Assessment of Efficacy and Mechanism of Resistance to Soil-Applied PPO Inhibitors in Amaranthus palmeri
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Response to Soil-Applied PPO Herbicides
2.3. Analyze the TSM Profile and Copy Number in Survivors from Soil-Applied Fomesafen
2.4. Analyze the Expression and Mutation Profile the Target-Site in of Survivors from Soil-Applied Fomesafen
2.5. Evaluate the Level of Resistance to Fomesafen in Transgenic Arabidopsis Lines Overexpressing A. palmeri PPO2
3. Results
3.1. Response to Soil-Applied PPO-Inhibiting Herbicides
3.2. Analysis of the TSM Profile and Copy Number in Survivors from Soil-Applied Fomesafen
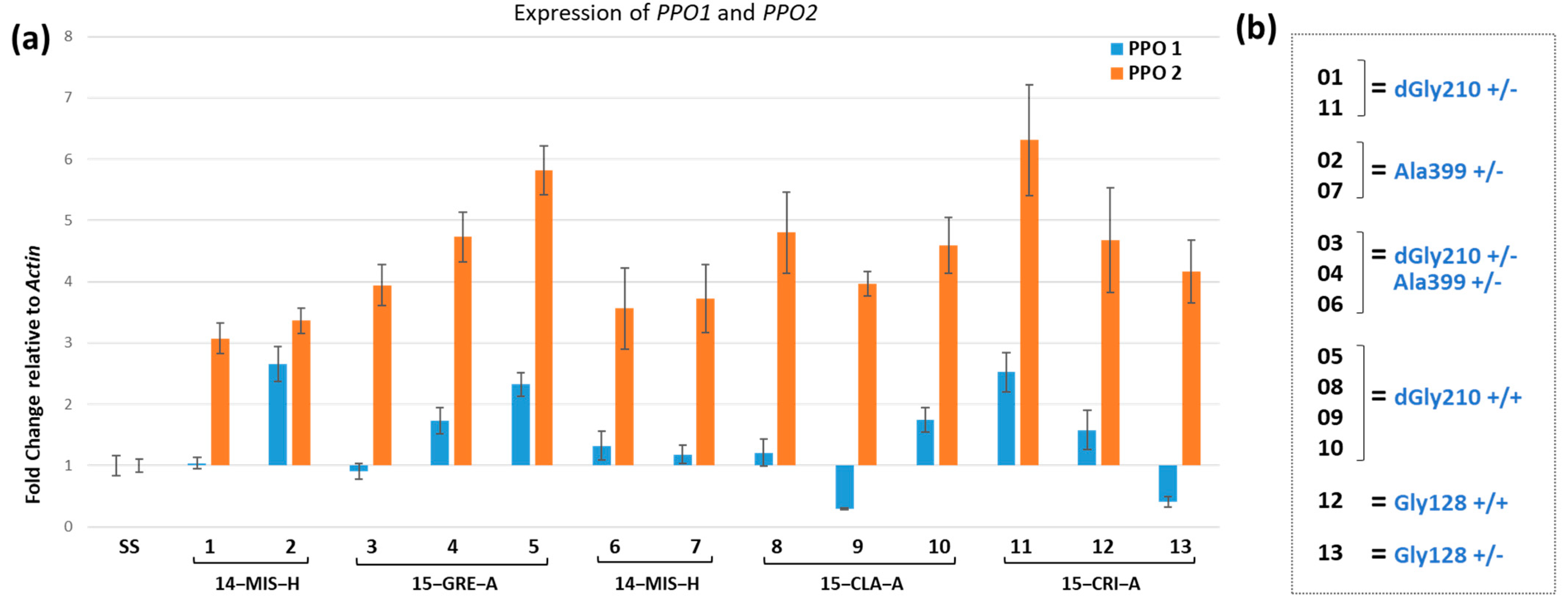
3.3. Analysis of Expression and Mutation Profile of the Target-Site in Survivors from Soil-Applied Fomesafen
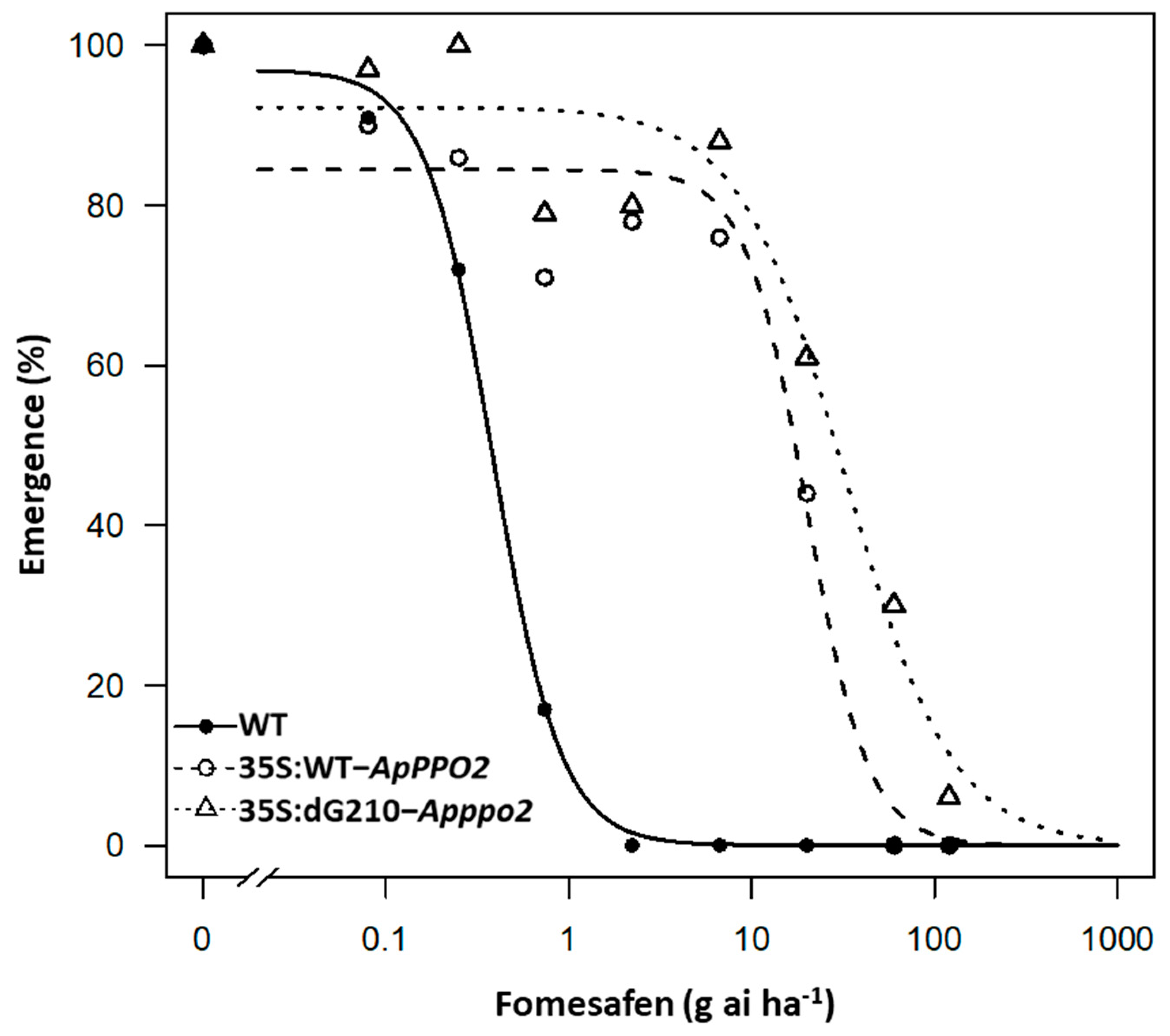
3.4. Evaluation of the Level of Resistance to Fomesafen in Transgenic Arabidopsis Lines Overexpressing A. palmeri PPO2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Heap, I. The International Herbicide-Resistant Weed Database. Available online: www.weedscience.org (accessed on 9 November 2022).
