilus genera were significantly enriched during the mulching treatment (Figure 2). Kaistobacter was previously reported to be a beneficial microbe [42]. Lysobacter and Ammoniphilus have been associated with plant disease resistance. Lin et al., 2021 showed that Lysobacter secreted small molecular toxins and considered Lysobacter an antifungal biocontrol agent [43]. Yang et al., 2023 found that Ammoniphilus was regulated by signaling pathways that confer disease resistance to plants [44]. Aquicella and Nitrospira correlated positively with the nitrification potential [23,45]. Alicyclobacillus showed significant differences between healthy and diseased konjac rhizospheres, where the difference decreased with increasing continuous cropping years [42]. The dominant phyla detected in this study were consistent with those detected previously [46]. The significantly enriched genera, Chaetomium and Microascus, can inhibit plant diseases. For example, Chaetomium promoted plant growth and inhibited replant diseases [47]. Previous data showed that Microascus can be used as a plant biocontrol strain [48,49]. However, the significantly enriched genera Trichocladium, Fusarium, and Gibberella in the control group have been correlated with plant disease. Gibberella and Trichocladium were more abundant in samples obtained after continuous cropping for years than in those obtained after 1 year [50]. Previous results showed that Fusarium and Gibberella act as plant pathogens [51,52].
The microbial co-occurrence pattern is a key indicator of soil health and has been strongly associated with microbial functions [53,54,55]. Plastic-film mulching decreases the complexity of co-occurring networks [56], which might be related to a favorable soil environment [57]. However, the results of this study showed that the complexity of both the bacterial and fungal co-occurrence networks increased after mulch treatment. These outcomes may have been related to the mulch materials; both organic and inorganic materials were used in this study. Otherwise, the bacterial and fungal co-occurrence networks became more stable during mulching. Despite the differences observed in the bacterial and fungal co-occurrence networks, the bacterial co-occurrence network showed greater clustering when the fungal co-occurrence network became looser. Recent findings showed that bacteria and fungi respond differently to the environment [58,59]. Generally, the practice of organic and inorganic mulching can improve soil health.
Few studies have focused on transcriptome variations in root systems after mulch treatment. In this study, we determined the transcriptome of grapevine roots. Our results revealed DEG enrichment for nitrogen-metabolism pathways (Figure 5C). Wang et al., 2022 also showed that plastic films promoted the expression of genes related to nitrogen metabolism in grapevine roots. We also identified differences in the microbes involved in the nitrogen cycle [60]. In addition, KEGG pathway analysis revealed enrichment for plant–pathogen interactions (Figure 5C). Significant variations in microbes between the control and mulching treatments were considered in terms of their functions as biocontrol microbes and pathogens. Additionally, phenylpropanoid metabolism-related pathways (phenylpropanoid biosynthesis and phenylalanine metabolism) differed significantly between the control and mulch treatments (Figure 5C). Previous findings showed that phenylpropanoid metabolism participates in plant–environment interactions [61] and that phenylpropanoids can act as signaling molecules in plant–microbe interactions [62]. An increase in phenylpropanoid biosynthesis can increase the resistance to fungal pathogens [63].
Further analysis of the correlations between microbes that were significantly enriched after the control and mulch treatments revealed an opposite correlation trend with the MEblue module. Transcriptomics analysis revealed that beneficial and anti-pathogenic microbes correlated positively with the root MEblue module, whereas potential pathogens correlated negatively with the root MEblue module. The results of several studies have shown similar findings with root-associated microbes [64,65]. Plant functions can be regulated directly or indirectly by microbes [66,67]. Our results showed that rhizosphere microbes contribute to some functions of grapevine roots under mulching treatment, although the underlying mechanisms remain unknown and should be studied in the future.
Circadian rhythm is an important factor in plant development [68] and is mainly regulated by light and temperature [69]. In the present study, the circadian rhythm pathway was detected in MEblack module (Figure 6A) and a soil temperature difference was observed (Table S1). Furthermore, recent reports found that complex rhizosphere microbial communities also had effects on plant circadian rhythm [12]. Certain fungi were found to have a strong effect on MEblack module (Figure 9B). However, their effects on plant growth and development are still unknown; thus, more research will be needed to clarify the relationship between microorganisms and plant function.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy13061656/s1, Figure S1: Number of taxa according to 16S rRNA amplicon sequencing; Figure S2: Number of taxa according to ITS rRNA amplicon sequencing; Figure S3: Modular gene-clustering tree based on WGCNA; Table S1: Air and soil temperatures during the treatment period; Table S2: Number of 16S rRNA-amplicon sequence reads; Table S3: Number of ITS rRNA-amplicon sequence reads; Table S4. Statistical analysis of all sample qualities after filtering.
