Seasonal Dynamics of Soil Bacterial Community under Long-Term Abandoned Cropland in Boreal Climate
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site and Sampling Workflow
2.2. Labile Carbon and Nitrogen Determination
2.3. DNA and RNA Extraction
2.4. RNA Treatment and Reverse Transcription
2.5. Quantitative PCR Analysis
2.6. Sequencing of 16S rRNA Gene Libraries
2.7. Processing of 16S rRNA Gene Data
2.8. Statistical Analyses
3. Results
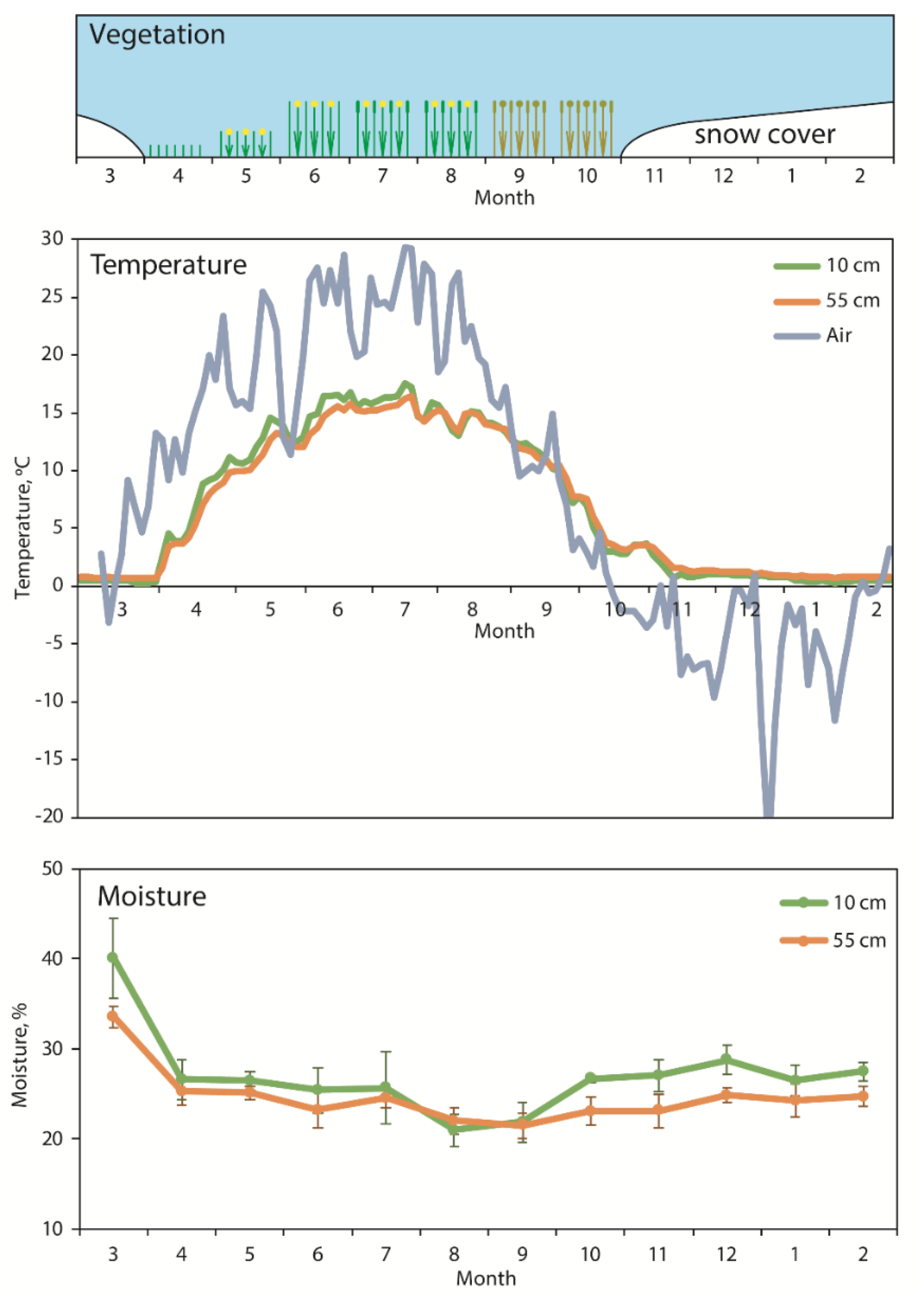
3.1. Dynamics of Soil Moisture and Temperature
3.2. Seasonal Changes in Soil DNA Yields, WEOC and WEON
