Characterization of the NGP4A Gene in Regulating Grain Number Per Panicle of Rice (Oryza sativa L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Agronomic Evaluation
2.2. Primer Development and Gene Mapping
2.3. Vector Construction and Plant Transformation
2.4. Transcriptome Sequencing
3. Results
3.1. Characterization of ngp4a Mutant
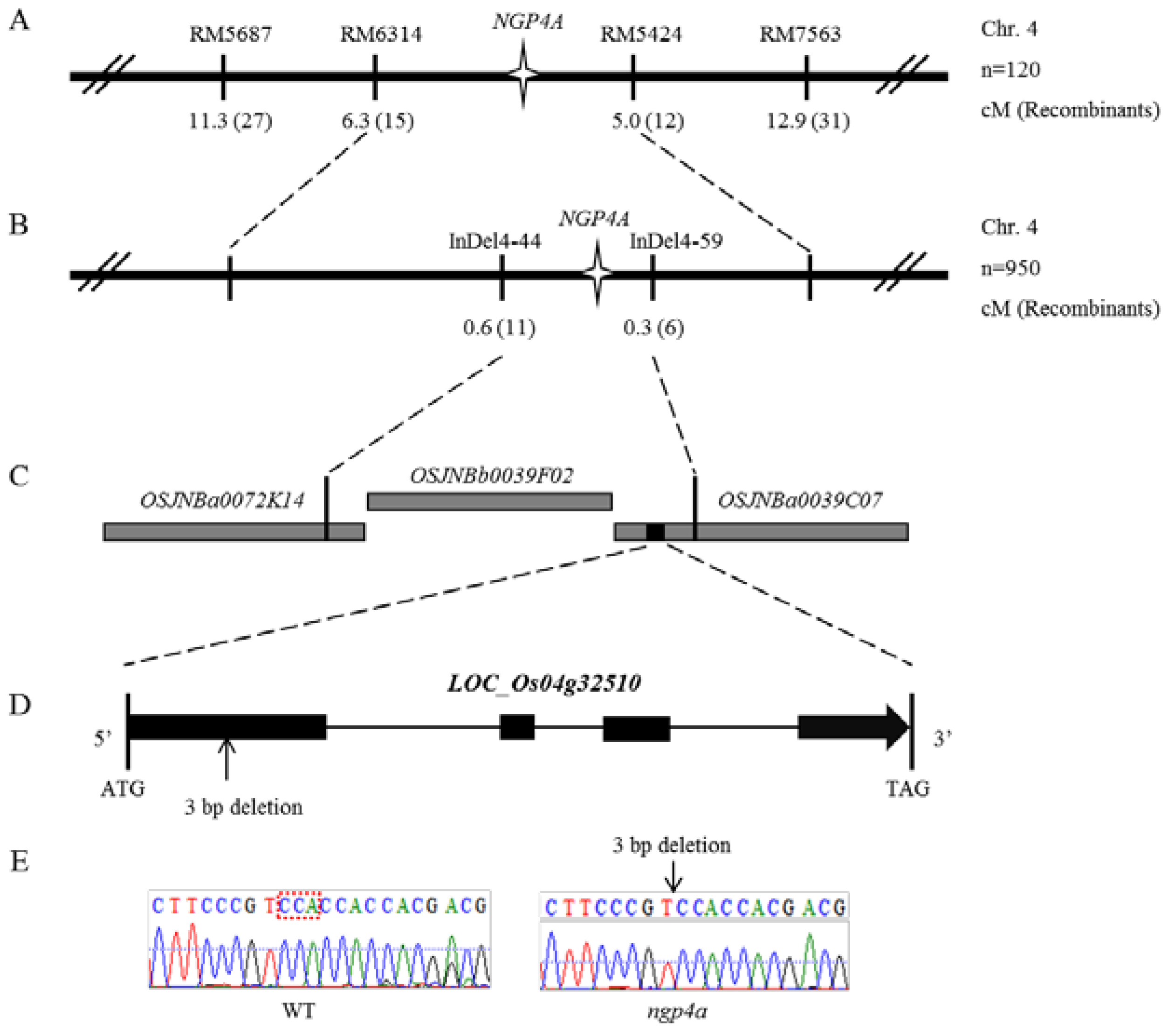
3.2. Fine Mapping of NGP4A
3.3. Sequence and Complementation Analysis
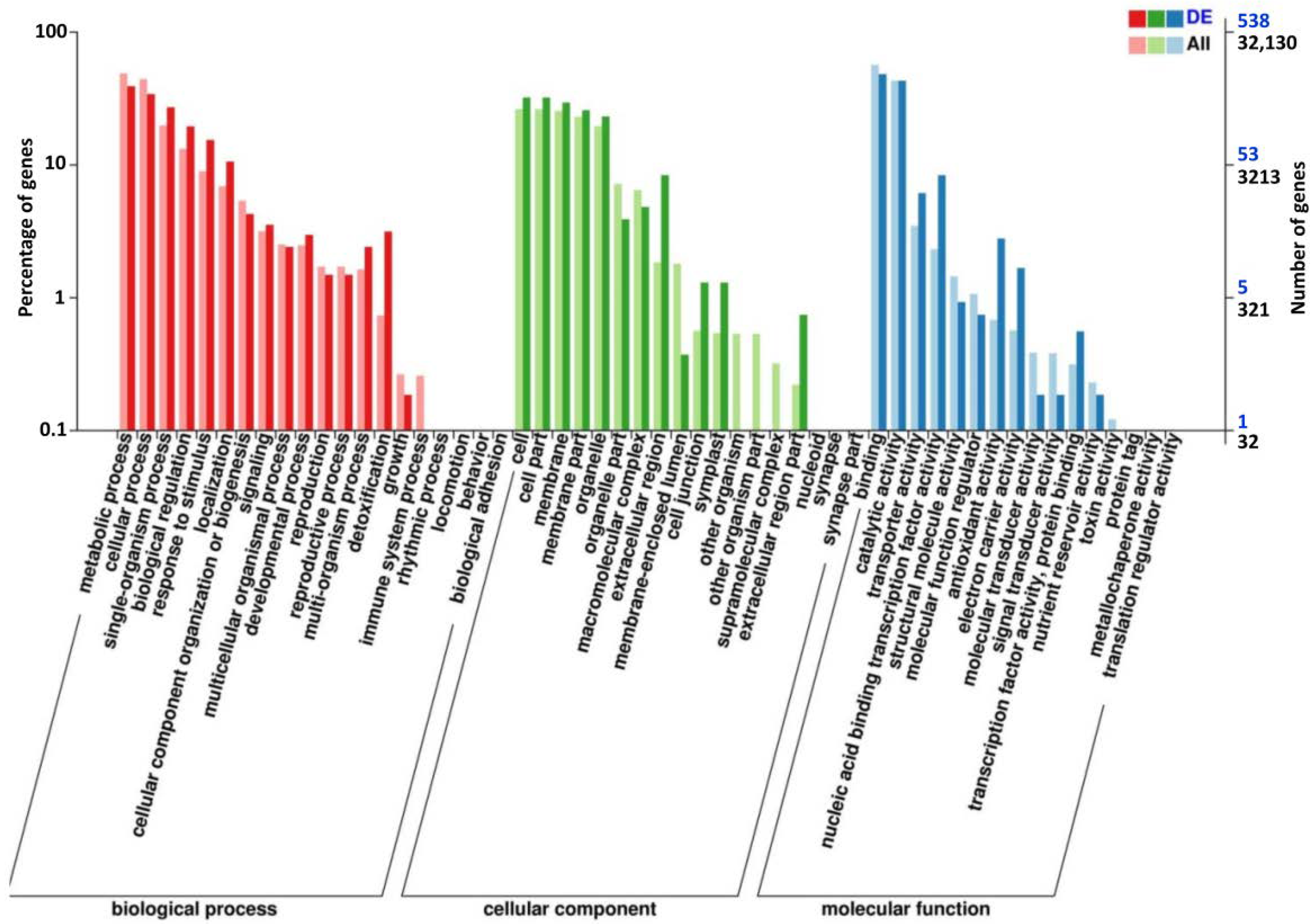
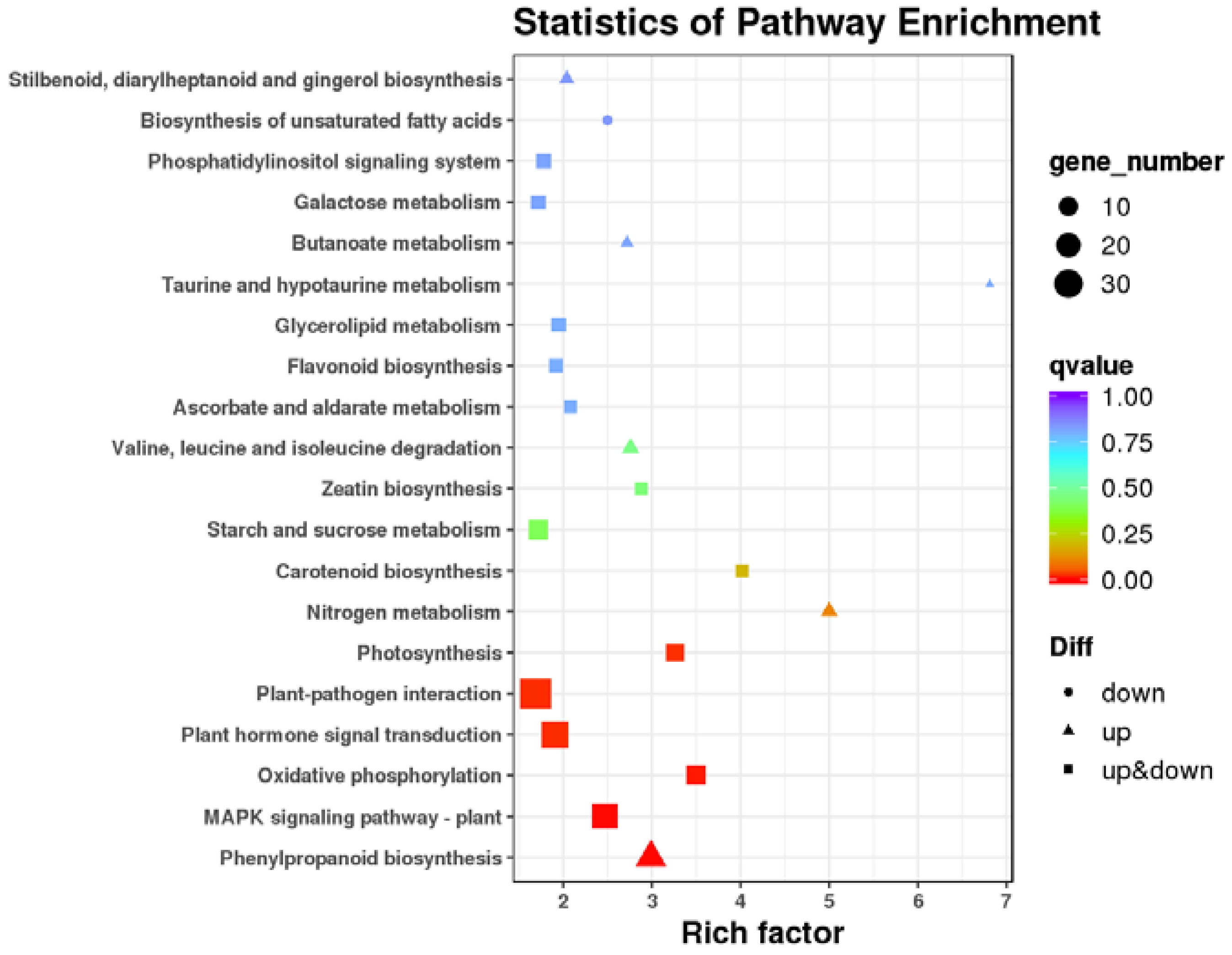
3.4. Transcriptome Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lu, Z.; Guo, X.; Huang, Z.; Xia, J.; Li, X.; Wu, J.; Yu, H.; Shahid, M.Q.; Liu, X. Transcriptome and gene editing analyses reveal MOF1a defect alters the expression of genes associated with tapetum development and chromosome behavior at meiosis stage resulting in low pollen fertility of tetraploid rice. Int. J. Mol. Sci. 2020, 21, 7489. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Xu, K.; Li, Z.; Hu, Y.; Hu, Z.; Zheng, X.; Song, S.; Tang, Z.; Li, L. Genome-wide association study and Mendelian randomization analysis provide insights for improving rice yield potential. Sci. Rep. 2021, 11, 6894. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Feng, F.; Zhang, Y.; Elesawi, I.E.; Xu, K.; Li, T.; Mei, H.; Liu, H.; Gao, N.; Chen, C.; et al. A novel rice grain size gene OsSNB was identified by genome-wide association study in natural population. PLoS Genet. 2019, 15, e1008191. [Google Scholar] [CrossRef]
- Kushibuchi, K. Historical changes in rice cultivars. In Science of the Rice Plant—Volume 3, Genetics; Matsuo, T., Futsuhara, Y., Kikuchi, F., Yamaguchi, H., Eds.; Food and Agriculture Policy Research Center: Tokyo, Japan, 1997; pp. 837–875. [Google Scholar]
- Yuan, L.P. Hybrid rice breeding for super high yield. Hybrid Rice 1997, 12, 1–6. (In Chinese) [Google Scholar]