Author Contributions
Conceptualization, B.W. and W.-M.W.; data curation, X.-C.W.; formal analysis, X.-C.W.; funding acquisition, W.-M.W.; investigation, Z.-X.C.; methodology, B.W. and Z.-X.C.; project administration, W.-M.W.; resources, X.-C.W.; software, B.W. and Z.-W.W.; supervision, X.-C.W.; validation, X.-C.W. and Z.-W.W.; visualization, X.-C.W.; writing—original draft, B.W.; writing—review and editing, B.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Jiangsu Agricultural Science and Technology Innovation Fund: CX(20)2022, Jiangsu Province Breeding Project of Agricultural Leading New Varieties: PZCZ201722, Xuzhou City Policy Guidance Plan (Industry-University-Research Collaboration): KC21332 and the China Agriculture Research System of MOF and MARA.
Data Availability Statement
The datasets for this study can be found in the NCBI SRA [PRJNA974977] [http://www.ncbi.nlm.nih.gov/sra/PRJNA974977] (accessed on 22 May 2023).
Acknowledgments
The data were analyzed with the help of Wekemo Tech Group Co., Ltd., Shenzhen, China.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Jones, J.; Savin, M.C.; Rom, C.R.; Gbur, E. Denitrifier community response to seven years of ground cover and nutrient management in an organic fruit tree orchard soil. Appl. Soil Ecol. 2017, 112, 60–70. [Google Scholar] [CrossRef]
- El-Beltagi, H.S.; Basit, A.; Mohamed, H.I.; Ali, I.; Ullah, S.; Kamel, E.A.R.; Shalaby, T.A.; Ramadan, K.M.A.; Alkhateeb, A.A.; Ghazzawy, H.S. Mulching as a sustainable water and soil saving practice in agriculture: A review. Agronomy 2022, 12, 1881. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, L.; Luo, Y.; Awasthi, M.K.; Yang, J.; Duan, Y.; Li, H.; Zhao, Z. Mulching practices alter the bacterial-fungal community and network in favor of soil quality in a semiarid orchard system. Sci. Total Environ. 2020, 725, 138527. [Google Scholar] [CrossRef]
- Li, B.; Chen, X.; Shi, X.; Liu, J.; Wei, Y.; Xiong, F. Effects of ridge tillage and straw mulching on cultivation the fresh faba beans. Agronomy 2021, 11, 1054. [Google Scholar] [CrossRef]
- Shirzadi, M.H.; Arvin, M.J.; Abootalebi, A.; Hasandokht, M.R. Effect of nylon mulch and some plant growth regulators on water use efficiency and some quantitative traits in onion (Allium cepa cv.) under water deficit stress. Cogent Food Agric. 2020, 6, 1779562. [Google Scholar] [CrossRef]
- Li, Y.; Li, T.; Wang, Z.; Wang, S.; Qin, X.; Liao, Y. Plastic film mulch changes the microbial community in maize root-associated compartments. Plant Soil 2022, 470, 5–20. [Google Scholar] [CrossRef]
- Shen, J.; Li, C.; Mi, G.; Li, L.; Yuan, L.; Jiang, R.; Zhang, F. Maximizing root/rhizosphere efficiency to improve crop productivity and nutrient use efficiency in intensive agriculture of China. J. Exp. Bot. 2013, 64, 1181–1192. [Google Scholar] [CrossRef]
- Moran, J.; McGrath, C. Comparison of methods for mapping rhizosphere processes in the context of their surrounding root and soil environments. Biotechniques 2021, 71, 604–614. [Google Scholar] [CrossRef]
- Aparicio, M.A.; Lucena, C.; Garcia, M.J.; Ruiz-Castilla, F.J.; Jimenez-Adrian, P.; Lopez-Berges, M.S.; Prieto, P.; Alcantara, E.; Perez-Vicente, R.; Ramos, J.; et al. The nonpathogenic strain of Fusarium oxysporum FO12 induces Fe deficiency responses in cucumber (Cucumis sativus L.) plants. Planta 2023, 257, 50. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Gao, J.; Wang, S.; Li, W.; Wang, A. Community differentiation of rhizosphere microorganisms and their responses to environmental factors at different development stages of medicinal plant Glehnia littoralis. PeerJ 2023, 11, e14988. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Wang, X.; Wang, Z.; Zhu, K.; Wu, W. Comparative metagenomic analysis reveals rhizosphere microbial community composition and functions help protect grapevines against salt stress. Front. Microbiol. 2023, 14, 1102547. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, D.; Sankar, A.; Devika, O.S.; Singh, S.; Shikha; Parihar, M.; Rakshit, A.; Sayyed, R.Z.; Gafur, A.; Ansari, M.J.; et al. Optimizing nutrient use efficiency, productivity, energetics, and economics of red cabbage following mineral fertilization and biopriming with compatible rhizosphere microbes. Sci. Rep. 2021, 11, 15680. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, C.J.; McMinn, R.; Weinig, C. Rhizosphere microbes influence host circadian clock function. Phytobiomes J. 2021, 5, 368–372. [Google Scholar] [CrossRef]
- Juhos, K.; Papdi, E.; Kovács, F.; Vasileiadis, V.P.; Veres, A. The effect of wool mulch on plant development in the context of the physical and biological conditions in soil. Plants 2023, 12, 684. [Google Scholar] [CrossRef]
- Xu, D.; Ling, J.; Qiao, F.; Xi, P.; Zeng, Y.; Zhang, J.; Lan, C.; Jiang, Z.; Peng, A.; Li, P. Organic mulch can suppress litchi downy blight through modification of soil microbial community structure and functional potentials. BMC Microbiol. 2022, 22, 155. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, Y.; Sun, L.; Qiu, C.; Ding, Y.; Gu, H.; Wang, L.; Wang, Z.; Ding, Z. Organic mulching positively regulates the soil microbial communities and ecosystem functions in tea plantation. BMC Microbiol. 2020, 20, 103. [Google Scholar] [CrossRef] [PubMed]
- Shan, X.; Zhang, W.; Dai, Z.L.; Li, J.B.; Mao, W.W.; Yu, F.W.; Ma, J.J.; Wang, S.Y.; Zeng, X.P. Comparative analysis of the effects of plastic mulch films on soil nutrient, yields and soil microbiome in three vegetable fields. Agronomy 2022, 12, 506. [Google Scholar] [CrossRef]
- Zhang, W.; Li, S.Q.; Shen, Y.F.; Yue, S.C. Film mulching affects root growth and function in dryland maize-soybean intercropping. Field Crops Res. 2021, 271, 108240. [Google Scholar] [CrossRef]
- Lee, O.N.; Park, H.Y. Effects of different colored film mulches on the growth and bolting time of radish (Raphanus sativus L.). Sci. Hortic. 2020, 266, 109271. [Google Scholar] [CrossRef]
- Yan, F.J.; Sun, Y.J.; Xu, H.; Yin, Y.Z.; Wang, H.Y.; Wang, C.Y.; Guo, C.C.; Yang, Z.Y.; Sun, Y.Y.; Ma, J. Effects of wheat straw mulch application and nitrogen management on rice root growth, dry matter accumulation and rice quality in soils of different fertility. Paddy Water Environ. 2018, 16, 507–518. [Google Scholar] [CrossRef]
- Burg, P.; Cizkova, A.; Masan, V.; Sedlar, A.; Matwijczuk, A.; Soucek, J. The effect of mulch materials on selected soil properties, yield and grape quality in vineyards under central european conditions. Agronomy 2022, 12, 1862. [Google Scholar] [CrossRef]
- Hu, J.G.; Bai, S.J.; Zhao, R.H.; Chen, G.; Cai, J.S. Effects of black geotextile mulch and grass mulch on the microclimate, fruit quality and anthocyanin components of ‘Xinyu’ table grape. N. Z. J. Crop Hort. 2022, 2108066. [Google Scholar] [CrossRef]
- Fraga, H.; Santos, J.A. Vineyard mulching as a climate change adaptation measure:fFuture simulations for Alentejo, Portugal. Agric. Sys. 2018, 164, 107–115. [Google Scholar] [CrossRef]
