Bean Common Mosaic Disease: Etiology, Resistance Resource, and Future Prospects
Abstract
1. Introduction
2. Biological Characteristics of BCMV
2.1. Genome Structure
2.2. Transmission
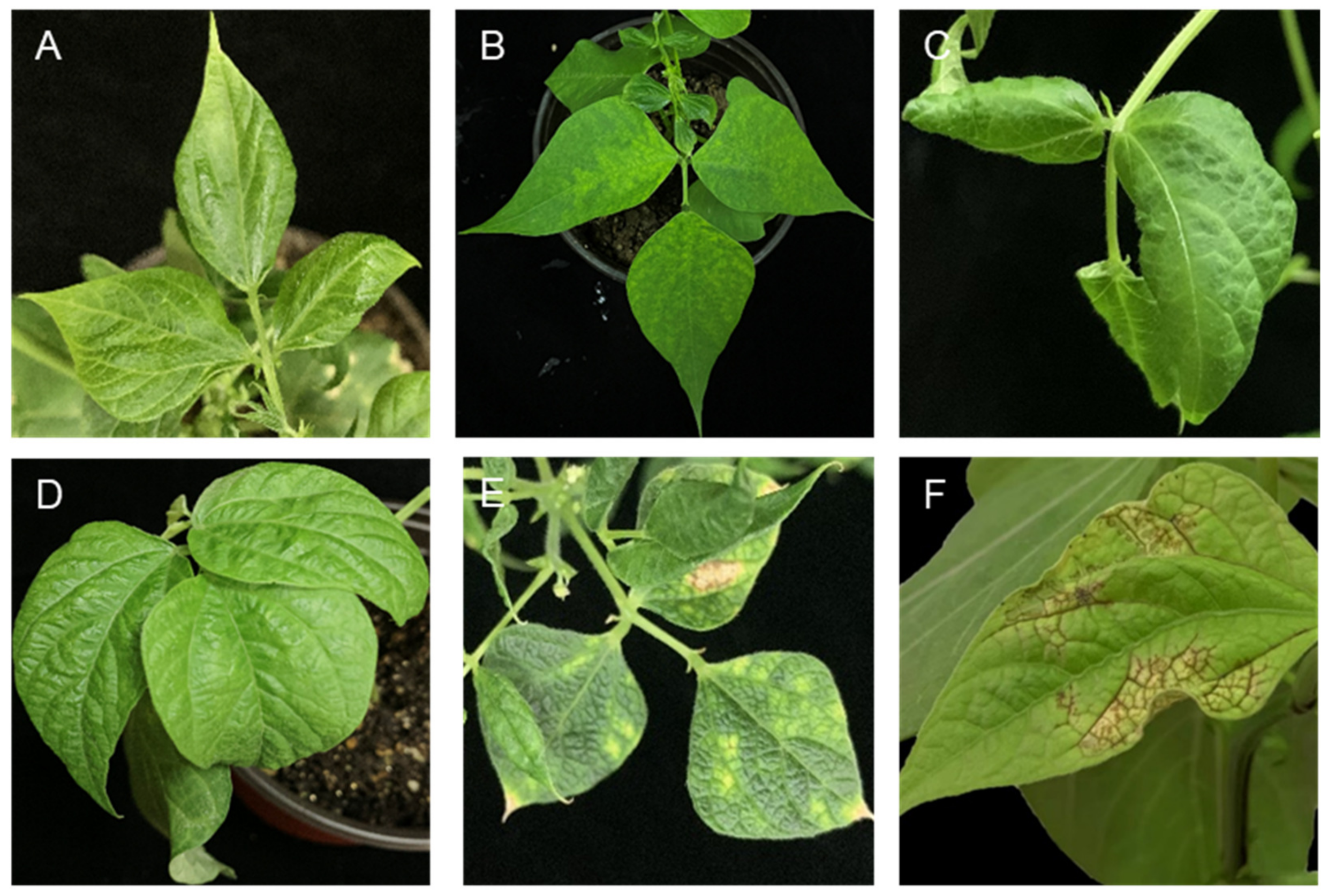
2.3. Host Range and Symptoms
3. Diversity and Evolution of BCMV and BCMNV
3.1. Strain Composition
3.2. Evolution
4. Resistance Resources
4.1. Dominate Resistance
4.2. Recessive Resistance
5. Future Prospects
| BCMV Resistance Alleles | ID Marker | Sequence | Reference | |
|---|---|---|---|---|
| I | SW-13 | F | CACAGCGACATTAATTTTCCTTTC | [176] |
| R | CACAGCGACAGGAGGAGCTTATTA | |||
| S02_48908259 | Fa | gcgggcCAAAGTGCTAGAGGCATGATCA | [19] | |
| R | TGGTTATCATTCATTGTGAAGTCAATG | |||
| Fb | gcgggcagggcggcAATCTTATGCTTGAAGCAGTGAAAGC | |||
| BCMV_48289723_ CAPS | F | AGGAGGAAGAACGGTGGTC | [15] | |
| R | TTTGGTGGTAATTTGAAAATGG | |||
| OW13690 | RAPD marker | CACAGCGACA | [177] | |
| bc-1 | SBD-5 | F | GTGCGGAGAGGCCATCCATTGGTG | [134] |
| R | GTGCGGAGAGTTTCAGTGTTGACA | |||
| S03_4203361 | Fa | gcgggcTGGTCAGTTTGTCTTCCCTAACT | [19] | |
| R | TGCAGAAGAGCTCAACTCGAAG | |||
| Fb | gcgggcagggcggcGGTCAGTTTGTCTTCCCTAACA | |||
| bc-2 | Pvvps4_del | Fa | AGACCGTTTGCTAGGTTCACAA | [18] |
| R | TGTAGGCAATAAGGCGACGTTT | |||
| Fb | AAATTATAAACATGTGTTGGCGAGC | |||
| Pvmit-2_C_del | Fa | gcgggcagggccATTTCTGCGTGATTGCCTCT | [18] | |
| R | CTTCAAAACGCACCTCAAGTATGA | |||
| Fb | gcgggcTCTGCGTGATTGCCTCC | |||
| bc-3 | ROC11 | F | CCAATTCTCTTTCACTTGTAACC | [177] |
| R | GCATGTTCCAGCAAACC | |||
| eIF4E | F | ACCGATGAGCAAAACCCTA | [17] | |
| R | CAACCAACTGGTATCGGATT | |||
| OAD19690 | RAPD marker | CTTGGCACGA | [178] | |
| OS13660 | RAPD marker | GTCGGTTCCTG | [178] | |
| PveIF4E1,3,4_ PveIF4E2 | Fa | gcgggcCAATCTTATGCTTGAAGCAGTGAAAGT | [19] | |
| R | ATTTACAATAACATTCACCACCCGAGCAA | |||
| Fb | gcgggcagggcggcAATCTTATGCTTGAAGCAGTGAAAGC | |||
| bc-4 | Pvmit1_T_G | Fa | gcgggcCGAAAGCGTTCCCTCTCTACAT | [18] |
| R | GCGTGATGGCTTCCTTGATCTT | |||
| Fb | gcgggcagggcggcCGAAAGCGTTCCCTCTCTACAG | |||
| bc-ud | Pvbzip1_A_C | Fa | gcgggcTAGGAGAACTTGGTTTGTCTGAGTA | [19] |
| R | GCACTCCATAAGGGATGTGGT | |||
| Fb | gcgggcagggcggcGGAGAACTTGGTTTGTCTGAGTC |
| BCMV Resistance Alleles | Location | Candidate Genes | Reference |
|---|---|---|---|
| I | Pv02: 48,908,259 | Phvul.002G323000~Phvul.002G323,500 Phvul.002G323800 | [15,21] |
| bc-1 | Pv03: 4,203,361 | Side-by-side receptor-like protein kinases (RLKs), Phvul.003G038700, Phvul.003G038800 | [19] |
| bc-2 | Pv11: 9,272,542 -9,262,459 | Phus.011G092700 | [18] |
| bc-3 | Pv06: 27,204,768 | Phvul.006G168400 | [176] |
| bc-4 | Pv05: 36,225,550 | Phus.005G125100 | [18] |
| bc-ud | Pv05: 36,114,516 | Phvul.005G124100 | [19] |
| Bean Varieties | Resistance Genes | PG-I | PG-II | PG-III | PG-IV | PG-V | PG-VI | PG-VII | PG-VIII |
|---|---|---|---|---|---|---|---|---|---|
| DW | / | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ |
| Poncho (DDP041) | bc-ud | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ |
| RGLC | bc-1 | -/- | +/+ | -/- | +/+ | +/+ | +/+ | +/+ | -/- |
| RGLB | bc-1, bc-? | -/- | -/- | -/- | +/+ | -/- | +/+ | +/+ | -/- |
| Sanilac | bc-2, bc-4 | -/- | -/- | +/+ | -/- | +/+ | +/+ | -/- | +/+ |
| UI35 | bc-ud, bc-1, bc-2 | -/- | -/- | -/- | -/- | -/- | -/- | +/+ | -/- |
| IVT7214 | bc-3, bc-4, bc-2? | -/- | -/- | -/- | -/- | -/- | -/- | -/- | +/+ |
| Jubila | I, bc-1 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- |
| Amanda | I, bc-1, bc-? | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- |
| US1006 | I, bc-ud, bc-2 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- |
| IVT7233 | I, bc-ud, bc-2 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- |
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Graham, P.; Ranalli, P. Common bean (Phaseolus vulgaris L.). Field Crops Res. 1997, 53, 131–146. [Google Scholar] [CrossRef]
- Karavidas, I.; Ntatsi, G.; Vougeleka, V.; Karkanis, A.; Ntanasi, T.; Saitanis, C.; Agathokleous, E.; Ropokis, A.; Sabatino, L.; Tran, F. Agronomic Practices to Increase the Yield and Quality of Common Bean (Phaseolus vulgaris L.): A Systematic Review. Agronomy 2022, 12, 271. [Google Scholar] [CrossRef]
- Singh, S.P. Broadening the genetic base of common bean cultivars: A review. Crop Sci. 2001, 41, 1659–1675. [Google Scholar] [CrossRef]
- Drijfhout, E.; Morales, F. Bean common mosaic. In Compendium of Bean Diseases, 2nd ed.; Schwartz, H.F., Steadman, J.R., Hall, R., Forster, R.L., Eds.; American Phytopathological Society: Saint Paul, MN, USA, 2005; pp. 60–62. [Google Scholar]
- Arli-Sokmen, M.; Deligoz, I.; Kutluk-Yilmaz, N.D. Characterization of Bean common mosaic virus and Bean common mosaic necrosis virus isolates in common bean growing areas in Turkey. Eur. J. Plant Pathol. 2016, 146, 1–16. [Google Scholar] [CrossRef]
- Chiquito-Almanza, E.; Acosta-Gallegos, J.A.; Garcia-Alvarez, N.C.; Garrido-Ramirez, E.R.; Montero-Tavera, V.; Guevara-Olvera, L.; Anaya-Lopez, J.L. Simultaneous Detection of Both RNA and DNA Viruses Infecting Dry Bean and Occurrence of Mixed Infections by BGYMV, BCMV and BCMNV in the Central-West Region of Mexico. Viruses 2017, 9, 63. [Google Scholar] [CrossRef]
- Mwaipopo, B.; Nchimbi-Msolla, S.; Njau, P.J.R.; Mark, D.; Mbanzibwa, D.R. Comprehensive Surveys of Bean common mosaic virus and Bean common mosaic necrosis virus and Molecular Evidence for Occurrence of Other Phaseolus vulgaris Viruses in Tanzania. Plant Dis. 2018, 102, 2361–2370. [Google Scholar] [CrossRef]
- Ac, U.; Nayaka, S.C.; Niranjana, S.R.; Prkash, H.S.; Mortensen, C.N. Bean Common Mosaic of French Bean; Danish Seed Health Centre for Developing Countries: Copenhagen, Denmark, 2020. [Google Scholar]
- Han, T.; Yang, C.X.; Jing-Jing, F.U.; Liao, Y.M.; Shuang-Yu, L.; Jia, R.; Shi, C.Y. Identification and Sequence Analysis of Bean common mosaic virus in Liaoning. Acta Agric. Univ. Jiangxiensis 2018, 40, 378–382. [Google Scholar]