- Yang, S.R.; Zhang, L.B.; Chen, W.F.; Xu, Z.J.; Wang, J.M. Theories and methods of rice breeding for maximum yield. Acta Agromomic Sin. 1995, 22, 295–304. (In Chinese) [Google Scholar]
- Zhou, K.D.; Wang, X.D.; Li, S.G.; Li, P.; Li, H.Y.; Huang, G.S.; Liu, T.Q.; Shen, M.S. The study on heavy panicle type of inter-subspecific hybrid rice (Oryza sativa L.). Sci. Agric. Sin. 1997, 30, 91–93. (In Chinese) [Google Scholar]
- Gouda, G.; Gupta, M.K.; Donde, R.; Mohapatra, T.; Vadde, R.; Behera, L. Marker-assisted selection for grain number and yield-related traits of rice (Oryza sativa L.). Physiol. Mol. Biol. Plants 2020, 26, 885–898. [Google Scholar] [CrossRef]
- Yin, C.; Zhu, Y.; Li, X.; Lin, Y. Molecular and genetic aspects of grain number determination in rice (Oryza sativa L.). Int. J. Mol. Sci. 2021, 22, 728. [Google Scholar] [CrossRef]
- Xie, J.; Li, F.; Khan, N.U.; Zhu, X.; Wang, X.; Zhang, Z.; Ma, X.; Zhao, Y.; Zhang, Q.; Zhang, S.; et al. Identifying natural genotypes of grain number per panicle in rice (Oryza sativa L.) by association mapping. Genes Genom. 2019, 41, 283–295. [Google Scholar] [CrossRef]
- Singh, V.K.; Ellur, R.K.; Singh, A.K.; Nagarajan, M.; Singh, B.D.; Singh, N.K. Effect of qGN4.1 QTL for grain number per panicle in genetic backgrounds of twelve different mega varieties of rice. Rice 2018, 11, 8. [Google Scholar] [CrossRef] [Green Version]
- Hu, Z.; Cao, L.; Sun, X.; Zhu, Y.; Zhang, T.; Jiang, L.; Liu, Y.; Dong, S.; Sun, D.; Yang, J.; et al. Fine mapping of a major quantitative trait locus, qgnp7(t), controlling grain number per panicle in African rice (Oryza glaberrima S.). Breed. Sci. 2018, 68, 606–613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Tan, L.; Zhu, Z.; Xiao, L.; Xie, D.; Sun, C. PAY1 improves plant architecture and enhances grain yield in rice. Plant J. 2015, 83, 528–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Qian, Q.; Fu, Z.; Wang, Y.; Xiong, G.; Zeng, D.; Wang, X.; Liu, X.; Teng, S.; Hiroshi, F.; et al. Control of tillering in rice. Nature 2003, 422, 618–621. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Li, J.J.; Yao, G.X.; Zhang, H.L.; Dou, H.J.; Shi, H.L.; Sun, X.M.; Li, Z.C. Fine mapping and cloning of the grain number per-panicle gene (Gnp4) on chromosome 4 in rice (Oryza sativa L.). J. Integr. Agric. 2011, 10, 1825–1833. [Google Scholar] [CrossRef]
- Tabuchi, H.; Zhang, Y.; Hattori, S.; Omae, M.; Shimizu-Sato, S.; Oikawa, T.; Qian, Q.; Nishimura, M.; Kitano, H.; Xie, H.; et al. LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell 2011, 23, 3276–3287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komatsu, M.; Maekawa, M.; Shimamoto, K.; Kyozuka, J. The LAX1 and FRIZZY PANICLE 2 genes determine the inflorescence architecture of rice by controlling rachis-branch and spikelet development. Dev. Biol. 2001, 231, 364–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Qian, Q.; Fu, Z.; Zeng, D.; Meng, X.; Kyozuka, J.; Maekawa, M.; Zhu, X.; Zhang, J.; Li, J.; et al. Short panicle1 encodes a putative PTR family transporter and determines rice panicle size. Plant J. 2009, 58, 592–605. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Qian, Q.; Liu, Z.; Sun, H.; He, S.; Luo, D.; Xia, G.; Chu, C.; Li, J.; Fu, X. Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 2009, 41, 494–497. [Google Scholar] [CrossRef]
- Yoshida, A.; Sasao, M.; Yasuno, N.; Takagi, K.; Daimon, Y.; Chen, R.; Yamazaki, R.; Tokunaga, H.; Kitaguchi, Y.; Sato, Y.; et al. TAWAWA1, a regulator of rice inflorescence architecture, functions through the suppression of meristem phase transition. Proc. Natl. Acad. Sci. USA 2013, 110, 767–772. [Google Scholar] [CrossRef] [Green Version]
- Mao, C.; Ding, W.; Wu, Y.; Yu, J.; He, X.; Shou, H.; Wu, P. Overexpression of a NAC-domain protein promotes shoot branching in rice. New Phytol. 2007, 176, 288–298. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, Y.; Mi, X.F.; Shan, J.X.; Li, X.M.; Xu, J.L.; Lin, H.X. The QTL GNP1 encodes GA20ox1, which increases grain number and yield by increasing cytokinin activity in rice panicle meristems. PLoS Genet. 