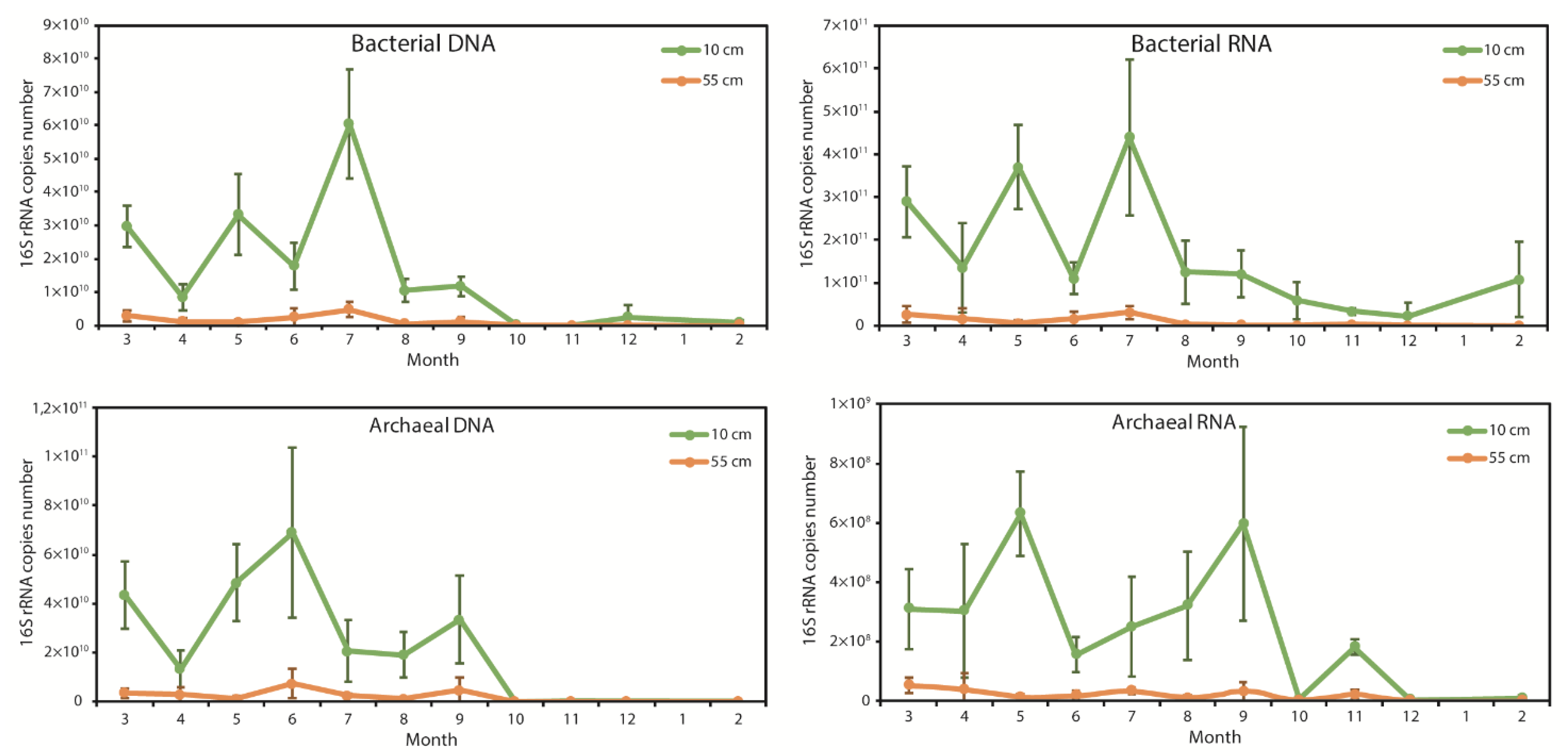
3.3. Seasonal Changes of Bacterial and Archaeal 16S rRNA Gene Copies in Total Soil DNA and RNA
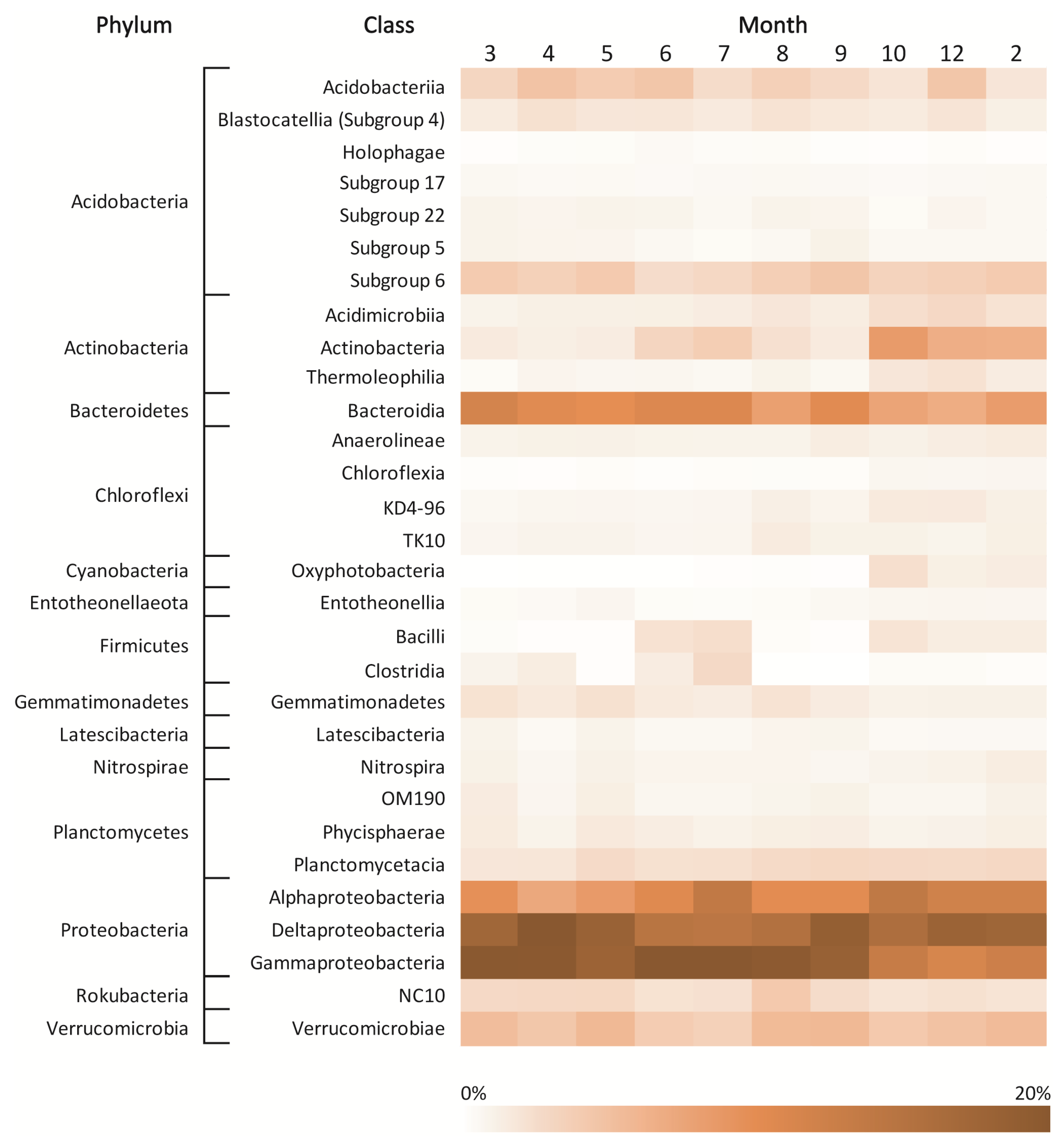
3.4. Taxonomic Structure of Bacterial Communities
3.5. Changes of α- and β-Diversity Indices throughout the Year
4. Discussion
4.1. Physical and Chemical Factors Affecting Dynamics of Soil Microbial Communities
4.2. Seasonal Dynamics of Soil Bacterial Community Structure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Henebry, G.M. Carbon in idle croplands. Nature 2009, 457, 1089–1090. [Google Scholar] [CrossRef] [PubMed]
- Kurganova, I.; Lopes de Gerenyu, V.; Kuzyakov, Y. Large-scale carbon sequestration in post-agrogenic ecosystems in Russia and Kazakhstan. Catena 2015, 133, 461–466. [Google Scholar] [CrossRef]
- Kurganova, I.; Lopes de Gerenyu, V.; Six, J.; Kuzyakov, Y. Carbon cost of collective farming collapse in Russia. Glob. Chang. Biol. 2014, 20, 938–947. [Google Scholar] [CrossRef] [PubMed]
- ROSSTAT. Regions of Russia, Socio-Economic Indicators. Retrieved from Russian Federal Service of State Statistics Website. Available online: www.gks.ru (accessed on 10 January 2022).