- Worrall, E.A.; Wamonje, F.O.; Mukeshimana, G.; Harvey, J.J.; Carr, J.P.; Mitter, N. Bean common mosaic virus and Bean common mosaic necrosis virus: Relationships, biology, and prospects for control. Adv. Virus Res. 2015, 93, 1–46. [Google Scholar]
- Kabeja, A. Gene Ecology of the Climbing Common Bean (Phaseolus vulgaris)-Bean Common Mosaic Virus/Bean Common Mosaic Necrosis Virus (BCMV/BCMNV) Relationship in Rwanda: A Key for the Development of Virus-Resistant Beans. Ph.D. Thesis, University of California, Davis, CA, USA, 2020. [Google Scholar]
- Valli, A.; García, J.A.; López-Moya, J.J. Potyviridae. In eLS; John Wiley & Sons: Chichester, UK, 2015; pp. 1–10. [Google Scholar] [CrossRef]
- Shukla, D.D.; Ward, C.W.; Brunt, A.A. The Potyviridae; CAB International: Wallingford, UK, 1994. [Google Scholar]
- Naderpour, M.; Johansen, I.E. Visualization of resistance responses in Phaseolus vulgaris using reporter tagged clones of Bean common mosaic virus. Virus Res. 2011, 159, 1–8. [Google Scholar] [CrossRef]
- Bello, M.H.; Moghaddam, S.M.; Massoudi, M.; Mcclean, P.E.; Cregan, P.B.; Miklas, P.N. Application of in silico bulked segregant analysis for rapid development of markers linked to Bean common mosaic virus resistance in common bean. BMC Genom. 2014, 15, 903. [Google Scholar] [CrossRef]
- Meziadi, C.; Richard, M.M.S.; Derquennes, A.; Thareau, V.; Blanchet, S.; Gratias, A.; Pflieger, S.; Geffroy, V. Development of molecular markers linked to disease resistance genes in common bean based on whole genome sequence. Plant Sci. 2016, 242, 351–357. [Google Scholar] [CrossRef]
- Schmutz, J.; McClean, P.E.; Mamidi, S.; Wu, G.A.; Cannon, S.B.; Grimwood, J.; Jenkins, J.; Shu, S.; Song, Q.; Chavarro, C. A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 2014, 46, 707–713. [Google Scholar] [CrossRef]
- Soler-Garzon, A.; McClean, P.E.; Miklas, P.N. Coding Mutations in Vacuolar Protein-Sorting 4 AAA+ ATPase Endosomal Sorting Complexes Required for Transport Protein Homologs Underlie bc-2 and New bc-4 Gene Conferring Resistance to Bean Common Mosaic Virus in Common Bean. Front. Plant Sci. 2021, 12, 769247. [Google Scholar] [CrossRef]
- Soler-Garzon, A.; McClean, P.E.; Miklas, P.N. Genome-Wide Association Mapping of bc-1 and bc-u Reveals Candidate Genes and New Adjustments to the Host-Pathogen Interaction for Resistance to Bean Common Mosaic Necrosis Virus in Common Bean. Front. Plant Sci. 2021, 12, 699569. [Google Scholar] [CrossRef]
- Strausbaugh, C.; Myers, J.; Forster, R.; McClean, P. bc-1 and bc-u—Two loci controlling bean common mosaic virus resistance in common bean are linked. J. Am. Soc. Hortic. Sci. 1999, 124, 644–648. [Google Scholar] [CrossRef]
- Vallejos, C.E.; Astua-Monge, G.; Jones, V.; Plyler, T.R.; Sakiyama, N.S.; Mackenzie, S.A. Genetic and molecular characterization of the I locus of Phaseolus vulgaris. Genetics 2006, 172, 1229–1242. [Google Scholar] [CrossRef]
- Larsen, R.C.; Miklas, P.N.; Druffel, K.L.; Wyatt, S.D. NL-3 K Strain Is a Stable and Naturally Occurring Interspecific Recombinant Derived from Bean common mosaic necrosis virus and Bean common mosaic virus. Phytopathology 2005, 95, 1037–1042. [Google Scholar] [CrossRef]
- Naderpour, M.; Lund, O.; Johansen, I. Sequence analysis of expressed cDNA of Bean common mosaic virus RU1 isolate. Iran. J. Virol. 2009, 3, 39–41. [Google Scholar] [CrossRef]
- Feng, X.; Poplawsky, A.R.; Karasev, A.V. A Recombinant of Bean common mosaic virus Induces Temperature-Insensitive Necrosis in an I Gene-Bearing Line of Common Bean. Phytopathology 2014, 104, 1251–1257. [Google Scholar] [CrossRef]
- Ivanov, K.I.; Eskelin, K.; Lohmus, A.; Makinen, K. Molecular and cellular mechanisms underlying potyvirus infection. J. Gen. Virol. 2014, 95, 1415–1429. [Google Scholar] [CrossRef]
- Hull, R. Plant Virology; Academic Press: Cambridge, MA, USA, 2013. [Google Scholar]
- Elsharkawy, M.M.; Shimizu, M.; Takahashi, H.; Ozaki, K.; Hyakumachi, M. Induction of systemic resistance against Cucumber mosaic virus in Arabidopsis thaliana by Trichoderma asperellum SKT-1. Plant Pathol. J. 2013, 29, 193. [Google Scholar] [CrossRef] [PubMed]
- Wylie, S.J.; Adams, M.; Chalam, C.; Kreuze, J.; Lopez-Moya, J.J.; Ohshima, K.; Praveen, S.; Rabenstein, F.; Stenger, D.; Wang, A.; et al. ICTV Virus Taxonomy Profile: Potyviridae. J. Gen. Virol. 2017, 98, 352–354. [Google Scholar] [CrossRef] [PubMed]
- Revers, F.; Garcia, J.A. Molecular biology of potyviruses. Adv. Virus Res. 2015, 92, 101–199. [Google Scholar] [CrossRef]
- Urcuqui-Inchima, S.; Haenni, A.L.; Bernardi, F. Potyvirus proteins: A wealth of functions. Virus Res. 2001, 74, 157–175. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.W.; Miller, W.A.; Atkins, J.F.; Firth, A.E. An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. USA 2008, 105, 5897–5902. [Google Scholar] [CrossRef] [PubMed]
- Olspert, A.; Chung, Y.W.; Atkins, J.F.; Carr, J.P.; Firth, A.E. Transcriptional slippage in the positive-sense RNA virus family Potyviridae. EMBO Rep. 2015, 16, 995–1004. [Google Scholar] [CrossRef]
- Rodamilans, B.; Valli, A.; Mingot, A.; San Leon, D.; Baulcombe, D.; Lopez-Moya, J.J.; Garcia, J.A. RNA polymerase slippage as a mechanism for the production of frameshift gene products in plant viruses of the potyviridae family. J. Virol. 2015, 89, 6965–6967. [Google Scholar] [CrossRef]
- Chai, M.; Wu, X.; Liu, J.; Fang, Y.; Luan, Y.; Cui, X.; Zhou, X.; Wang, A.; Cheng, X. P3N-PIPO Interacts with P3 via the Shared N-Terminal Domain to Recruit Viral Replication Vesicles for Cell-to-Cell Movement. J. Virol. 2020, 94, e01898-19. [Google Scholar] [CrossRef]
- Revers, F.; García, J.A. Molecular biology of potyviruses. In Advances in Virus Research; Karl, M., Thomas, C.M., Murphy, F.A., Eds.; Academic Press: Waltham, MA, USA, 2015; Volume 92, pp. 101–199. [Google Scholar]