2016, 12, e1006386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Zhao, B.; Yuan, D.; Duan, M.; Qian, Q.; Tang, L.; Wang, B.; Liu, X.; Zhang, J.; Wang, J.; et al. Rice zinc finger protein DST enhances grain production through controlling Gn1a/OsCKX2 expression. Proc. Natl. Acad. Sci. USA 2013, 110, 3167–3172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, L.; Li, X.; Liu, F.; Sun, X.; Li, C.; Zhu, Z.; Fu, Y.; Cai, H.; Wang, X.; Xie, D.; et al. Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 2008, 40, 1360–1364. [Google Scholar] [CrossRef] [PubMed]
- McCouch, S.R.; Teytelman, L.; Xu, Y.; Lobos, K.B.; Clare, K.; Walton, M.; Fu, B.; Maghirang, R.; Li, Z.; Xing, Y.; et al. Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res. 2002, 9, 199–207. [Google Scholar] [CrossRef]
- Liu, X.Q.; Bai, X.Q.; Wang, X.J.; Chu, C.C. OsWRKY71, a rice transcription factor, is involved in rice defense response. J. Plant Physiol. 2007, 164, 969–979. [Google Scholar] [CrossRef]
- Maignan, V.; Géliot, P.; Avice, J.C. Glutacetine® biostimulant applied on wheat under contrasting field conditions improves grain number leading to better yield, upgrades N-related traits and changes grain ionome. Plants 2021, 10, 456. [Google Scholar] [CrossRef]
- Dampanaboina, L.; Jiao, Y.; Chen, J.; Gladman, N.; Chopra, R.; Burow, G.; Hayes, C.; Christensen, S.A.; Burke, J.; Ware, D.; et al. Sorghum MSD3 encodes an ω-3 fatty acid desaturase that increases grain number by reducing jasmonic acid levels. Int. J. Mol. Sci. 2019, 20, 5359. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Tao, Y.; Zhu, J.; Miao, J.; Liu, J.; Liu, Y.; Yi, C.; Yang, Z.; Gong, Z.; Liang, G. GNS4, a novel allele of DWARF11, regulates grain number and grain size in a high-yield rice variety. Rice 2017, 10, 34. [Google Scholar] [CrossRef] [Green Version]