- Kalinina, O.; Goryachkin, S.V.; Lyuri, D.I.; Giani, L. Post-agrogenic development of vegetation, soils, and carbon stocks under self-restoration in different climatic zones of European Russia. Catena 2015, 129, 18–29. [Google Scholar] [CrossRef]
- Tardy, V.; Spor, A.; Mathieu, O.; Lévèque, J.; Terrat, S.; Plassart, P.; Regnier, T.; Bardgett, R.D.; van der Putten, W.H.; Roggero, P.P.; et al. Shifts in microbial diversity through land use intensity as drivers of carbon mineralization in soil. Soil Biol. Biochem. 2015, 90, 204–213. [Google Scholar] [CrossRef]
- Semenov, V.M.; Pautova, N.B.; Lebedeva, T.N.; Khromychkina, D.P. Plant Residues Decomposition and Formation of Active Organic Matter in the Soil of the Incubation Experiments. Eurasian Soil Sci. 2019, 52, 1183–1194. [Google Scholar] [CrossRef]
- Liang, C.; Schimel, J.P.; Jastrow, J.D. The importance of anabolism in microbial control over soil carbon storage. Nat. Microbiol. 2017, 2, 17105. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, S.; Semenov, M.V.; Yao, F.; Ye, J.; Bu, R.; Ma, R.; Lin, J.; Kurganova, I.; Wang, X.; et al. Temperature sensitivity of SOM decomposition is linked with a K-selected microbial community. Glob. Chang. Biol. 2021, 27, 2763–2779. [Google Scholar] [CrossRef]
- Waldrop, M.P.; Firestone, M.K. Seasonal dynamics of microbial community composition and function in oak canopy and open grassland soils. Microb. Ecol. 2006, 52, 470–479. [Google Scholar] [CrossRef]
- Lauber, C.L.; Ramirez, K.S.; Aanderud, Z.; Lennon, J.; Fierer, N. Temporal variability in soil microbial communities across land-use types. ISME J. 2013, 7, 1641–1650. [Google Scholar] [CrossRef]
- Lacerda-Júnior, G.V.; Noronha, M.F.; Cabral, L.; Delforno, T.P.; de Sousa, S.T.P.; Fernandes-Júnior, P.I.; Melo, I.S.; Oliveira, V.M. Land Use and Seasonal Effects on the Soil Microbiome of a Brazilian Dry Forest. Front. Microbiol. 2019, 10, 648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuzyakov, Y.; Blagodatskaya, E. Microbial hotspots and hot moments in soil: Concept & review. Soil Biol. Biochem. 2015, 83, 184–199. [Google Scholar] [CrossRef]
- Chapin, F.S.; Matson, P.A.; Vitousek, P.M. Principles of Terrestrial Ecosystem Ecology; Springer: New York, NY, USA, 2011; ISBN 978-1-4419-9503-2. [Google Scholar]
- Medeiros, P.M.; Fernandes, M.F.; Dick, R.P.; Simoneit, B.R.T. Seasonal variations in sugar contents and microbial community in a ryegrass soil. Chemosphere 2006, 65, 832–839. [Google Scholar] [CrossRef]
- Isobe, K.; Oka, H.; Watanabe, T.; Tateno, R.; Urakawa, R.; Liang, C.; Senoo, K.; Shibata, H. High soil microbial activity in the winter season enhances nitrogen cycling in a cool-temperate deciduous forest. Soil Biol. Biochem. 2018, 124, 90–100. [Google Scholar] [CrossRef]
- Xu, Z.; Dai, X.; Chai, X. Effect of different carbon sources on denitrification performance, microbial community structure and denitrification genes. Sci. Total Environ. 2018, 634, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Wang, J.; Wu, N.; Wu, Y.; Shi, F. Seasonal and interannual dynamics of soil microbial biomass and available nitrogen in an alpine meadow in the eastern part of Qinghai-Tibet Plateau, China. Biogeosciences 2018, 15, 567–579. [Google Scholar] [CrossRef] [Green Version]
- Žifčáková, L.; Větrovský, T.; Howe, A.; Baldrian, P. Microbial activity in forest soil reflects the changes in ecosystem properties between summer and winter. Environ. Microbiol. 2016, 18, 288–301. [Google Scholar] [CrossRef] [PubMed]
- IUSS Working Group WRB. World Reference Base for Soil Resources 2014. International Soil Classification System for Naming Soils and Creating Legends for Soil Maps; IUSS Working Group WRB: Rome, Italy, 2014; ISBN 9789251083697. [Google Scholar]