- Carvalho-Moore, P.; Norsworthy, J.K.; González-Torralva, F.; Hwang, J.-I.; Patel, J.D.; Barber, L.T.; Butts, T.R.; McElroy, J.S. Unraveling the Mechanism of Resistance in a Glufosinate-Resistant Palmer Amaranth (Amaranthus palmeri) Accession. Weed Sci. 2022, 70, 370–379. [Google Scholar] [CrossRef]
- Noguera, M.M.; Porri, A.; Werle, I.S.; Heiser, J.; Brändle, F.; Lerchl, J.; Murphy, B.; Betz, M.; Gatzmann, F.; Penkert, M.; et al. Involvement of Glutamine Synthetase 2 (GS2) Amplification and Overexpression in Amaranthus Palmeri Resistance to Glufosinate. Planta 2022, 256, 57. [Google Scholar] [CrossRef]
- Shyam, C.; Borgato, E.A.; Peterson, D.E.; Dille, J.A.; Jugulam, M. Predominance of Metabolic Resistance in a Six-Way-Resistant Palmer Amaranth (Amaranthus Palmeri) Population. Front. Plant Sci. 2021, 11, 614618. [Google Scholar] [CrossRef]
- Hao, G.-F.; Zuo, Y.; Yang, S.-G.; Yang, G.-F. Protoporphyrinogen Oxidase Inhibitor: An Ideal Target for Herbicide Discovery. Chimia 2011, 65, 961. [Google Scholar] [CrossRef] [PubMed]
- Sherman, T.D.; Becerril, J.M.; Matsumoto, H.; Duke, M.V.; Jacobs, J.M.; Jacobs, N.J.; Duke, S.O. Physiological Basis for Differential Sensitivities of Plant Species to Protoporphyrinogen Oxidase-Inhibiting Herbicides 1. Plant Physiol. 1991, 97, 280–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lermontova, I.; Kruse, E.; Mock, H.-P.; Grimm, B. Cloning and Characterization of a Plastidal and a Mitochondrial Isoform of Tobacco Protoporphyrinogen IX Oxidase. Proc. Natl. Acad. Sci. USA 1997, 94, 8895–8900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patzoldt, W.L.; Hager, A.G.; McCormick, J.S.; Tranel, P.J. A Codon Deletion Confers Resistance to Herbicides Inhibiting Protoporphyrinogen Oxidase. Proc. Natl. Acad. Sci. USA 2006, 103, 12329–12334. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, N.; Che, F.-S.; Iwano, M.; Takayama, S.; Yoshida, S.; Isogai, A. Dual Targeting of Spinach Protoporphyrinogen Oxidase II to Mitochondria and Chloroplasts by Alternative Use of Two In-Frame Initiation Codons *. J. Biol. Chem. 2001, 276, 20474–20481. [Google Scholar] [CrossRef] [Green Version]
- Becerril, J.M.; Duke, S.O. Protoporphyrin IX Content Correlates with Activity of Photobleaching Herbicides. Plant Physiol. 1989, 90, 1175–1181. [Google Scholar] [CrossRef] [Green Version]
- Jacobs, J.M.; Jacobs, N.J.; Sherman, T.D.; Duke, S.O. Effect of Diphenyl Ether Herbicides on Oxidation of Protoporphyrinogen to Protoporphyrin in Organellar and Plasma Membrane Enriched Fractions of Barley 1. Plant Physiol. 1991, 97, 197–203. [Google Scholar] [CrossRef]
