Genotypic and Phenotypic Characterization of Two Triticum aestivum L.—Dasypyrum villosum Translocations Lines in the Same Wheat Genetic Background
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Genotyping
2.3. Field Experimental Design and Investigation of Agronomic Characters
2.4. Statistical Analysis
2.5. Evaluation of PM Resistance of the Translocation Lines and Wheat Wan7107
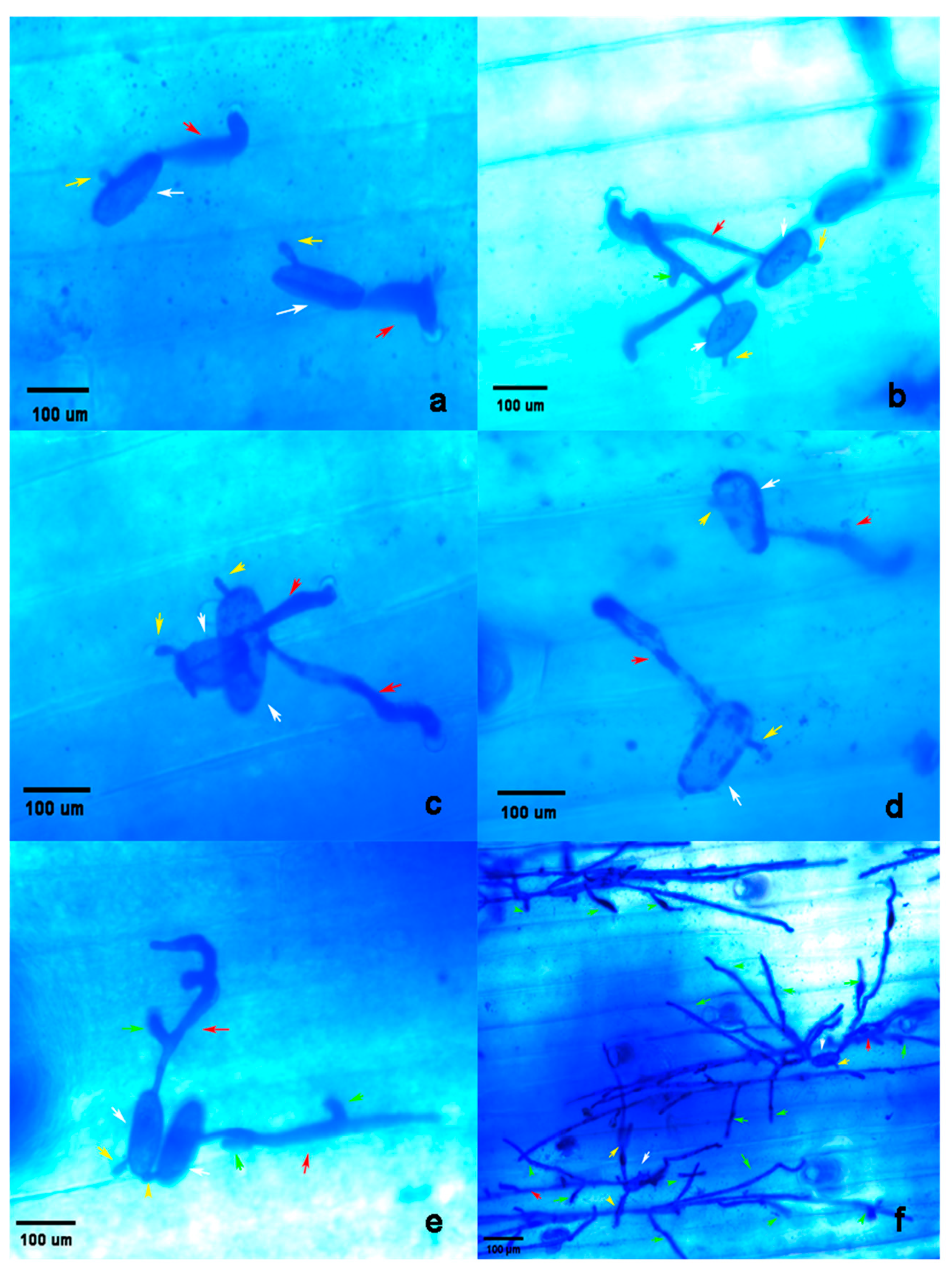
2.5.1. Development of Conidia of Bgt on Plant Leaves
2.5.2. Reaction to Different Isolates of the Pathogen
3. Results