- Ananyeva, N.D.; Susyan, E.A.; Chernova, O.V.; Chernov, I.Y.; Makarova, O.L. The ratio of fungi and bacteria in the biomass of different types of soil determined by selective inhibition. Microbiology 2006, 75, 702–707. [Google Scholar] [CrossRef]
- Ananyeva, N.D.; Susyan, E.A.; Ryzhova, I.M.; Bocharnikova, E.O.; Stolnikova, E.V. Microbial biomass carbon and the microbial carbon dioxide production by soddy-podzolic soils in postagrogenic biogeocenoses and in native spruce forests of the southern taiga (Kostroma oblast). Eurasian Soil Sci. 2009, 42, 1029–1037. [Google Scholar] [CrossRef]
- Sazonov, S.N.; Manucharova, N.A.; Gorlenko, M.V.; Umarov, M.M. Natural restoration of the microbiological properties of soddy-podzolic soil of fallow land. Eurasian Soil Sci. 2005, 38, 511–515. [Google Scholar]
- Loeppmann, S.; Semenov, M.; Kuzyakov, Y.; Blagodatskaya, E. Shift from dormancy to microbial growth revealed by RNA:DNA ratio. Ecol. Indic. 2018, 85, 603–612. [Google Scholar] [CrossRef]
- Ananyeva, N.D.; Susyan, E.A.; Chernova, O.V.; Wirth, S. Microbial respiration activities of soils from different climatic regions of European Russia. Eur. J. Soil Biol. 2008, 44, 147–157. [Google Scholar] [CrossRef]
- Dobrovol’skaya, T.G.; Zvyagintsev, D.G.; Chernov, I.Y.; Golovchenko, A.V.; Zenova, G.M.; Lysak, L.V.; Manucharova, N.A.; Marfenina, O.E.; Polyanskaya, L.M.; Stepanov, A.L.; et al. The role of microorganisms in the ecological functions of soils. Eurasian Soil Sci. 2015, 48, 959–967. [Google Scholar] [CrossRef]
- Lednev, A.V.; Dmitriev, A.V. Overgrowing of fallow soddy podzolic soils as a factor of modern pedogenesis. Russ. Agric. Sci. 2017, 43, 482–485. [Google Scholar] [CrossRef]
- Telesnina, V.M.; Zhukov, M.A. The Influence of Agricultural Land Use on the Dynamics of Biological Cycling and Soil Properties in the Course of Postagrogenic Succession (Kostroma Oblast). Eurasian Soil Sci. 2019, 52, 1122–1136. [Google Scholar] [CrossRef]
- Blagodatskaya, E.; Kuzyakov, Y. Active microorganisms in soil: Critical review of estimation criteria and approaches. Soil Biol. Biochem. 2013, 67, 192–211. [Google Scholar] [CrossRef]
- Blazewicz, S.J.; Barnard, R.L.; Daly, R.A.; Firestone, M.K. Evaluating rRNA as an indicator of microbial activity in environmental communities: Limitations and uses. ISME J. 2013, 7, 2061–2068. [Google Scholar] [CrossRef] [Green Version]
- Bowsher, A.W.; Kearns, P.J.; Shade, A. 16S rRNA/rRNA Gene Ratios and Cell Activity Staining Reveal Consistent Patterns of Microbial Activity in Plant-Associated Soil. mSystems 2019, 4, e00003-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carini, P.; Marsden, P.J.; Leff, J.W.; Morgan, E.E.; Strickland, M.S.; Fierer, N. Relic DNA is abundant in soil and obscures estimates of soil microbial diversity. Nat. Microbiol. 2016, 2, 16242. [Google Scholar] [CrossRef]
- Levy-Booth, D.J.; Campbell, R.G.; Gulden, R.H.; Hart, M.M.; Powell, J.R.; Klironomos, J.N.; Pauls, K.P.; Swanton, C.J.; Trevors, J.T.; Dunfield, K.E. Cycling of extracellular DNA in the soil environment. Soil Biol. Biochem. 2007, 39, 2977–2991. [Google Scholar] [CrossRef]
- Semenov, M.; Blagodatskaya, E.; Stepanov, A.; Kuzyakov, Y. DNA-based determination of soil microbial biomass in alkaline and carbonaceous soils of semi-arid climate. J. Arid Environ. 2018, 150, 54–61. [Google Scholar] [CrossRef]
- Anderson, T.H.; Martens, R. DNA determinations during growth of soil microbial biomasses. Soil Biol. Biochem. 2013, 57, 487–495. [Google Scholar] [CrossRef]
- Wang, Y.; Hayatsu, M.; Fujii, T. Extraction of Bacterial RNA from Soil: Challenges and Solutions. Microbes Environ. 2012, 27, 111–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mills, H.J.; Martinez, R.J.; Story, S.; Sobecky, P. Identification of Members of the Metabolically Active Microbial Populations Associated with Beggiatoa Species Mat Communities from Gulf of Mexico Cold-Seep Sediments. Appl. Environ. Microbiol. 2004, 70, 5447–5458. [Google Scholar] [CrossRef] [Green Version]