- Mäkinen, K.; Paulin, L.; Saarma, M. The nucleotide sequence of potato virus A genomic RNA and its sequence similarities with other potyviruses. J. Gen. Virol. 1994, 75, 457–461. [Google Scholar]
- Kekarainen, T.; Merits, A.; Oruetxebarria, I.; Rajamäki, M.-L.; Valkonen, J. Comparison of the complete sequences of five different isolates of Potato virus A (PVA), genus Potyvirus. Arch. Virol. 1999, 144, 2355–2366. [Google Scholar] [CrossRef]
- Shi, Y.; Chen, J.; Hong, X.; Chen, J.; Adams, M.J. A potyvirus P1 protein interacts with the Rieske Fe/S protein of its host. Mol. Plant Pathol. 2007, 8, 785–790. [Google Scholar] [CrossRef]
- Li, M.-J.; Kim, J.-K.; Seo, E.-Y.; Hong, S.M.; Hwang, E.-I.; Moon, J.-K.; Domier, L.L.; Hammond, J.; Youn, Y.-N.; Lim, H.-S. Sequence variability in the HC-Pro coding regions of Korean soybean mosaic virus isolates is associated with differences in RNA silencing suppression. Arch. Virol. 2014, 159, 1373–1383. [Google Scholar] [CrossRef]
- Lim, H.-S.; Ko, T.-S.; Lambert, K.N.; Kim, H.-G.; Korban, S.S.; Hartman, G.L.; Domier, L.L. Soybean mosaic virus helper component-protease enhances somatic embryo production and stabilizes transgene expression in soybean. Plant Physiol. Biochem. 2005, 43, 1014–1021. [Google Scholar] [CrossRef]
- Lim, H.-S.; Jang, C.-Y.; Bae, H.-H.; Kim, J.-K.; Lee, C.-H.; Hong, J.-S.; Ju, H.-J.; Kim, H.-G.; Domier, L.L. Soybean mosaic virus infection and helper component-protease enhance accumulation of bean pod mottle virus-specific siRNAs. Plant Pathol. J. 2011, 27, 315–323. [Google Scholar] [CrossRef][Green Version]
- Jossey, S.; Hobbs, H.A.; Domier, L.L. Role of Soybean mosaic virus–encoded proteins in seed and aphid transmission in soybean. Phytopathology 2013, 103, 941–948. [Google Scholar] [CrossRef]
- Senda, M.; Masuta, C.; Ohnishi, S.; Goto, K.; Kasai, A.; Sano, T.; Hong, J.-S.; MacFarlane, S. Patterning of virus-infected Glycine max seed coat is associated with suppression of endogenous silencing of chalcone synthase genes. Plant Cell 2004, 16, 807–818. [Google Scholar] [CrossRef]
- Lu, L.; Wu, G.; Xu, X.; Luan, H.; Zhi, H.; Cui, J.; Cui, X.; Chen, X. Soybean actin-depolymerizing factor 2 interacts with Soybean mosaic virus-encoded P3 protein. Virus Genes 2015, 50, 333–339. [Google Scholar] [CrossRef]
- Ahangaran, A.; Habibi, M.K.; Mohammadi, G.-H.M.; Winter, S.; García-Arenal, F. Analysis of Soybean mosaic virus genetic diversity in Iran allows the characterization of a new mutation resulting in overcoming Rsv4-resistance. J. Gen. Virol. 2013, 94, 2557–2568. [Google Scholar] [CrossRef]
- Chowda-Reddy, R.; Sun, H.; Chen, H.; Poysa, V.; Ling, H.; Gijzen, M.; Wang, A. Mutations in the P3 protein of Soybean mosaic virus G2 isolates determine virulence on Rsv4-genotype soybean. Mol. Plant Microbe Interact. 2011, 24, 37–43. [Google Scholar] [CrossRef]
- Eggenberger, A.; Hajimorad, M.; Hill, J. Gain of virulence on Rsv1-genotype soybean by an avirulent Soybean mosaic virus requires concurrent mutations in both P3 and HC-Pro. Mol. Plant Microbe Interact. 2008, 21, 931–936. [Google Scholar] [CrossRef]
- Hajimorad, M.; Eggenberger, A.; Hill, J. Evolution of Soybean mosaic virus-G7 molecularly cloned genome in Rsv1-genotype soybean results in emergence of a mutant capable of evading Rsv1-mediated recognition. Virology 2003, 314, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Hajimorad, M.; Eggenberger, A.; Hill, J. Loss and gain of elicitor function of Soybean mosaic virus G7 provoking Rsv1-mediated lethal systemic hypersensitive response maps to P3. J. Virol. 2005, 79, 1215–1222. [Google Scholar] [CrossRef] [PubMed]
- Hajimorad, M.; Eggenberger, A.; Hill, J. Strain-specific P3 of Soybean mosaic virus elicits Rsv1-mediated extreme resistance, but absence of P3 elicitor function alone is insufficient for virulence on Rsv1-genotype soybean. Virology 2006, 345, 156–166. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hajimorad, M.; Eggenberger, A.; Hill, J. Adaptation of Soybean mosaic virus avirulent chimeras containing P3 sequences from virulent strains to Rsv1-genotype soybeans is mediated by mutations in HC-Pro. Mol. Plant Microbe Interact. 2008, 21, 937–946. [Google Scholar] [CrossRef] [PubMed]
- Hajimorad, M.; Wen, R.-H.; Eggenberger, A.; Hill, J.; Maroof, M.S. Experimental adaptation of an RNA virus mimics natural evolution. J. Virol. 2011, 85, 2557–2564. [Google Scholar] [CrossRef]
- Khatabi, B.; Fajolu, O.; Wen, R.H.; Hajimorad, M. Evaluation of North American isolates of Soybean mosaic virus for gain of virulence on Rsv-genotype soybeans with special emphasis on resistance-breaking determinants on Rsv4. Mol. Plant Pathol. 2012, 13, 1077–1088. [Google Scholar] [CrossRef]
- Khatabi, B.; Wen, R.H.; Hajimorad, M. Fitness penalty in susceptible host is associated with virulence of Soybean mosaic virus on R sv1-genotype soybean: A consequence of perturbation of HC-P ro and not P 3. Mol. Plant Pathol. 2013, 14, 885–897. [Google Scholar] [CrossRef]
- Lim, H.S.; Ko, T.S.; Hobbs, H.A.; Lambert, K.N.; Yu, J.M.; McCoppin, N.K.; Korban, S.S.; Hartman, G.L.; Domier, L.L. Soybean mosaic virus helper component-protease alters leaf morphology and reduces seed production in transgenic soybean plants. Phytopathology 2007, 97, 366–372. [Google Scholar] [CrossRef]
- Seo, J.-K.; Sohn, S.-H.; Kim, K.-H. A single amino acid change in HC-Pro of soybean mosaic virus alters symptom expression in a soybean cultivar carrying Rsv1 and Rsv3. Arch. Virol. 2011, 156, 135–141. [Google Scholar] [CrossRef]
- Wang, Y.; Hajimorad, M. Gain of virulence by Soybean mosaic virus on Rsv4-genotype soybeans is associated with a relative fitness loss in a susceptible host. Mol. Plant Pathol. 2016, 17, 1154–1159. [Google Scholar] [CrossRef]
- Wang, Y.; Khatabi, B.; Hajimorad, M. Amino acid substitution in P 3 of Soybean mosaic virus to convert avirulence to virulence on R sv4-genotype soybean is influenced by the genetic composition of P 3. Mol. Plant Pathol. 2015, 16, 301–307. [Google Scholar] [CrossRef]