- Lee, H.J.; Duke, M.V.; Duke, S.O. Cellular Localization of Protoporphyrinogen-Oxidizing Activities of Etiolated Barley (Hordeum vulgare L.) Leaves (Relationship to Mechanism of Action of Protoporphyrinogen Oxidase-Inhibiting Herbicides). Plant Physiol. 1993, 102, 881–889. [Google Scholar] [CrossRef] [Green Version]
- Adhikari, P.; Goodrich, E.; Fernandes, S.B.; Lipka, A.E.; Tranel, P.; Brown, P.; Jamann, T.M. Genetic Variation Associated with PPO-Inhibiting Herbicide Tolerance in Sorghum. PLoS ONE 2020, 15, e0233254. [Google Scholar] [CrossRef]
- Saballos, A.; Soler-Garzón, A.; Brooks, M.; Hart, J.P.; Lipka, A.E.; Miklas, P.; Peachey, R.E.; Tranel, P.J.; Williams, M.M. Multiple Genomic Regions Govern Tolerance to Sulfentrazone in Snap Bean (Phaseolus vulgaris L.). Front. Agron. 2022, 4, 869770. [Google Scholar] [CrossRef]
- Giacomini, D.A.; Umphres, A.M.; Nie, H.; Mueller, T.C.; Steckel, L.E.; Young, B.G.; Scott, R.C.; Tranel, P.J. Two New PPX2 Mutations Associated with Resistance to PPO-Inhibiting Herbicides in Amaranthus Palmeri. Pest Manag. Sci. 2017, 73, 1559–1563. [Google Scholar] [CrossRef] [PubMed]
- Rangani, G.; Salas-Perez, R.A.; Aponte, R.A.; Knapp, M.; Craig, I.R.; Mietzner, T.; Langaro, A.C.; Noguera, M.M.; Porri, A.; Roma-Burgos, N. A Novel Single-Site Mutation in the Catalytic Domain of Protoporphyrinogen Oxidase IX (PPO) Confers Resistance to PPO-Inhibiting Herbicides. Front Plant Sci. 2019, 10, 568. [Google Scholar] [CrossRef]
- Bi, B.; Wang, Q.; Coleman, J.J.; Porri, A.; Peppers, J.M.; Patel, J.D.; Betz, M.; Lerchl, J.; McElroy, J.S. A Novel Mutation A212T in Chloroplast Protoporphyrinogen Oxidase (PPO1) Confers Resistance to PPO Inhibitor Oxadiazon in Eleusine indica. Pest Manag. Sci. 2020, 76, 1786–1794. [Google Scholar] [CrossRef] [PubMed]
- Salas, R.A.; Burgos, N.R.; Tranel, P.J.; Singh, S.; Glasgow, L.; Scott, R.C.; Nichols, R.L. Resistance to PPO-Inhibiting Herbicide in Palmer Amaranth from Arkansas. Pest Manag. Sci. 2016, 72, 864–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noguera, M.M.; Rangani, G.; Heiser, J.; Bararpour, T.; Steckel, L.E.; Betz, M.; Porri, A.; Lerchl, J.; Zimmermann, S.; Nichols, R.L.; et al. Functional PPO2 Mutations: Co-Occurrence in One Plant or the Same Ppo2 Allele of Herbicide-Resistant Amaranthus Palmeri in the US Mid-South. Pest Manag. Sci. 2021, 77, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
- Wuerffel, R.J.; Young, J.M.; Matthews, J.L.; Young, B.G. Characterization of PPO-Inhibitor–Resistant Waterhemp (Amaranthus Tuberculatus) Response to Soil-Applied PPO-Inhibiting Herbicides. Weed Sci. 2015, 63, 511–521. [Google Scholar] [CrossRef]
- Houston, M.M.; Norsworthy, J.K.; Barber, T.; Brabham, C. Field Evaluation of Preemergence and Postemergence Herbicides for Control of Protoporphyrinogen Oxidase-Resistant Palmer Amaranth (Amaranthus Palmeri S. Watson). Weed Technol. 2019, 33, 610–615. [Google Scholar] [CrossRef] [Green Version]