- Skvortsova, E.B.; Shein, E.V.; Abrosimov, K.N.; Romanenko, K.A.; Yudina, A.V.; Klyueva, V.V.; Khaidapova, D.D.; Rogov, V.V. The Impact of Multiple Freeze–Thaw Cycles on the Microstructure of Aggregates from a Soddy-Podzolic Soil: A Microtomographic Analysis. Eurasian Soil Sci. 2018, 51, 190–198. [Google Scholar] [CrossRef]
- Nikitin, D.A.; Chernov, T.V.; Zhelezova, A.D.; Tkhakakhova, A.K.; Nikitina, S.A.; Semenov, M.V.; Xenofontova, N.A.; Kutovaya, O.V. Seasonal Dynamics of Microbial Biomass in Soddy-Podzolic Soil. Eurasian Soil Sci. 2019, 52, 1414–1421. [Google Scholar] [CrossRef]
- Makarov, M.I.; Shuleva, M.S.; Malysheva, T.I.; Menyailo, O.V. Solubility of the labile forms of soil carbon and nitrogen in K2SO4 of different concentrations. Eurasian Soil Sci. 2013, 46, 369–374. [Google Scholar] [CrossRef]
- Fierer, N.; Jackson, J.A.; Vilgalys, R.; Jacksson, R.B. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl. Environ. Microbiol. 2005, 71, 4117. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Lee, C.; Hwang, S. Analysis of community structures in anaerobic processes using a quantitative real-time PCR method. Water Sci. Technol. 2005, 52, 85–91. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Aronesty, E. Comparison of Sequencing Utility Programs. Open Bioinforma. J. 2013, 7, 1–8. [Google Scholar] [CrossRef]
- Quince, C.; Nichols, B.; Rognes, T.; Flouri, T.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Lozupone, C.; Lladser, M.E.; Knights, D.; Stombaugh, J.; Knight, R. UniFrac: An effective distance metric for microbial community comparison. ISME J. 2011, 5, 169–172. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Xiong, D.; Xiao, L.; Zhang, S.; Yuan, Y.; Su, Z.; Zhang, B.; Yang, D. Effects of vegetation coverage and seasonal change on soil microbial biomass and community structure in the dry-hot valley region. J. Mt. Sci. 2018, 15, 1546–1558. [Google Scholar] [CrossRef]
- Devi, N.B.; Yadava, P.S. Seasonal dynamics in soil microbial biomass C, N and P in a mixed-oak forest ecosystem of Manipur, North-east India. Appl. Soil Ecol. 2006, 31, 220–227. [Google Scholar] [CrossRef]
- Price, P.B.; Sowers, T. Temperature dependence of metabolic rates for microbial growth, maintenance, and survival. Proc. Natl. Acad. Sci. USA 2004, 101, 4631–4636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Regan, K.M.; Nunan, N.; Boeddinghaus, R.S.; Baumgartner, V.; Berner, D.; Boch, S.; Oelmann, Y.; Overmann, J.; Prati, D.; Schloter, M.; et al. Seasonal controls on grassland microbial biogeography: Are they governed by plants, abiotic properties or both? Soil Biol. Biochem. 2014, 71, 21–30. [Google Scholar] [CrossRef]
- Rasche, F.; Knapp, D.; Kaiser, C.; Koranda, M.; Kitzler, B.; Zechmeister-Boltenstern, S.; Richter, A.; Sessitsch, A. Seasonality and resource availability control bacterial and archaeal communities in soils of a temperate beech forest. ISME J. 2011, 5, 389–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldrian, P. Microbial activity and the dynamics of ecosystem processes in forest soils. Curr. Opin. Microbiol. 2017, 37, 128–134. [Google Scholar] [CrossRef]
- Lipson, D.A. Relationships between temperature responses and bacterial community structure along seasonal and altitudinal gradients. FEMS Microbiol. Ecol. 2007, 59, 418–427. [Google Scholar] [CrossRef]
- Moche, M.; Gutknecht, J.; Schulz, E.; Langer, U.; Rinklebe, J. Monthly dynamics of microbial community structure and their controlling factors in three floodplain soils. Soil Biol. Biochem. 2015, 90, 169–178. [Google Scholar] [CrossRef]