- Wen, R.-H.; Khatabi, B.; Ashfield, T.; Maroof, M.S.; Hajimorad, M. The HC-Pro and P3 cistrons of an avirulent Soybean mosaic virus are recognized by different resistance genes at the complex Rsv1 locus. Mol. Plant Microbe Interact. 2013, 26, 203–215. [Google Scholar] [CrossRef]
- Hong, X.-Y.; Chen, J.; Shi, Y.-H.; Chen, J.-P. The ‘6K1′protein of a strain of Soybean mosaic virus localizes to the cell periphery. Arch. Virol. 2007, 152, 1547–1551. [Google Scholar] [CrossRef]
- Moreno, M.; Bernal, J.J.; Jiménez, I.; Rodrńuez-Cerezo, I. Resistance in plants transformed with the P1 or P3 gene of tobacco vein mottling potyvirus. J. Gen. Virol. 1998, 79, 2819–2827. [Google Scholar] [CrossRef]
- Verchot, J.; Carrington, J.C. Debilitation of plant potyvirus infectivity by P1 proteinase-inactivating mutations and restoration by second-site modifications. J. Virol. 1995, 69, 1582–1590. [Google Scholar] [CrossRef]
- Blanc, S.; López-Moya, J.-J.; Wang, R.; García-Lampasona, S.; Thornbury, D.W.; Pirone, T.P. A specific interaction between coat protein and helper component correlates with aphid transmission of a potyvirus. Virology 1997, 231, 141–147. [Google Scholar] [CrossRef]
- Rojas, M.R.; Zerbini, F.M.; Allison, R.F.; Gilbertson, R.L.; Lucas, W.J. Capsid protein and helper component-proteinase function as potyvirus cell-to-cell movement proteins. Virology 1997, 237, 283–295. [Google Scholar] [CrossRef]
- Jenner, C.E.; Tomimura, K.; Ohshima, K.; Hughes, S.L.; Walsh, J.A. Mutations in Turnip mosaic virus P3 and cylindrical inclusion proteins are separately required to overcome two Brassica napus resistance genes. Virology 2002, 300, 50–59. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rodriguez-Cerezo, E.; Ammar, E.; Pirone, T.; Shaw, J. Association of the non-structural P3 viral protein with cylindrical inclusions in potyvirus-infected cells. J. Gen. Virol. 1993, 74, 1945–1949. [Google Scholar] [CrossRef]
- Merits, A.; Guo, D.; Järvekülg, L.; Saarma, M. Biochemical and genetic evidence for interactions between potato A potyvirus-encoded proteins P1 and P3 and proteins of the putative replication complex. Virology 1999, 263, 15–22. [Google Scholar] [CrossRef]
- Wei, T.; Wang, A. Biogenesis of cytoplasmic membranous vesicles for plant potyvirus replication occurs at endoplasmic reticulum exit sites in a COPI-and COPII-dependent manner. J. Virol. 2008, 82, 12252–12264. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Huang, T.-S.; McNeil, J.; Laliberté, J.-F.; Hong, J.; Nelson, R.S.; Wang, A. Sequential recruitment of the endoplasmic reticulum and chloroplasts for plant potyvirus replication. J. Virol. 2010, 84, 799–809. [Google Scholar] [CrossRef] [PubMed]
- Lellis, A.D.; Kasschau, K.D.; Whitham, S.A.; Carrington, J.C. Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF (iso) 4E during potyvirus infection. Curr. Biol. 2002, 12, 1046–1051. [Google Scholar] [CrossRef]
- El-Sawy, M.A.; Mohamed, H.A.E.; Elsharkawy, M.M. Serological and molecular characterisations of the Egyptian isolate of Bean common mosaic virus. Arch. Phytopathol. Plant Prot. 2014, 47, 1431–1443. [Google Scholar] [CrossRef]
- Pasev, G.; Kostova, D.; Sofkova, S. Identification of Genes for Resistance to Bean Common Mosaic Virus and Bean Common Mosaic Necrosis Virus in Snap Bean (Phaseolus vulgaris L.) Breeding Lines Using Conventional and Molecular Methods. J. Phytopathol. 2013, 162, 19–25. [Google Scholar] [CrossRef]
- Aishwarya, P.; Rangaswamy, K.T.; Basavaraju, S.; Achari, R.; Prameela, H.A. Evaluation of the Seed-borne Nature of Bean Common Mosaic Virus (BCMV) in Cowpea. Int. J. Curr. Microbiol. Appl. Sci. 2020, 9, 239–245. [Google Scholar] [CrossRef]
- Aishwarya, P. Identification of Genotypes and Tagging of Available SSR Markers Linked to BCMV (Bean Common Mosaic Virus) Resistance in Early Segregating Generation of Cowpea [Vigna unguiculata (L.) Walp.] and Evaluation of Nature of Seed Transmission. Ph. D. Thesis, University of Agricultural Sciences, Bangalore, India, 2017. [Google Scholar]
- Su, W. Molecular Identification and Analysis of the Complete Genome Sequences of Bean Common Mosaic Virus from Three Legumes. Master’s Thesis, Shandong Agricultural University, Taian, China, 2013. [Google Scholar]
- Edwardson, J.; Christie, R.; Ko, N. Potyvirus cylindrical inclusions—Subdivision-IV. Phytopathology 1984, 74, 1111–1114. [Google Scholar] [CrossRef]
- Klein, R.; Wyatt, S.; Kaiser, W. Incidence of bean common mosaic virus in USDA Phaseolus germ plasm collection. Plant Dis. 1988, 72, 301–302. [Google Scholar] [CrossRef]
- Kuhn, C.; Nutter, F., Jr.; Padgett, G. Multiple levels of resistance to tobacco etch virus in pepper. Phytopathology 1989, 79, 814–818. [Google Scholar] [CrossRef]
- Kachroo, A.; Wang, X.W.; Valdes-Lopez, O.; Carr, J.P.; Gilligan, C.A. Three Aphid-Transmitted Viruses Encourage Vector Migration from Infected Common Bean (Phaseolus vulgaris) Plants Through a Combination of Volatile and Surface Cues. Front. Plant Sci. 2020, 11, 613772. [Google Scholar]
- Mhlanga, N.M. Effects of Plant Viral Pathogens on Plant-Pollinator Relationships. Ph.D. Thesis, University of Cambridge, Cambridge, UK, 2020. [Google Scholar]
- Westwood, J.H.; Lewsey, M.G.; Murphy, A.M.; Tungadi, T.; Bates, A.; Gilligan, C.A.; Carr, J.P. Interference with jasmonic acid-regulated gene expression is a general property of viral suppressors of RNA silencing but only partly explains virus-induced changes in plant–aphid interactions. J. Gen. Virol. 2014, 95, 733. [Google Scholar] [CrossRef] [PubMed]
- Sade, D.; Shriki, O.; Cuadros-Inostroza, A.; Tohge, T.; Semel, Y.; Haviv, Y.; Willmitzer, L.; Fernie, A.R.; Czosnek, H.; Brotman, Y. Comparative metabolomics and transcriptomics of plant response to Tomato yellow leaf curl virus infection in resistant and susceptible tomato cultivars. Metabolomics 2015, 11, 81–97. [Google Scholar] [CrossRef]