- Wang, B.; Zhou, T.; Li, K.; Guo, X.W.; Guo, Y.S.; Liu, Z.D.; Xie, H.G. Bacterial communities that metabolize 4-Hydroxybenzoic acid in grape (Vitis vinifera L.) rhizosphere soil. Allelopath. J. 2019, 46, 41–53. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Meth. 2016, 13, 581. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Caporaso, J.G. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2′s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Grillakis, M.G.; Koutroulis, A.G.; Papadimitriou, L.V.; Daliakopoulos, I.N.; Tsanis, I.K. Climate-induced shifts in global soil temperature regimes. Soil Sci. 2016, 181, 264–272. [Google Scholar] [CrossRef]
- Arai-Sanoh, Y.; Ishimaru, T.; Ohsumi, A.; Kondo, M. Effects of soil temperature on growth and root function in rice. Plant Prod. Sci. 2010, 13, 235–242. [Google Scholar] [CrossRef]
- Goel, L.; Shankar, V.; Sharma, R.K. Influence of different organic mulches on soil hydrothermal and plant growth parameters in potato crop (Solanum tuberosum L.). J. Agrometeorol. 2020, 22, 56–59. [Google Scholar] [CrossRef]
- Zhang, X.; You, S.; Tian, Y.; Li, J. Comparison of plastic film, biodegradable paper and bio-based film mulching for summer tomato production: Soil properties, plant growth, fruit yield and fruit quality. Sci. Hortic. 2019, 249, 38–48. [Google Scholar] [CrossRef]
- Chen, S.Y.; Zhang, X.Y.; Pei, D.; Sun, H.Y.; Chen, S.L. Effects of straw mulching on soil temperature, evaporation and yield of winter wheat: Field experiments on the North China Plain. Ann. Appl. Biol. 2007, 150, 261–268. [Google Scholar] [CrossRef]
- Javed, A.; Iqbal, M.; Farooq, M.; Lal, R.; Shehzadi, R. Plastic film and straw mulch effects on maize yield and water use efficiency under different irrigation levels in Punjab, Pakistan. Int. J. Agric. Biol. 2019, 21, 767–774. [Google Scholar]
- Han, W.; Cao, P.; Sun, Y.; Dang, G.; Xue, S.; America, L. Mechanized mulching practices with plastic filmand wheat straw in dryland wheat planting. AMA Agric. Mech. Asia Afr. Lat. Am. 2015, 46, 82–88. [Google Scholar]
- Sas-Paszt, L.; Pruski, K.; Zurawicz, E.; Sumorok, B.; Derkowska, E.; Gluszek, A. The effect of organic mulches and mycorrhizal substrate on growth, yield and quality of Gold Milenium apples on M.9 rootstock. Can. J. Plant Sci. 2014, 94, 281–291. [Google Scholar] [CrossRef]
- Tang, W.Z.; Yang, H.S.; Wang, W.E.; Wang, C.X.; Pang, Y.Y.; Chen, D.Y.; Hu, X.T. Effects of living grass mulch on soil properties and assessment of soil quality in Chinese apple orchards: A meta-analysis. Agronomy 2022, 12, 1974. [Google Scholar] [CrossRef]
- Saikia, R.; Sharma, S.; Thind, H.S.; Sidhu, H.S.; Yadvinder, S. Temporal changes in biochemical indicators of soil quality in response to tillage, crop residue and green manure management in a rice-wheat system. Ecol. Indic. 2019, 103, 383–394. [Google Scholar] [CrossRef]
- Zhang, M.; Zhao, G.; Li, Y.; Wang, Q.; Dang, P.; Qin, X.; Zou, Y.; Chen, Y.; Siddique, K.H.M. Straw incorporation with ridge–furrow plastic film mulch alters soil fungal community and increases maize yield in a semiarid region of China. Appl. Soil Ecol. 2021, 167, 104038. [Google Scholar] [CrossRef]
- Shi, M.F.; Kang, Y.C.; Zhang, W.N.; Yang, X.Y.; Fan, Y.L.; Yu, H.F.; Zhang, R.Y.; Guo, A.X.; Qin, S.H. Plastic film mulching with ridge planting alters soil chemical and biological properties to increase potato yields in semiarid Northwest China. Chem. Biol. Technol. Agri. 2022, 9, 16. [Google Scholar] [CrossRef]
- Wang, Y.P.; Li, X.G.; Hai, L.; Siddique, K.H.M.; Gan, Y.T.; Li, F.M. Film fully-mulched ridge-furrow cropping affects soil biochemical properties and maize nutrient uptake in a rainfed semi-arid environment. Soil Sci. Plant Nutr. 2014, 60, 486–498. [Google Scholar] [CrossRef]