- Hao, J.; Wang, D.; Wu, Y.; Huang, K.; Duan, P.; Li, N.; Xu, R.; Zeng, D.; Dong, G.; Zhang, B.; et al. The GW2-WG1-OsbZIP47 pathway controls grain size and weight in rice. Mol. Plant 2021, 14, 1266–1280. [Google Scholar] [CrossRef]
- Chen, H.; Tang, Y.; Liu, J.; Tan, L.; Jiang, J.; Wang, M.; Zhu, Z.; Sun, X.; Sun, C. Emergence of a novel chimeric gene underlying grain number in rice. Genetics 2017, 205, 993–1002. [Google Scholar] [CrossRef]
- Nguyen, K.L.; Grondin, A.; Courtois, B.; Gantet, P. Next-generation sequencing accelerates crop gene discovery. Trends Plant Sci. 2019, 24, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Dueñas, C.J.; Slamet-Loedin, I.; Macovei, A. Transcriptomics view over the germination landscape in biofortified rice. Genes 2021, 12, 2013. [Google Scholar] [CrossRef] [PubMed]
- dos Santos, A.B.; Bottcher, A.; Kiyota, E.; Mayer, J.L.; Vicentini, R.; Brito Mdos, S.; Creste, S.; Landell, M.G.; Mazzafera, P. Water stress alters lignin content and related gene expression in two sugarcane genotypes. J. Agric. Food Chem. 2015, 63, 4708–4720. [Google Scholar] [CrossRef] [PubMed]
- Ksouri, N.; Jiménez, S.; Wells, C.E.; Contreras-Moreira, B.; Gogorcena, Y. Transcriptional responses in root and leaf of Prunus persica under drought stress using RNA sequencing. Front. Plant Sci. 2016, 7, 1715. [Google Scholar] [CrossRef]
- Li, J.; Fan, F.; Wang, L.; Zhan, Q.; Wu, P.; Du, J.; Yang, X.; Liu, Y. Cloning and expression analysis of cinnamoyl-CoA reductase (CCR) genes in sorghum. PeerJ 2016, 4, e2005. [Google Scholar] [CrossRef]
- Jardim-Messeder, D.; Felix-Cordeiro, T.; Barzilai, L.; de Souza-Vieira, Y.; Galhego, V.; Bastos, G.A.; Valente-Almeida, G.; Aiube, Y.R.A.; Faria-Reis, A.; Corrêa, R.L.; et al. Genome-wide analysis of general phenylpropanoid and monolignol-specific metabolism genes in sugarcane. Funct. Integr. Genom. 2021, 21, 73–99. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, Y.; Naeem, M.; Zhu, M.; Li, J.; Yu, X.; Hu, Z.; Chen, G. An AGAMOUS MADS-box protein, SlMBP3, regulates the speed of placenta liquefaction and controls seed formation in tomato. J. Exp. Bot. 2019, 70, 909–924. [Google Scholar] [CrossRef]