- Moreno-Delafuente, A.; Garzo, E.; Moreno, A.; Fereres, A. A plant virus manipulates the behavior of its whitefly vector to enhance its transmission efficiency and spread. PLoS ONE 2013, 8, e61543. [Google Scholar] [CrossRef]
- Li, Y.; Cui, H.; Cui, X.; Wang, A. The altered photosynthetic machinery during compatible virus infection. Curr. Opin. Virol. 2016, 17, 19–24. [Google Scholar] [CrossRef]
- Hodge, S.; Powell, G. Do plant viruses facilitate their aphid vectors by inducing symptoms that alter behavior and performance? Environ. Entomol. 2008, 37, 1573–1581. [Google Scholar] [CrossRef]
- Isaacs, R.; Willis, M.A.; Byrne, D.N. Modulation of whitefly take-off and flight orientation by wind speed and visual cues. Physiol. Entomol. 1999, 24, 311–318. [Google Scholar] [CrossRef]
- Hajimorad, M.R.; Domier, L.L.; Tolin, S.A.; Whitham, S.A.; Saghai Maroof, M.A. Soybean mosaic virus: A successful potyvirus with a wide distribution but restricted natural host range. Mol. Plant Pathol. 2018, 19, 1563–1579. [Google Scholar] [CrossRef]
- Yan, L.; Xu, Z.; Liao, B. Molecular identification of peanut stripe virus isolated from Sesame (Sesamun indicium L.). Chin. J. Oil Crop Sci. 2009, 31, 4. [Google Scholar]
- Yadav, D.L.; Jaisani, P.; Pandey, R.N.; Chalam, V.C. Detection and molecular characterization of Bean Common mosaic virus in mungbean. Int. J. Chem. Stud. 2021, 9, 2996–3001. [Google Scholar] [CrossRef]
- Smith, M.R.; Rao, I.M. Chapter 12—Common bean. In Crop Physiology Case Histories for Major Crops; Sadras, V.O., Calderini, D.F., Eds.; Academic Press: Waltham, MA, USA, 2021; pp. 384–406. [Google Scholar]
- Cadle-Davidson, M.M.; Jahn, M.M. Resistance conferred against bean common mosaic virus by the incompletely dominant I locus of Phaseolus vulgaris is active at the single cell level. Arch. Virol. 2005, 150, 2601–2608. [Google Scholar] [CrossRef]
- Drijfhout, E. Genetic interaction between Phaseolus vulgaris and bean common mosaic virus with implications for strain identification and breeding for resistance. Verslagen Van Landbouwkundige Onderzoekingen; Centre for Agricultural Publishing and Documentation: Wageningen, The Netherlands, 1978. [Google Scholar]
- Hart, J.P.; Griffiths, P.D. A series of eIF4E alleles at the Bc-3 locus are associated with recessive resistance to Clover yellow vein virus in common bean. Theor. Appl. Genet. 2013, 126, 2849–2863. [Google Scholar] [CrossRef] [PubMed]
- Kelly, J.D.; Afanador, L.; Haley, S.D. Pyramiding genes for resistance to bean common mosaic virus. Euphytica 1995, 82, 207–212. [Google Scholar] [CrossRef]
- Naderpour, M.; Lund, O.S.; Larsen, R.; Johansen, E. Potyviral resistance derived from cultivars of Phaseolus vulgaris carrying bc-3 is associated with the homozygotic presence of a mutated eIF4E allele. Mol. Plant. Pathol. 2010, 11, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Collmer, C.W.; Marston, M.F.; Taylor, J.C.; Jahn, M. The I gene of bean: A dosage-dependent allele conferring extreme resistance, hypersensitive resistance, or spreading vascular necrosis in response to the potyvirus Bean common mosaic virus. Mol. Plant Microbe Interact. 2000, 13, 1266. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.J.; Zerbini, F.M.; French, R.; Rabenstein, F.; Valkonen, J. Family Potyviridae. In Virus Taxonomy: 9th Report of the International Committee on Taxonomy of Viruses; Elsevier: Amsterdam, The Netherlands, 2012. [Google Scholar]
- Singh, S.P.; Schwartz, H.F. Breeding common bean for resistance to diseases: A review. Crop Science 2010, 50, 2199–2223. [Google Scholar] [CrossRef]
- Mangeni, B.C.; Were, H.K.; Ndong’a, M.; Mukoye, B. Incidence and severity of bean common mosaic disease and resistance of popular bean cultivars to the disease in western Kenya. J. Phytopathol. 2020, 168, 501–515. [Google Scholar] [CrossRef]
- Pardina, P.R. Viral diseases in common bean crops in Argentina. Curr. Top. Virol. 2021, 17, 95–110. [Google Scholar]
- Morales, F. Common beans. In Natural Resistance Mechanisms of Plants to Viruses; Loebenstein, G., Carr, J.P., Eds.; Springer: Dordrecht, The Netherlands, 2006. [Google Scholar]
- Feng, X.; Myers, J.R.; Karasev, A.V. Bean common mosaic virus isolate exhibits a novel pathogenicity profile in common bean, overcoming the bc-3 resistance allele coding for the mutated eIF4E translation initiation factor. Phytopathology 2015, 105, 1487–1495. [Google Scholar] [CrossRef]
- Feng, X.; Orellana, G.E.; Myers, J.R.; Karasev, A.V. Recessive Resistance to Bean common mosaic virus Conferred by the bc-1 and bc-2 Genes in Common Bean (Phaseolus vulgaris) Affects Long-Distance Movement of the Virus. Phytopathology 2018, 108, 1011–1018. [Google Scholar] [CrossRef]
- Feng, X.; Orellana, G.E.; Green, J.C.; Melzer, M.J.; Hu, J.S.; Karasev, A.V. A New Strain of Bean Common Mosaic Virus from Lima Bean (Phaseolus lunatus): Biological and Molecular Characterization. Plant Dis. 2019, 103, 1220–1227. [Google Scholar] [CrossRef]
- Feng, X.; Guzmán, P.; Myers, J.R.; Karasev, A.V. Resistance to Bean common mosaic necrosis virus Conferred by the bc-1 Gene Affects Systemic Spread of the Virus in Common Bean. Phytopathology 2017, 107, 893–900. [Google Scholar] [CrossRef]
- McKern, N.; Shukla, D.; Barnett, O.; Vetten, H.; Dijkstra, J.; Whittaker, L.; Ward, C. Coat protein properties suggest that azuki bean mosaic virus, blackeye cowpea mosaic virus, peanut stripe virus, and three isolates from soybean are all strains of the same potyvirus. Intervirology 1992, 33, 121–134. [Google Scholar] [CrossRef]
- Berger, P.; Wyatt, S.; Shiel, P.; Silbernagel, M.; Druffel, K.; Mink, G. Phylogenetic analysis of the Potyviridae with emphasis on legume-infecting potyviruses. Arch. Virol. 1997, 142, 1979–1999. [Google Scholar] [CrossRef]
- Sáiz, M.; Dopazo, J.; Castro, S.; Romero, J. Evolutionary relationships among bean common mosaic virus strains and closely related potyviruses. Virus Res. 1994, 31, 39–48. [Google Scholar] [CrossRef]