- Wei, Y.J.; Wu, Y.; Yan, Y.Z.; Zou, W.; Xue, J.; Ma, W.R.; Wang, W.; Tian, G.; Wang, L.Y. High-throughput sequencing of microbial community diversity in soil, grapes, leaves, grape juice and wine of grapevine from China. PLoS ONE 2018, 13, e0193097. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, N.; Guo, X.Y.; Zhang, Y.; Ye, B.P. Comparative analysis of bacterial community structure in the rhizosphere of maize by high-throughput pyrosequencing. PLoS ONE 2017, 12, e0178425. [Google Scholar] [CrossRef]
- Wu, W.; Wu, J.; Liu, X.; Chen, X.; Wu, Y.; Yu, S. Inorganic phosphorus fertilizer ameliorates maize growth by reducing metal uptake, improving soil enzyme activity and microbial community structure. Ecotoxicol. Environ. Saf. 2017, 143, 322–329. [Google Scholar] [CrossRef]
- Lin, L.; Xu, K.; Shen, D.; Chou, S.H.; Gomelsky, M.; Qian, G. Antifungal weapons of Lysobacter, a mighty biocontrol agent. Environ. Microbiol. 2021, 23, 5704–5715. [Google Scholar] [CrossRef]
- Yang, B.; Zheng, M.; Dong, W.; Xu, P.; Zheng, Y.; Yang, W.; Luo, Y.; Guo, J.; Niu, D.; Yu, Y.; et al. Plant disease resistance-related pathways recruit beneficial bacteria by remodeling root exudates upon Bacillus cereus AR156 treatment. Microbiol. Spectr. 2023, 11, e0361122. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Zhao, Y.; Yao, X.; Wang, J.; Zheng, P.; Xi, C.; Hu, B. Dominance of comammox Nitrospira in soil nitrification. Sci. Total Environ. 2021, 780, 146558. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Yang, H.; Chu, M.; Niu, X.; Wang, N.; Lin, Q.; Lou, K.; Zuo, C.; Wang, J.; Zou, Q.; et al. Differentiation and variability in the rhizosphere and endosphere microbiomes of healthy and diseased cotton (Gossypium sp.). Front. Microbiol. 2021, 12, 765269. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Elrys, A.S.; Liu, L.; Iqbal, M.; Zhao, J.; Huang, X.; Cai, Z. Cover plants-mediated suppression of Fusarium Wilt and root-knot incidence of cucumber is associated with the changes of rhizosphere fungal microbiome structure-under plastic shed system of North China. Front. Microbiol. 2022, 13, 697815. [Google Scholar] [CrossRef] [PubMed]
- Cao, P.; Wei, X.; Wang, G.; Chen, X.; Han, J.; Li, Y. Microbial inoculants and garbage fermentation liquid reduced root-knot nematode disease and as uptake in Panax quinquefolium cultivation by modulating rhizosphere microbiota community. Chin. Herb. Med. 2022, 14, 58–69. [Google Scholar] [CrossRef]
- Fotso, S.; Graupner, P.; Xiong, Q.; Gilbert, J.R.; Hahn, D.; Avila-Adame, C.; Davis, G.; Sumiyoshi, K. Alveolarides: Antifungal peptides from Microascus alveolaris active against phytopathogenic fungi. J. Nat. Prod. 2018, 81, 10–15. [Google Scholar] [CrossRef]
- Song, X.; Pan, Y.; Li, L.; Wu, X.; Wang, Y. Composition and diversity of rhizosphere fungal community in Coptis chinensis Franch. continuous cropping fields. PLoS ONE 2018, 13, e0193811. [Google Scholar] [CrossRef]
- Kebede, A.Z.; Johnston, A.; Schneiderman, D.; Bosnich, W.; Harris, L.J. Transcriptome profiling of two maize inbreds with distinct responses to Gibberella ear rot disease to identify candidate resistance genes. BMC Genom. 2018, 19, 131. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, L.; Wang, L.; Wu, Q.; Li, K.; Guo, X. Autotoxin affects the rhizosphere microbial community structure by influencing the secretory characteristics of grapevine roots. Front. Microbiol. 2022, 13, 953424. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Zhao, K.; Lv, X.; Su, W.; Dai, Z.; Gilbert, J.A.; Brookes, P.C.; Faust, K.; Xu, J. Genetic correlation network prediction of forest soil microbial functional organization. ISME J. 2018, 12, 2492–2505. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Kirkby, C.A.; Schmutter, D.; Bissett, A.; Kirkegaard, J.A.; Richardson, A.E. Network analysis reveals functional redundancy and keystone taxa amongst bacterial and fungal communities during organic matter decomposition in an arable soil. Soil Biol. Biochem. 2016, 97, 188–198. [Google Scholar] [CrossRef]