- Edwards, K.A.; Jefferies, R.L. Inter-annual and seasonal dynamics of soil microbial biomass and nutrients in wet and dry low-Arctic sedge meadows. Soil Biol. Biochem. 2013, 57, 83–90. [Google Scholar] [CrossRef]
- Vořiškova, J.; Brabcová, V.; Cajthaml, T.; Baldrian, P. Seasonal dynamics of fungal communities in a temperate oak forest soil. New Phytol. 2014, 201, 269–278. [Google Scholar] [CrossRef]
- Xu, R.; Zhang, K.; Liu, P.; Han, H.; Zhao, S.; Kakade, A.; Khan, A.; Du, D.; Li, X. Lignin depolymerization and utilization by bacteria. Bioresour. Technol. 2018, 269, 557–566. [Google Scholar] [CrossRef]
- Landesman, W.J.; Freedman, Z.B.; Nelson, D.M. Seasonal, sub-seasonal and diurnal variation of soil bacterial community composition in a temperate deciduous forest. FEMS Microbiol. Ecol. 2019, 95, fiz002. [Google Scholar] [CrossRef] [Green Version]
- Shigyo, N.; Umeki, K.; Hirao, T. Seasonal dynamics of soil fungal and bacterial communities in cool-temperate montane forests. Front. Microbiol. 2019, 10, 1944. [Google Scholar] [CrossRef] [Green Version]
- Thomas, F.; Hehemann, J.H.; Rebuffet, E.; Czjzek, M.; Michel, G. Environmental and gut Bacteroidetes: The food connection. Front. Microbiol. 2011, 2, 93. [Google Scholar] [CrossRef] [Green Version]
- Uroz, S.; Ioannidis, P.; Lengelle, J.; Cébron, A.; Morin, E.; Buée, M.; Martin, F. Functional assays and metagenomic analyses reveals differences between the microbial communities inhabiting the soil horizons of a Norway spruce plantation. PLoS ONE 2013, 8, e55929. [Google Scholar] [CrossRef]
- Gao, G.F.; Li, P.F.; Zhong, J.X.; Shen, Z.J.; Chen, J.; Li, Y.T.; Isabwe, A.; Zhu, X.Y.; Ding, Q.S.; Zhang, S.; et al. Spartina alterniflora invasion alters soil bacterial communities and enhances soil N 2 O emissions by stimulating soil denitrification in mangrove wetland. Sci. Total Environ. 2019, 653, 231–240. [Google Scholar] [CrossRef]
- Taubert, M.; Stähly, J.; Kolb, S.; Küsel, K. Divergent microbial communities in groundwater and overlying soils exhibit functional redundancy for plant-polysaccharide degradation. PLoS ONE 2019, 14, e0212937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siles, J.A.; Margesin, R. Insights into microbial communities mediating the bioremediation of hydrocarbon-contaminated soil from an Alpine former military site. Appl. Microbiol. Biotechnol. 2018, 102, 4409–4421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crowe, M.A.; Power, J.F.; Morgan, X.C.; Dunfield, P.F.; Lagutin, K.; Rijpstra, W.I.C.; Rijpstra, I.C.; Sinninghe Damste, J.S.; Houghton, K.M.; Ryan, J.L.J.; et al. Pyrinomonas methylaliphatogenes gen. nov., sp. nov., a novel group 4 thermophilic member of the phylum Acidobacteria from geothermal soils. Int. J. Syst. Evol. Microbiol. 2014, 64, 220–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tank, M.; Bryant, D.A. Chloracidobacterium thermophilum gen. nov., sp. nov.: An anoxygenic microaerophilic chlorophotoheterotrophic acidobacterium. Int. J. Syst. Evol. Microbiol. 2015, 65, 1426–1430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kant, R.; van Passe, M.W.J.; Palva, A.; Lucas, S.; Lapidus, A.; del Rio, T.G.; Dalin, E.; Tice, H.; Bruce, D.; Goodwin, L.; et al. Genome sequence of Chthoniobacter flavus Ellin428, an aerobic heterotrophic soil bacterium. J. Bacteriol. 2011, 193, 2902–2903. [Google Scholar] [CrossRef] [Green Version]
- Okazaki, K.; Iino, T.; Kuroda, Y.; Taguchi, K.; Takahashi, H.; Ohwada, T.; Tsurumaru, H.; Okubo, T.; Minamisawa, K.; Ikeda, S. An assessment of the diversity of culturable bacteria from main root of sugar beet. Microbes Environ. 2014, 29, 220–223. [Google Scholar] [CrossRef] [Green Version]