- Gibbs, A.; Ohshima, K. Potyviruses and the digital revolution. Annu. Rev. Phytopathol. 2010, 48, 205–223. [Google Scholar] [CrossRef]
- Gibbs, A.J.; Trueman, J.; Gibbs, M.J. The bean common mosaic virus lineage of potyviruses: Where did it arise and when? Arch. Virol. 2008, 153, 2177–2187. [Google Scholar] [CrossRef]
- Gibbs, A.J.; Ohshima, K.; Phillips, M.J.; Gibbs, M.J. The prehistory of potyviruses: Their initial radiation was during the dawn of agriculture. PLoS ONE 2008, 3, e2523. [Google Scholar] [CrossRef]
- Fang, G.W.; Allison, R.F.; Zambolim, E.M.; Maxwell, D.P.; Gilbertson, R.L. The complete nucleotide sequence and genome organization of bean common mosaic virus (NL3 strain). Virus Res. 1995, 39, 13–23. [Google Scholar] [CrossRef]
- Bousalem, M.; Douzery, E.J.; Fargette, D. High genetic diversity, distant phylogenetic relationships and intraspecies recombination events among natural populations of Yam mosaic virus: A contribution to understanding potyvirus evolution. J. Gen. Virol. 2000, 81, 243. [Google Scholar] [CrossRef]
- Cuevas, J.M.; Delaunay, A.; Visser, J.C.; Bellstedt, D.U.; Jacquot, E.; Elena, S.F. Phylogeography and molecular evolution of potato virus Y. PLoS ONE 2012, 7, e37853. [Google Scholar] [CrossRef]
- Moreno, I.M.; Malpica, J.M.; Díaz-Pendón, J.A.; Moriones, E.; Fraile, A.; GarcíA-Arenal, F. Variability and genetic structure of the population of watermelon mosaic virus infecting melon in Spain. Virology 2004, 318, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Ohshima, K.; Tomitaka, Y.; Wood, J.T.; Minematsu, Y.; Kajiyama, H.; Tomimura, K.; Gibbs, A.J. Patterns of recombination in turnip mosaic virus genomic sequences indicate hotspots of recombination. J. Gen. Virol. 2007, 88, 298–315. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.K.; Lee, S.H.; Kim, K.H. Strain-specific cylindrical inclusion protein of soybean mosaic virus elicits extreme resistance and a lethal systemic hypersensitive response in two resistant soybean cultivars. Mol. Plant Microbe Interact. 2009, 22, 1151. [Google Scholar] [CrossRef] [PubMed]
- Abadkhah, M.; Hajizadeh, M.; Koolivand, D. Global population genetic structure of Bean common mosaic virus. Arch. Phytopathol. Plant Prot. 2020, 53, 266–281. [Google Scholar] [CrossRef]
- Karasev, A.V.; Gray, S.M. Continuous and emerging challenges of Potato virus Y in potato. Annu. Rev. Phytopathol. 2013, 51, 571–586. [Google Scholar] [CrossRef]
- Karasev, A.V.; Gray, S.M. Genetic diversity of Potato virus Y complex. Am. J. Potato Res. 2013, 90, 7–13. [Google Scholar] [CrossRef]
- Silbernagel, M.J.; Mink, G.I.; Zhao, R.L.; Zheng, G.Y. Phenotypic recombination between bean common mosaic and bean common mosaic necrosis potyviruses in vivo. Arch. Virol. 2001, 146, 1007–1020. [Google Scholar] [CrossRef]
- Cordero, T.; Mohamed, M.A.; Lopez-Moya, J.-J.; Daròs, J.-A. A recombinant potato virus y infectious clone tagged with the rosea1 visual marker (pvy-ros1) facilitates the analysis of viral infectivity and allows the production of large amounts of anthocyanins in plants. Front. Microbiol. 2017, 8, 611. [Google Scholar] [CrossRef]
- Simon-Loriere, E.; Holmes, E.C. Why do RNA viruses recombine? Nat. Rev. Microbiol. 2011, 9, 617–626. [Google Scholar] [CrossRef]
- Feng, X.; Poplawsky, A.R.; Nikolaeva, O.V.; Myers, J.R.; Karasev, A.V. Recombinants of bean common mosaic virus (BCMV) and genetic determinants of BCMV involved in overcoming resistance in common bean. Phytopathology 2014, 104, 786–793. [Google Scholar] [CrossRef]
- Salvador, B.; Saenz, P.; Yanguez, E.; Quiot, J.B.; Quiot, L.; Delgadillo, M.O.; Garcia, J.A.; Simon-Mateo, C. Host-specific effect of P1 exchange between two potyviruses. Mol. Plant Pathol. 2008, 9, 147–155. [Google Scholar] [CrossRef]
- Mao, C.; Shan, S.; Huang, Y.; Jiang, C.; Zhang, H.; Li, Y.; Chen, J.; Wei, Z.; Sun, Z. The hypervariable N-terminal of soybean mosaic virus P1 protein influences its pathogenicity and host defense responses. Phytopathol. Res. 2022, 4, 10. [Google Scholar] [CrossRef]
- Jiang, H.; Li, K.; Dou, D.; Gai, J. Characterization of a soybean mosaic virus variant causing different diseases in Glycine max and Nicotiana benthamiana. Arch. Virol. 2017, 162, 549–553. [Google Scholar] [CrossRef]
- Ali, M.A. Genetics of resistance to the common bean mosaic virus (bean virus 1) in the bean (Phaseolus vulgaris L.). Phytopathology 1950, 40, 69–79. [Google Scholar]
- Hart, J.P.; Griffiths, P.D. Genotyping-by-Sequencing Enabled Mapping and Marker Development for the By-2 Potyvirus Resistance Allele in Common Bean. Plant Genome 2015, 8, plantgenome2014.09.0058. [Google Scholar] [CrossRef]
- Wu, M.; Wu, W.P.; Liu, C.C.; Liu, Y.N.; Wu, X.Y.; Ma, F.F.; Zhu, A.Q.; Yang, J.Y.; Wang, B.; Chen, J.Q. A bean common mosaic virus (BCMV)-resistance gene is fine-mapped to the same region as Rsv1-h in the soybean cultivar Suweon 97. TAG. Theoretical and applied genetics. Theor. Angew. Genet. 2018, 131, 1851–1860. [Google Scholar] [CrossRef]
- Wu, M.; Liu, Y.N.; Zhang, C.; Liu, X.T.; Liu, C.C.; Guo, R.; Niu, K.X.; Zhu, A.Q.; Yang, J.Y.; Chen, J.Q.; et al. Molecular mapping of the gene(s) conferring resistance to Soybean mosaic virus and Bean common mosaic virus in the soybean cultivar Raiden. TAG. Theoretical and applied genetics. Theor. Angew. Genet. 2019, 132, 3101–3114. [Google Scholar] [CrossRef]
- Liu, X.-T.; Wu, X.-Y.; Wu, W.-P.; Wu, M.; Chen, J.-Q.; Wang, B. A bean common mosaic virus-resistance gene in the soybean variant V94-5152 was mapped to the Rsv4 locus conferring resistance to soybean mosaic virus. Theor. Appl. Genet. 2021, 134, 2367–2377. [Google Scholar] [CrossRef]
- Ishibashi, K.; Saruta, M.; Shimizu, T.; Shu, M.; Anai, T.; Komatsu, K.; Yamada, N.; Katayose, Y.; Ishikawa, M.; Ishimoto, M.; et al. Soybean antiviral immunity conferred by dsRNase targets the viral replication complex. Nat. Commun. 2019, 10, 4033. [Google Scholar] [CrossRef]