- Hao, M.; Hu, H.; Liu, Z.; Dong, Q.; Sun, K.; Feng, Y.; Li, G.; Ning, T. Shifts in microbial community and carbon sequestration in farmland soil under long-term conservation tillage and straw returning. Appl. Soil Ecol. 2019, 136, 43–54. [Google Scholar] [CrossRef]
- Liu, J.; Li, S.; Yue, S.; Tian, J.; Chen, H.; Jiang, H.; Siddique, K.H.M.; Zhan, A.; Fang, Q.; Yu, Q. Soil microbial community and network changes after long-term use of plastic mulch and nitrogen fertilization on semiarid farmland. Geoderma 2021, 396, 115086. [Google Scholar] [CrossRef]
- Luo, S.S.; Wang, S.J.; Yao, P.W.; Guo, D.; Li, X.J.; Li, S.Q.; Tian, C.J. Soil microbial communities under film mulching and N fertilization in semiarid farmland. Nutr. Cycl. Agroecosyst. 2019, 114, 157–170. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Maestre, F.T.; Reich, P.B.; Jeffries, T.C.; Gaitan, J.J.; Encinar, D.; Berdugo, M.; Campbell, C.D.; Singh, B.K. Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat. Commun. 2016, 7, 10541. [Google Scholar] [CrossRef]
- de Vries, F.T.; Griffiths, R.I.; Bailey, M.; Craig, H.; Girlanda, M.; Gweon, H.S.; Hallin, S.; Kaisermann, A.; Keith, A.M.; Kretzschmar, M.; et al. Soil bacterial networks are less stable under drought than fungal networks. Nat. Commun. 2018, 9, 3033. [Google Scholar] [CrossRef]
- Wang, R.; Sun, Q.; Xing, X. Root transcriptome reveals responses to plastic film mulching and grass cover in wine grape ‘Cabernet Sauvignon’ root and berry. Vitis 2020, 59, 1–8. [Google Scholar] [CrossRef]
- Dong, N.Q.; Lin, H.X. Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. J. Integr. Plant Biol. 2021, 63, 180–209. [Google Scholar] [CrossRef] [PubMed]
- Naoumkina, M.A.; Zhao, Q.A.; Gallego-Giraldo, L.; Dai, X.B.; Zhao, P.X.; Dixon, R.A. Genome-wide analysis of phenylpropanoid defence pathways. Mol. Plant Pathol. 2010, 11, 829–846. [Google Scholar] [CrossRef]
- Xoca-Orozco, L.; Aguilera-Aguirre, S.; Vega-Arreguín, J.; Acevedo-Hernández, G.; Tovar-Pérez, E.; Stoll, A.; Herrera-Estrella, L.; Chacón-López, A. Activation of the phenylpropanoid biosynthesis pathway reveals a novel action mechanism of the elicitor effect of chitosan on avocado fruit epicarp. Food Res. Int. 2019, 121, 586–592. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Cheng, L.; Nian, H.; Jin, J.; Lian, T. Linking plant functional genes to rhizosphere microbes: A review. Plant Biotechnol. J. 2023, 21, 902–917. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, R.L.; Vismans, G.; Yu, K.; Song, Y.; de Jonge, R.; Burgman, W.P.; Burmølle, M.; Herschend, J.; Bakker, P.; Pieterse, C.M.J. Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 2018, 12, 1496–1507. [Google Scholar] [CrossRef]
- Hacquard, S.; Spaepen, S.; Garrido-Oter, R.; Schulze-Lefert, P. Interplay between innate immunity and the plant microbiota. Annu. Rev. Phytopathol. 2017, 55, 565–589. [Google Scholar] [CrossRef]
- Netzker, T.; Shepherdson, E.M.F.; Zambri, M.P.; Elliot, M.A. Bacterial volatile compounds: Functions in communication, cooperation, and competition. Annu. Rev. Microbiol. 2020, 74, 409–430. [Google Scholar] [CrossRef]
- Inoue, K.; Araki, T.; Endo, M. Circadian clock during plant development. J. Plant Res. 2018, 131, 59–66. [Google Scholar] [CrossRef]
- McClung, C.R. Plant circadian rhythms. Plant Cell 2006, 18, 792–803. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).