- Doyle, J.J.; Doyle, J.L. A Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Porri, A.; Noguera, M.M.; Betz, M.; Sälinger, D.; Brändle, F.; Bowe, S.J.; Lerchl, J.; Meyer, L.; Knapp, M.; Roma-Burgos, N. Can Double PPO Mutations Exist in the Same Allele and Are Such Mutants Functional? Pest Manag. Sci. 2022, 78, 2258–2264. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Singh, V.; Lawton-Rauh, A.; Bagavathiannan, M.V.; Roma-Burgos, N. EPSPS Gene Amplification Primarily Confers Glyphosate Resistance among Arkansas Palmer Amaranth (Amaranthus Palmeri) Populations. Weed Sci. 2018, 66, 293–300. [Google Scholar] [CrossRef]
- Carvalho-Moore, P.; Rangani, G.; Langaro, A.C.; Srivastava, V.; Porri, A.; Bowe, S.J.; Lerchl, J.; Roma-Burgos, N. Field-Evolved ΔG210-Ppo2 from Palmer Amaranth Confers Pre-Emergence Tolerance to PPO-Inhibitors in Rice and Arabidopsis. Genes 2022, 13, 1044. [Google Scholar] [CrossRef]
- R Core Team. A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2019. [Google Scholar]
- Ritz, C.; Baty, F.; Streibig, J.C.; Gerhard, D. Dose-Response Analysis Using R. PLoS ONE 2015, 10, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salas-Perez, R.A.; Burgos, N.R.; Rangani, G.; Singh, S.; Refatti, J.P.; Piveta, L.; Tranel, P.J.; Mauromoustakos, A.; Scott, R.C. Frequency of Gly-210 Deletion Mutation among Protoporphyrinogen Oxidase Inhibitor–Resistant Palmer Amaranth (Amaranthus Palmeri) Populations. Weed Sci. 2017, 65, 718–731. [Google Scholar] [CrossRef]
- Falk, J.S.; Shoup, D.E.; Al-Khatib, K.; Peterson, D.E. Protox-Resistant Common Waterhemp (Amaranthus Rudis) Response to Herbicides Applied at Different Growth Stages. Weed Sci. 2006, 54, 793–799. [Google Scholar] [CrossRef]
- Shoup, D.E.; Al-Khatib, K.; Peterson, D.E. Common Waterhemp (Amaranthus Rudis) Resistance to Protoporphyrinogen Oxidase-Inhibiting Herbicides. Weed Sci. 2003, 51, 145–150. [Google Scholar] [CrossRef]
- Dayan, F.E.; Barker, A.; Tranel, P.J. Origins and Structure of Chloroplastic and Mitochondrial Plant Protoporphyrinogen Oxidases: Implications for the Evolution of Herbicide Resistance. Pest Manag. Sci. 2018, 74, 2226–2234. [Google Scholar] [CrossRef]
- Bailly, C. Active Oxygen Species and Antioxidants in Seed Biology. Seed Sci. Res. 2004, 14, 93–107. [Google Scholar] [CrossRef]
- Bailly, C.; El-Maarouf-Bouteau, H.; Corbineau, F. From Intracellular Signaling Networks to Cell Death: The Dual Role of Reactive Oxygen Species in Seed Physiology. Comptes Rendus Biol. 2008, 331, 806–814. [Google Scholar] [CrossRef]