- Miklas, P.N.; Larsen, R.C.; Riley, R.; Kelly, J.D. Potential marker-assisted selection for bc-12 resistance to bean common mosaic potyvirus in common bean. Euphytica 2000, 116, 211–219. [Google Scholar] [CrossRef]
- Obita, T.; Saksena, S.; Ghazi-Tabatabai, S.; Gill, D.J.; Perisic, O.; Emr, S.D.; Williams, R.L. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature 2007, 449, 735–739. [Google Scholar] [CrossRef] [PubMed]
- Amano, M.; Mochizuki, A.; Kawagoe, Y.; Iwahori, K.; Niwa, K.; Svoboda, J.; Maeda, T.; Imura, Y. High-resolution mapping of zym, a recessive gene for Zucchini yellow mosaic virus resistance in cucumber. Theor. Appl. Genet. 2013, 126, 2983–2993. [Google Scholar] [CrossRef] [PubMed]
- Barajas, D.; Martín, I.F.d.C.; Pogany, J.; Risco, C.; Nagy, P.D. Noncanonical role for the host Vps4 AAA+ ATPase ESCRT protein in the formation of Tomato bushy stunt virus replicase. PLoS Pathog. 2014, 10, e1004087. [Google Scholar] [CrossRef] [PubMed]
- Carberry, S.E.; Goss, D.J.; Darzynkiewicz, E. A comparison of the binding of methylated cap analogs to wheat germ protein synthesis initiation factors 4F and (iso) 4F. Biochemistry 1991, 30, 1624–1627. [Google Scholar] [CrossRef] [PubMed]
- Leonard, S.; Plante, D.; Wittmann, S.; Daigneault, N.; Fortin, M.G.; Laliberté, J.F. Complex formation between potyvirus VPg and translation eukaryotic initiation factor 4E correlates with virus infectivity. J. Virol. 2000, 74, 7730–7737. [Google Scholar] [CrossRef]
- Beauchemin, C.; Boutet, N.; Laliberte, J.F. Visualization of the interaction between the precursors of VPg, the viral protein linked to the genome of turnip mosaic virus, and the translation eukaryotic initiation factor iso 4E in Planta. J. Virol. 2007, 81, 775–782. [Google Scholar] [CrossRef]
- Leonard, S.; Viel, C.; Beauchemin, C.; Daigneault, N.; Fortin, M.G.; Laliberte, J.-F. Interaction of VPg-Pro of Turnip mosaic virus with the translation initiation factor 4E and the poly (A)-binding protein in planta. J. Gen. Virol. 2004, 85, 1055–1063. [Google Scholar] [CrossRef]
- Schaad, M.C.; Anderberg, R.J.; Carrington, J.C. Strain-specific interaction of the tobacco etch virus NIa protein with the translation initiation factor eIF4E in the yeast two-hybrid system. Virology 2000, 273, 300–306. [Google Scholar] [CrossRef]
- Udvardi, M.K.; Kakar, K.; Wandrey, M.; Montanari, O.; Murray, J.; Andriankaja, A.; Zhang, J.Y.; Benedito, V.; Hofer, J.M.; Chueng, F.; et al. Legume transcription factors: Global regulators of plant development and response to the environment. Plant Physiol. 2007, 144, 538–549. [Google Scholar] [CrossRef]
- Walter, J.; Barra, A.; Charon, J.; Tavert-Roudet, G.; Michon, T. Spectroscopic Investigation of the Kinetic Mechanism Involved in the Association of Potyviral VPg with the Host Plant Translation Initiation Factor eIF4E. Int. J. Mol. Sci. 2020, 21, 5618. [Google Scholar] [CrossRef]
- Saha; Mkinen. Insights into the Functions of eIF4E-Binding Motif of VPg in Potato Virus A Infection. Viruses 2020, 12, 197. [Google Scholar] [CrossRef]
- Wittmann, S.; Chatel, H.; Fortin, M.G.; Laliberté, J.-F. Interaction of the viral protein genome linked of turnip mosaic potyvirus with the translational eukaryotic initiation factor (iso) 4E of Arabidopsis thaliana Using the yeast two-hybrid system. Virology 1997, 234, 84–92. [Google Scholar] [CrossRef]
- Ala-Poikela, M.; Rajamaki, M.L.; Valkonen, J.P.T. A Novel Interaction Network Used by Potyviruses in Virus-Host Interactions at the Protein Level. Viruses 2019, 11, 1158. [Google Scholar] [CrossRef]
- Bastet, A.; Robaglia, C.; Gallois, J.-L. eIF4E resistance: Natural variation should guide gene editing. Trends Plant Sci. 2017, 22, 411–419. [Google Scholar] [CrossRef]
- Coutinho de Oliveira, L.; Volpon, L.; Rahardjo, A.K.; Osborne, M.J.; Culjkovic-Kraljacic, B.; Trahan, C.; Oeffinger, M.; Kwok, B.H.; Borden, K.L. Structural studies of the eIF4E–VPg complex reveal a direct competition for capped RNA: Implications for translation. Proc. Natl. Acad. Sci. USA 2019, 116, 24056–24065. [Google Scholar] [CrossRef]
- Zafirov, D.; Giovinazzo, N.; Bastet, A.; Gallois, J. When a knockout is an Achilles’ heel: Resistance to one potyvirus species triggers hypersusceptibility to another one in Arabidopsis thaliana. Mol. Plant Pathol. 2021, 22, 334–347. [Google Scholar] [CrossRef]
- Bruun-Rasmussen, M.; Moller, I.S.; Tulinius, G.; Hansen, J.; Johansen, I.E. The same allele of translation initiation factor 4E mediates resistance against two Potyvirus spp. in Pisum sativum. Mol. Plant Microbe Interact. 2007, 20, 1075. [Google Scholar] [CrossRef]
- Singh, K.B.; Foley, R.C.; Oñate-Sánchez, L. Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 2002, 5, 430–436. [Google Scholar] [CrossRef]
- Cichy, K.A.; Porch, T.G.; Beaver, J.S.; Cregan, P.; Fourie, D.; Glahn, R.P.; Grusak, M.A.; Kamfwa, K.; Katuuramu, D.N.; McClean, P. A Phaseolus vulgaris diversity panel for Andean bean improvement. Crop Sci. 2015, 55, 2149–2160. [Google Scholar] [CrossRef]
- Kaminaka, H.; Näke, C.; Epple, P.; Dittgen, J.; Schütze, K.; Chaban, C.; Holt III, B.F.; Merkle, T.; Schäfer, E.; Harter, K. bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO J. 2006, 25, 4400–4411. [Google Scholar] [CrossRef]
- Liao, Y.; Zhang, J.S.; Chen, S.Y.; Zhang, W.K. Role of soybean GmbZIP132 under abscisic acid and salt stresses. J. Integr. Plant Biol. 2008, 50, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Alves, M.S.; Dadalto, S.P.; Gonçalves, A.B.; De Souza, G.B.; Barros, V.A.; Fietto, L.G. Plant bZIP transcription factors responsive to pathogens: A review. Int. J. Mol. Sci. 2013, 14, 7815–7828. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Zhang, H.; Guo, C.; Cheng, C.; Guo, R.; Mao, L.; Fei, Z.; Wang, X. Evolutionary and expression analyses of basic zipper transcription factors in the highly homozygous model grape PN40024 (Vitis vinifera L.). Plant Mol. Biol. Report. 2014, 32, 1085–1102. [Google Scholar] [CrossRef]