- Semenov, M.V.; Chernov, T.I.; Tkhakakhova, A.K.; Zhelezova, A.D.; Ivanova, E.A.; Kolganova, T.V.; Kutovaya, O.V. Distribution of prokaryotic communities throughout the Chernozem profiles under different land uses for over a century. Appl. Soil Ecol. 2018, 127, 8–18. [Google Scholar] [CrossRef]
- Chernov, T.I.; Zhelezova, A.D.; Kutovaya, O.V.; Makeev, A.O.; Tkhakakhova, A.K.; Bgazhba, N.A.; Kurbanova, F.G.; Rusakov, A.V.; Puzanova, T.A.; Khokhlova, O.S. Comparative Analysis of the Structure of Buried and Surface Soils by Analysis of Microbial DNA. Microbiol. Russ. Fed. 2018, 87, 833–841. [Google Scholar] [CrossRef]
- Zhang, B.; Wu, X.; Zhang, G.; Li, S.; Zhang, W.; Chen, X.; Sun, L.; Zhang, B.; Liu, G.; Chen, T. The diversity and biogeography of the communities of Actinobacteria in the forelands of glaciers at a continental scale. Environ. Res. Lett. 2016, 11, 054012. [Google Scholar] [CrossRef]
- Ji, M.; van Dorst, J.; Bissett, A.; Brown, M.V.; Palmer, A.S.; Snape, I.; Siciliano, S.D.; Ferrari, B.C. Microbial diversity at Mitchell Peninsula, Eastern Antarctica: A potential biodiversity “hotspot”. Polar Biol. 2016, 39, 237–249. [Google Scholar] [CrossRef]
- Fricke, W.; Barnum, T.; Davila, A.F.; Robinson, C.K.; DiRuggiero, J.; Jedynak, B.; McKay, C.P.; Crits-Christoph, A. Colonization patterns of soil microbial communities in the Atacama Desert. Microbiome 2013, 1, 28. [Google Scholar] [CrossRef] [Green Version]
- Houfani, A.A.; Větrovský, T.; Navarrete, O.U.; Štursová, M.; Tláskal, V.; Beiko, R.G.; Boucherba, N.; Baldrian, P.; Benallaoua, S.; Jorquera, M.A. Cellulase−Hemicellulase Activities and Bacterial Community Composition of Different Soils from Algerian Ecosystems. Microb. Ecol. 2019, 77, 713–725. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Zang, Y.; Mao, Y.; Gao, B. Identification of Molecular Markers That Are Specific to the Class Thermoleophilia. Front. Microbiol. 2019, 10, 1185. [Google Scholar] [CrossRef] [PubMed]
- Preece, C.; Verbruggen, E.; Liu, L.; Weedon, J.T.; Peñuelas, J. Effects of past and current drought on the composition and diversity of soil microbial communities. Soil Biol. Biochem. 2019, 131, 28–39. [Google Scholar] [CrossRef]
- Schlatter, D.C.; Reardon, C.L.; Johnson-Maynard, J.; Brooks, E.; Kahl, K.; Norby, J.; Huggins, D.; Paulitz, T.C. Mining the Drilosphere: Bacterial Communities and Denitrifier Abundance in a No-Till Wheat Cropping System. Front. Microbiol. 2019, 10, 1339. [Google Scholar] [CrossRef] [Green Version]
- Lipson, D.A.; Schmidt, S.K. Seasonal changes in an alpine soil bacterial community in the Colorado Rocky Mountains. Appl. Environ. Microbiol. 2004, 70, 2867–2879. [Google Scholar] [CrossRef] [Green Version]
- Žifčáková, L.; Větrovský, T.; Lombard, V.; Henrissat, B.; Howe, A.; Baldrian, P. Feed in summer, rest in winter: Microbial carbon utilization in forest topsoil. Microbiome 2017, 5, 122. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhelezova, A.; Chernov, T.; Nikitin, D.; Tkhakakhova, A.; Ksenofontova, N.; Zverev, A.; Kutovaya, O.; Semenov, M. Seasonal Dynamics of Soil Bacterial Community under Long-Term Abandoned Cropland in Boreal Climate. Agronomy 2022, 12, 519. https://doi.org/10.3390/agronomy12020519
Zhelezova A, Chernov T, Nikitin D, Tkhakakhova A, Ksenofontova N, Zverev A, Kutovaya O, Semenov M. Seasonal Dynamics of Soil Bacterial Community under Long-Term Abandoned Cropland in Boreal Climate. Agronomy. 2022; 12(2):519. https://doi.org/10.3390/agronomy12020519
Chicago/Turabian StyleZhelezova, Alena, Timofey Chernov, Dmitry Nikitin, Azida Tkhakakhova, Natalia Ksenofontova, Aleksei Zverev, Olga Kutovaya, and Mikhail Semenov. 2022. "Seasonal Dynamics of Soil Bacterial Community under Long-Term Abandoned Cropland in Boreal Climate" Agronomy 12, no. 2: 519. https://doi.org/10.3390/agronomy12020519