- Oracz, K.; El-Maarouf Bouteau, H.; Farrant, J.M.; Cooper, K.; Belghazi, M.; Job, C.; Job, D.; Corbineau, F.; Bailly, C. ROS Production and Protein Oxidation as a Novel Mechanism for Seed Dormancy Alleviation. Plant J. 2007, 50, 452–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailly, C. The Signalling Role of ROS in the Regulation of Seed Germination and Dormancy. Biochem. J. 2019, 476, 3019–3032. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R. Oxidative Stress, Antioxidants and Stress Tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Corbineau, F.; Gay-Mathieu, C.; Vinel, D.; Côme, D. Decrease in Sunflower (Helianthus annuus) Seed Viability Caused by High Temperature as Related to Energy Metabolism, Membrane Damage and Lipid Composition. Physiol. Plant. 2002, 116, 489–496. [Google Scholar] [CrossRef]
- Macías, F.A.; Varela, R.M.; Torres, A.; Galindo, J.L.G.; Molinillo, J.M.G. Allelochemicals from Sunflowers: Chemistry, Bioactivity and Applications. In Chemical Ecology of Plants: Allelopathy in Aquatic and Terrestrial Ecosystems; Mallik, A.U., Inderjit, Eds.; Birkhäuser: Basel, Switzerland, 2002; pp. 73–87. ISBN 978-3-0348-8109-8. [Google Scholar]
- Oracz, K.; Bailly, C.; Gniazdowska, A.; Côme, D.; Corbineau, F.; Bogatek, R. Induction of Oxidative Stress by Sunflower Phytotoxins in Germinating Mustard Seeds. J. Chem. Ecol. 2007, 33, 251–264. [Google Scholar] [CrossRef] [Green Version]
- Gaines, T.A.; Zhang, W.; Wang, D.; Bukun, B.; Chisholm, S.T.; Shaner, D.L.; Nissen, S.J.; Patzoldt, W.L.; Tranel, P.J.; Culpepper, A.S.; et al. Gene Amplification Confers Glyphosate Resistance in Amaranthus palmeri. Proc. Natl. Acad. Sci. USA 2010, 107, 1029–1034. [Google Scholar] [CrossRef] [Green Version]
| Population a | Seedling Emergence without Herbicide Treatment | Seedling Emergence Reduction b | ||||
|---|---|---|---|---|---|---|
| Flum | Fom | Saf | Sulf | Oxy | ||
| plants 380 cm−2 | -----------------------------%---------------------- | |||||
| 14-CLA-D | 53 | 100 | 100 | 100 | 100 | 96 |
| 14-CRI-C | 44 | 100 | 100 | 100 | 100 | 99 |
| 14-CRI-G | 40 | 100 | 99 | 98 | 98 | 96 |
| 14-MIS-E | 50 | 100 | 99 | 98 | 99 | 99 |
| 14-MIS-H | 39 | 91 | 74 | 96 | 96 | 77 |
| 15-CLA-A | 64 | 98 | 63 | 97 | 91 | 77 |
| 15-CLA-B | 60 | 100 | 100 | 100 | 100 | 100 |
| 15-CON-A | 50 | 100 | 99 | 100 | 100 | 100 |
| 15-CRI-A | 62 | 100 | 89 | 98 | 98 | 87 |
| 15-CRI-B | 50 | 100 | 99 | 100 | 100 | 99 |
| 15-CRI-C | 58 | 100 | 93 | 100 | 98 | 94 |
| 15-CRI-D | 57 | 97 | 92 | 100 | 86 | 96 |
| 15-GRE-A | 68 | 97 | 83 | 100 | 95 | 68 |
| 15-IND-A | 38 | 100 | 98 | 100 | 100 | 98 |
| 15-LAW-A | 68 | 100 | 100 | 100 | 100 | 100 |
| 15-LAW-B | 46 | 100 | 100 | 100 | 100 | 100 |
| 15-LAW-C | 52 | 99 | 98 | 100 | 100 | 100 |