- Liu, Y.; Schiff, M.; Marathe, R.; Dinesh-Kumar, S. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 2002, 30, 415–429. [Google Scholar] [CrossRef] [PubMed]
- Engalycheva, I.A. Development pecularities of bean common mosaic virus(Potyvirus, Potyviridae) in moscow region and initial material for resistance breeding. Sel’skokhozyaistvennaya Biol. 2020, 55, 901–919. [Google Scholar] [CrossRef]
- Osorno, J.M.; Wal, A.; Posch, J.; Simons, K.; Grafton, K.F.; Pasche, J.S. ‘ND Whitetail’, a new white kidney bean with high seed yield and intermediate resistance to white mold and bacterial blights. J. Plant Regist. 2020, 14, 102–109. [Google Scholar] [CrossRef]
- Parker, T. Development of Phenotyping Methods and Genetic Resources for Common Bean Improvement. Ph.D. Thesis, University of California, Davis, CA, USA, 2020. [Google Scholar]
- Chiquito-Almanza, E.; Caballero-Pérez, J.; Acosta-Gallegos, J.A.; Montero-Tavera, V.; Mariscal-Amaro, L.A.; Anaya-López, J. Diversity and Distribution of Viruses Infecting Wild and Domesticated Phaseolus spp. in the Mesoamerican Center of Domestication. Viruses 2021, 13, 1153. [Google Scholar] [CrossRef]
- Amir, W.; Bhat, M.A.; Mir, A.; Dar, A.; Sofi, P.A. Screening of Genotypes for Identification of Resistant Genotypes for BCMV. Res. J. Agric. Sci. 2017, 8, 320–323. [Google Scholar]
- Mukeshimana, G.; Pañeda, A.; Rodríguez-Suárez, C.; Ferreira, J.J.; Giraldez, R.; Kelly, J.D. Markers linked to the bc-3 gene conditioning resistance to bean common mosaic potyviruses in common bean. Euphytica 2005, 144, 291–299. [Google Scholar] [CrossRef]
- Portilla, A.E.; Mayor-Duran, V.M.; Buendia, H.F.; Blair, M.W.; Cichy, K.; Raatz, B. Climbing bean breeding for disease resistance and grain quality traits. Legume Sci. 2021, 4, e122. [Google Scholar] [CrossRef]
- Urrea, C.A.; Hurtado-Gonzales, O.P.; Pastor-Corrales, M.A.; Steadman, J.R. Registration of Great Northern Common Bean Cultivar ‘Panhandle Pride’ with Enhanced Disease Resistance to Bean Rust and Common Bacterial Blight. J. Plant Regist. 2019, 13, 311–315. [Google Scholar] [CrossRef]
- Manjunatha, N.; Rangaswamy, K.; Sah, R.; Nagaraju, N.; Rudraswamy, P. Characterization and identification of SSR markers for screening of cowpea genotypes against Bean common mosaic virus (BCMV) disease resistance. Legume Res. 2017, 40, 878–883. [Google Scholar] [CrossRef]
- Bruckner, F.P.; Andrade, P.O.; de Souza Cascardo, R.; Laliberté, J.-F.; Alfenas-Zerbini, P. Translationally controlled tumour protein: A protein necessary for potyvirus intracellular multiplication that supports plant infection by unrelated viruses. Ann. Appl. Biol. 2022, 180, 90–98. [Google Scholar] [CrossRef]
- Bruckner, F.P.; da Silva Xavier, A.; de Souza Cascardo, R.; Otoni, W.C.; Zerbini, F.M.; Alfenas-Zerbini, P. Translationally controlled tumor protein (TCTP) from tomato and Nicotiana benthamiana is necessary for successful infection by A Potyvirus. Mol. Plant Pathol. 2017, 18, 672–683. [Google Scholar] [CrossRef]
- Kyrychenko, A. Seed transmission of bean common mosaic virus in Phaseolus vulgaris cv Chervona Shapochka. Biol. Syst. Theory Innov. 2020, 11, 587–597. [Google Scholar] [CrossRef]
- Worrall, E.A.; Bravo-Cazar, A.; Nilon, A.T.; Fletcher, S.J.; Robinson, K.E.; Carr, J.P.; Mitter, N. Exogenous application of RNAi-inducing double-stranded RNA inhibits aphid-mediated transmission of a plant virus. Front. Plant Sci. 2019, 10, 265. [Google Scholar] [CrossRef]
- Megasari, D.; Damayanti, T.A.; Santoso, S. Pengendalian Aphis craccivora Koch. dengan kitosan dan pengaruhnya terhadap penularan Bean common mosaic virus strain Black eye cowpea (BCMV-BlC) pada kacang panjang. J. Entomol. Indones. 2014, 11, 72–80. [Google Scholar] [CrossRef]
- Elsharkawy, M.M.; Shimizu, M.; Takahashi, H.; Hyakumachi, M. Induction of systemic resistance against Cucumber mosaic virus by Penicillium simplicissimum GP17-2 in Arabidopsis and tobacco. Plant Pathol. 2012, 61, 964–976. [Google Scholar] [CrossRef]
- Elsharkawy, M.M.; Shimizu, M.; Takahashi, H. The plant growth-promoting fungus Fusarium equiseti and the arbuscular mycorrhizal fungus Glomus mosseae induce systemic resistance against Cucumber mosaic virus in cucumber plants. Plant Soil 2012, 361, 397–409. [Google Scholar] [CrossRef]
- Roossinck, M.J. Symbiosis versus competition in plant virus evolution. Nat. Rev. Microbiol. 2005, 3, 917–924. [Google Scholar] [CrossRef]
- Melotto, M.; Afanador, L.; Kelly, J. Development of a SCAR marker linked to the I gene in common bean. Genome 1996, 39, 1216–1219. [Google Scholar] [CrossRef] [PubMed]
- Johnson, W.C.; Guzmán, P.; Mandala, D.; Mkandawire, A.B.; Temple, S.; Gilbertson, R.; Gepts, P. Molecular tagging of the bc-3 gene for introgression into Andean common bean. Crop Sci. 1997, 37, 248–254. [Google Scholar] [CrossRef]
- Haley, S.D.; Afanador, L.; Kelly, J.D. Selection for monogenic pest resistance traits with coupling-and repulsion-phase RAPD markers. Crop Sci. 1994, 34, 1061–1066. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, M.; Feng, X. Bean Common Mosaic Disease: Etiology, Resistance Resource, and Future Prospects. Agronomy 2023, 13, 58. https://doi.org/10.3390/agronomy13010058
Tang M, Feng X. Bean Common Mosaic Disease: Etiology, Resistance Resource, and Future Prospects. Agronomy. 2023; 13(1):58. https://doi.org/10.3390/agronomy13010058
Chicago/Turabian StyleTang, Muning, and Xue Feng. 2023. "Bean Common Mosaic Disease: Etiology, Resistance Resource, and Future Prospects" Agronomy 13, no. 1: 58. https://doi.org/10.3390/agronomy13010058
APA StyleTang, M., & Feng, X. (2023). Bean Common Mosaic Disease: Etiology, Resistance Resource, and Future Prospects. Agronomy, 13(1), 58. https://doi.org/10.3390/agronomy13010058