- Zhang, Y.M.; Yu, H.X.; Ye, W.W.; Shan, J.X.; Dong, N.Q.; Guo, T.; Kan, Y.; Xiang, Y.H.; Zhang, H.; Yang, Y.B.; et al. A rice QTL GS3.1 regulates grain size through metabolic-flux distribution between flavonoid and lignin metabolons without affecting stress tolerance. Commun. Biol. 2021, 4, 1171. [Google Scholar] [CrossRef]
- Li, S.; Han, X.; Lu, Z.; Qiu, W.; Yu, M.; Li, H.; He, Z.; Zhuo, R. MAPK cascades and transcriptional factors: Regulation of heavy metal tolerance in plants. Int. J. Mol. Sci. 2022, 23, 4463. [Google Scholar] [CrossRef]
- Wen, X.; Jiao, L.; Tan, H. MAPK/ERK pathway as a central regulator in vertebrate organ regeneration. Int. J. Mol. Sci. 2022, 23, 1464. [Google Scholar] [CrossRef]
- Wang, N.; Liu, Y.; Dong, C.; Zhang, Y.; Bai, S. MdMAPKKK1 regulates apple resistance to Botryosphaeria dothidea by interacting with MdBSK1. Int. J. Mol. Sci. 2022, 23, 4415. [Google Scholar] [CrossRef]
- Ma, H.; Gao, Y.; Wang, Y.; Dai, Y.; Ma, H. Regulatory mechanisms of mitogen-activated protein kinase cascades in plants: More than sequential phosphorylation. Int. J. Mol. Sci. 2022, 23, 3572. [Google Scholar] [CrossRef]
- Guo, T.; Chen, K.; Dong, N.Q.; Shi, C.L.; Ye, W.W.; Gao, J.P.; Shan, J.X.; Lin, H.X. GRAIN SIZE AND NUMBER1 negatively regulates the OsMKKK10-OsMKK4-OsMPK6 cascade to coordinate the trade-off between grain number per panicle and grain size in rice. Plant Cell 2018, 30, 871–888. [Google Scholar] [CrossRef] [Green Version]
- Hong, Y.; Liu, Q.; Cao, Y.; Zhang, Y.; Chen, D.; Lou, X.; Cheng, S.; Cao, L. The OsMPK15 negatively regulates Magnaporthe oryza and Xoo disease resistance via SA and JA signaling pathway in rice. Front. Plant Sci. 2019, 10, 752. [Google Scholar] [CrossRef]