| 15-LEE-A | 35 | 100 | 100 | 100 | 99 | 97 |
| 15-LEE-B | 75 | 100 | 100 | 100 | 100 | 100 |
| 15-MIS-A | 55 | 100 | 100 | 100 | 100 | 100 |
| 15-MIS-B | 48 | 99 | 99 | 100 | 100 | 100 |
| 15-MIS-C | 60 | 100 | 94 | 100 | 100 | 92 |
| 15-MIS-D | 65 | 97 | 89 | 100 | 100 | 92 |
| 15-MIS-E | 47 | 97 | 90 | 100 | 99 | 88 |
| 15-MIS-F | 49 | 98 | 95 | 99 | 100 | 94 |
| 15-PHI-A | 58 | 99 | 95 | 99 | 100 | 81 |
| 15-PRA-A | 43 | 100 | 100 | 100 | 100 | 96 |
| SS c | 120 | 100 | 100 | 100 | 100 | 100 |
| LSD0.05 d | NS | 9 | NS | 5 | 7 | |
| Population | Number of Plants Genotyped | Genotype of Each Mutation | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| dGly210 | Gly128 | Ala399 | dGly210 Gly128 | dGly210 Ala399 | Gly128 Ala399 | |||||
| +/+ | +/− | +/+ | +/− | +/+ | +/− | + −/− + | + −/− + | + −/− + | ||
| 15-GRE-A | 17 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 0 |
| 14-MIS-H | 13 | 0 | 2 | 0 | 0 | 1 | 0 | 8 | 1 | 1 |
| 15-CLA-A | 12 | 9 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| 15-CRI-A | 14 | 0 | 0 | 0 | 2 | 3 | 5 | 2 | 0 | 2 |
| 15-MIS-D | 11 | 0 | 1 | 0 | 3 | 1 | 3 | 0 | 2 | 1 |
| 15-MIS-E | 14 | 2 | 3 | 1 | 2 | 1 | 2 | 2 | 0 | 1 |
| SS | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| SUM | 81 | 20 | 8 | 1 | 7 | 6 | 10 | 13 | 11 | 5 |
| 52 | 29 | |||||||||
| Population | B 1 (±SE) | D 2 (±SE) | LD50 (g ai/ha) | 95% CI 3 (g ai/ha) | RI 4 | p-Value 5 Compared to WT | p-Value 6 Compare between PPO2 | |
|---|---|---|---|---|---|---|---|---|
| Upper | Lower | |||||||
| WT | 2.3 (0.7) | 97.0 (6.1) | 0.4 | 0.3 | 0.5 | - | - | - |
| 35S:WT-ApPPO2 | 2.6 (1.3) | 84.5 (84.5) | 20.0 | 13.6 | 26.3 | 51.8 | 3.0 × 10−4 | - |
| 35S:dG210-Apppo2 | 1.5 (0.4) | 92.3 (3.6) | 32.4 | 19.4 | 45.4 | 84.0 | 8.9 × 10−4 | 0.134 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rangani, G.; Porri, A.; Salas-Perez, R.A.; Lerchl, J.; Karaikal, S.K.; Velásquez, J.C.; Roma-Burgos, N. Assessment of Efficacy and Mechanism of Resistance to Soil-Applied PPO Inhibitors in Amaranthus palmeri. Agronomy 2023, 13, 592. https://doi.org/10.3390/agronomy13020592
Rangani G, Porri A, Salas-Perez RA, Lerchl J, Karaikal SK, Velásquez JC, Roma-Burgos N. Assessment of Efficacy and Mechanism of Resistance to Soil-Applied PPO Inhibitors in Amaranthus palmeri. Agronomy. 2023; 13(2):592. https://doi.org/10.3390/agronomy13020592
Chicago/Turabian StyleRangani, Gulab, Aimone Porri, Reiofeli A. Salas-Perez, Jens Lerchl, Srikanth Kumar Karaikal, Juan Camilo Velásquez, and Nilda Roma-Burgos. 2023. "Assessment of Efficacy and Mechanism of Resistance to Soil-Applied PPO Inhibitors in Amaranthus palmeri" Agronomy 13, no. 2: 592. https://doi.org/10.3390/agronomy13020592