- Nadolska-Orczyk, A.; Rajchel, I.K.; Orczyk, W.; Gasparis, S. Major genes determining yield-related traits in wheat and barley. Theor. Appl. Genet. 2017, 130, 1081–1098. [Google Scholar] [CrossRef] [Green Version]
- Wilkinson, S.; Kudoyarova, G.R.; Veselov, D.S.; Arkhipova, T.N.; Davies, W.J. Plant hormone interactions: Innovative targets for crop breeding and management. J. Exp. Bot. 2012, 63, 3499–3509. [Google Scholar] [CrossRef]
- Deveshwar, P.; Prusty, A.; Sharma, S.; Tyagi, A.K. Phytohormone-mediated molecular mechanisms involving multiple genes and QTL govern grain number in rice. Front. Genet. 2020, 11, 586462. [Google Scholar] [CrossRef]
- Shirley, N.J.; Aubert, M.K.; Wilkinson, L.G.; Bird, D.C.; Lora, J.; Yang, X.; Tucker, M.R. Translating auxin responses into ovules, seeds and yield: Insight from Arabidopsis and the cereals. J. Integr. Plant Biol. 2019, 61, 310–336. [Google Scholar] [CrossRef] [Green Version]
- Tu, B.; Tao, Z.; Wang, S.; Zhou, L.; Zheng, L.; Zhang, C.; Li, X.; Zhang, X.; Yin, J.; Zhu, X.; et al. Loss of Gn1a/OsCKX2 confers heavy-panicle rice with excellent lodging resistance. J. Integr. Plant Biol. 2022, 64, 23–38. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, J.; Peng, C.; Zhu, X.; Yin, J.; Li, W.; He, M.; Wang, J.; Chern, M.; Yuan, C.; et al. Four receptor-like cytoplasmic kinases regulate development and immunity in rice. Plant Cell Environ. 2016, 39, 1381–1392. [Google Scholar] [CrossRef]


| Traits | Wild Type (WT) | ngp4a | Compared with WT (%) |
|---|---|---|---|
| Plant height (cm) | 95.7 ± 1.5 | 84.0 ± 0.8 | −12.2 ** |
| Tiller number | 23.7 ± 1.5 | 16.0 ± 0.5 | −32.5 ** |
| Main panicle length (cm) | 29.3 ± 1.1 | 26.9 ± 1.0 | −8.2 * |
| Primary branches of main panicle | 10.7 ± 0.6 | 10.0 ± 0.3 | −6.5 |
| Secondary branches of main panicle | 36.7 ± 2.1 | 10.0 ± 0.5 | −72.8 ** |
| Total spikelets of main panicle | 166.7 ± 4.7 | 106.3 ± 2.5 | −36.2 ** |
| Seed setting rate (%) | 85.0 ± 1.4 | 88.01 ± 1.8 | +3.5 |
| 1000-grain weight (g) | 22.8 ± 0.4 | 26.09 ± 0.7 | +14.4 * |
| Grain length (mm) | 7.3 ± 0.1 | 8.0 ± 0.1 | +9.6 * |
| Grain width (mm) | 3.8 ± 0.1 | 3.9 ± 0.1 | +2.6 |
| Traits | Wild Type (WT) | ngp4a/ NGP4A | Compared with WT (%) |
|---|---|---|---|
| Plant height (cm) | 95.6 ± 0.8 | 93.0 ± 1.1 | −2.7 |
| Tiller number | 22.7 ± 1.5 | 23.3 ± 0.9 | +2.8 |
| Main panicle length (cm) | 30.0 ± 0.4 | 28.8 ± 0.7 | −4.0 |
| Primary branches of main panicle | 10.7 ± 0.6 | 10.3 ± 0.6 | −3.1 |
| Secondary branches of main panicle | 35.7 ± 1.7 | 33.8 ± 2.5 | −5.2 |
| Total spikelets of main panicle | 172.0 ± 5.3 | 162.7 ± 3.7 | −5.4 |
| Seed setting rate (%) | 85.5 ± 0.7 | 87.2 ± 0.5 | +2.0 |
| 1000-grain weight (g) | 22.8 ± 0.9 | 23.4 ± 0.8 | +2.8 |
| Grain length (mm) | 7.3 ± 0.1 | 7.4 ± 0.1 | +0.5 |
| Grain width (mm) | 3.8 ± 0.1 | 3.9 ± 0.1 | +0.9 |
| Sample | Wild Type (WT) | ngp4a |
|---|---|---|
| Clean reads | 20,593,555 | 20,481,813 |
| GC content | 52.83% | 53.10% |
| Clean bases | 6,150,165,034 | 6,126,925,284 |
| Total mapped reads (%) | 39,836,670 (96.72%) | 39,516,745 (96.47%) |
| Multiple mapped reads (%) | 1,184,323 (2.88%) | 1,130,576 (2.76%) |
| Unique mapped reads (%) | 38,652,348 (93.85%) | 38,386,169 (93.71%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Yang, W.; Zhao, M.; Ding, G.; Zhou, Y.; Xie, J.; Zhang, F. Characterization of the NGP4A Gene in Regulating Grain Number Per Panicle of Rice (Oryza sativa L.). Agronomy 2022, 12, 1549. https://doi.org/10.3390/agronomy12071549
Chen Y, Yang W, Zhao M, Ding G, Zhou Y, Xie J, Zhang F. Characterization of the NGP4A Gene in Regulating Grain Number Per Panicle of Rice (Oryza sativa L.). Agronomy. 2022; 12(7):1549. https://doi.org/10.3390/agronomy12071549
Chicago/Turabian StyleChen, Yanhong, Wanling Yang, Minmin Zhao, Gumu Ding, Yi Zhou, Jiankun Xie, and Fantao Zhang. 2022. "Characterization of the NGP4A Gene in Regulating Grain Number Per Panicle of Rice (Oryza sativa L.)" Agronomy 12, no. 7: 1549. https://doi.org/10.3390/agronomy12071549