3.1. Characteristics of Base Composition in the Subgenome by Genotyping the Three Lines
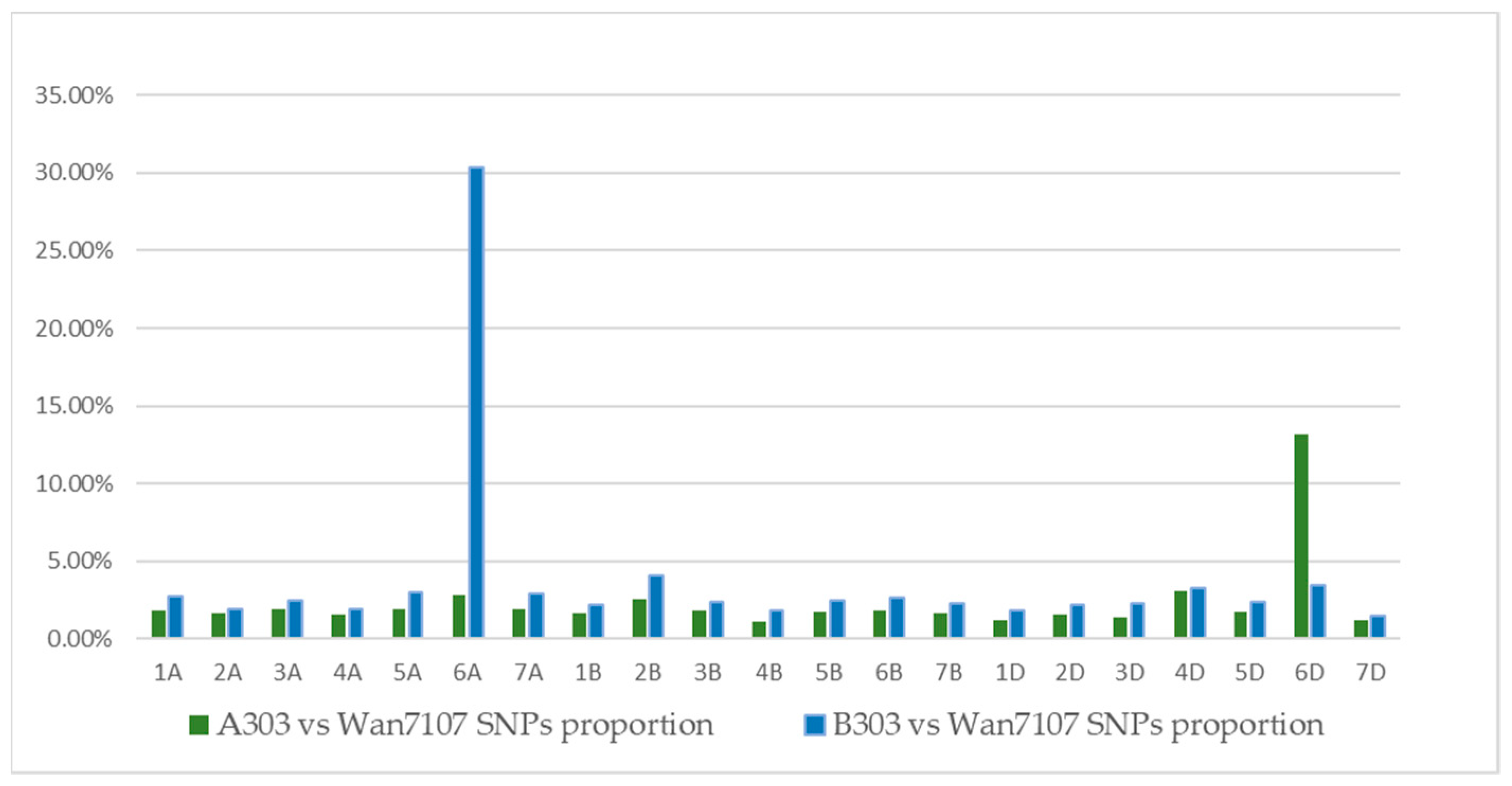
3.2. Snps between Wheat Wan7107 and Translocation Lines
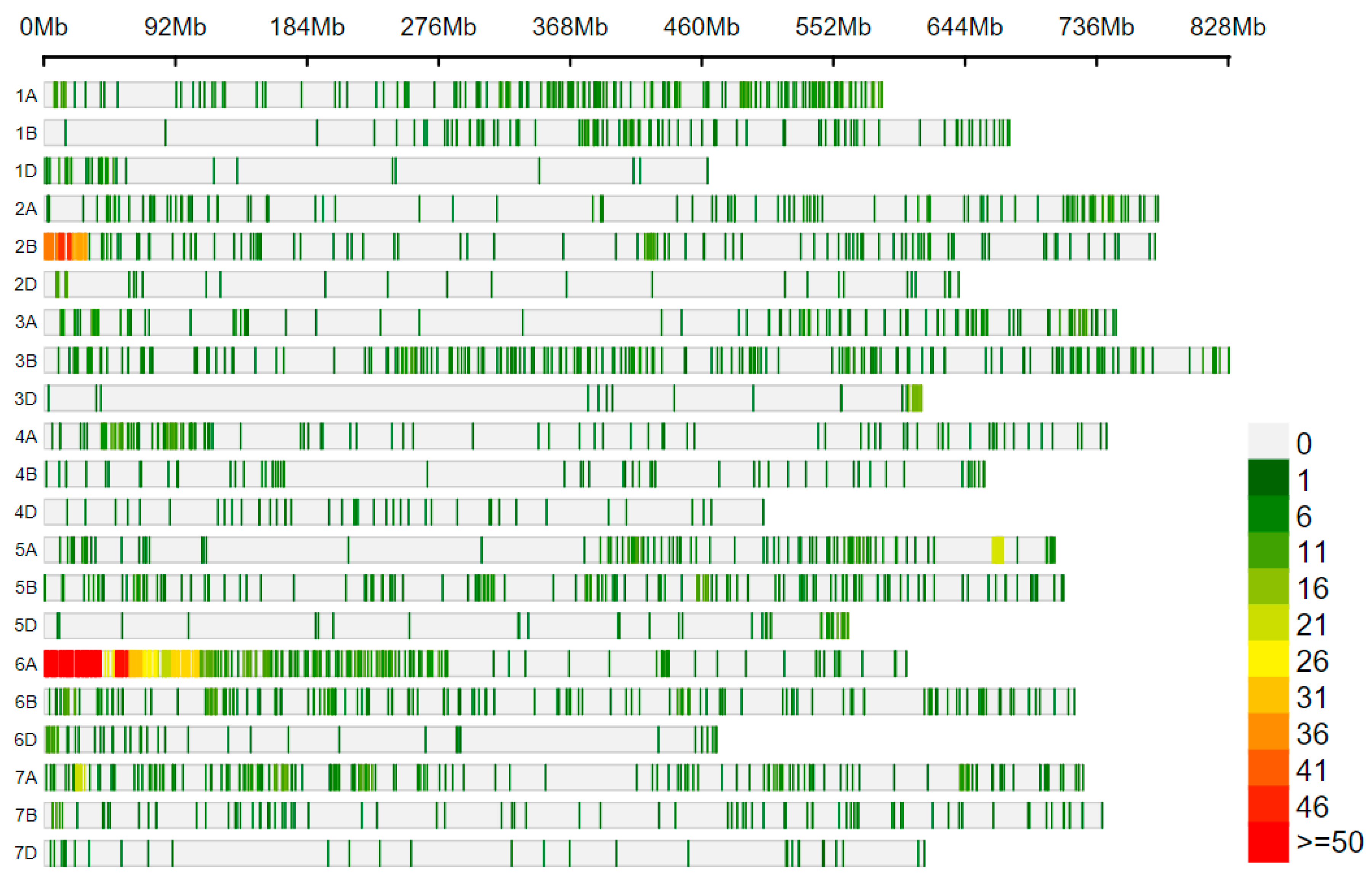
3.3. Distribution of SNPs on Chromosomes between Wan7107 and Translocation Lines
3.4. Differences in Powdery Mildew Resistance between Two Translocation Lines
3.5. Effects of Different Chromosome Translocations on Agronomic Traits
4. Discussion
4.1. Differences in SNP Values between Translocation Lines and Recurrent Parent
4.2. SNP Distribution between Translocation Lines and Recurrent Parent
4.3. 6VS Association with PM Resistance
4.4. 6VS Association with Agronomic Characters
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| SNP | Single Nucleotide Polymorphism |
| QTL | quantitative trait loci |
| EN | ear number |
| RIL | recombinant inbred line |
| SFN | sterile floret number |
| TGW | thousand grain weight |
| EL | ear length |
| SNPS | spikelet number per spike |
| SN | spike number |
| FN | floret number |
| PH | plant height |
| GNPS | grain number per spike |
| KASP | Kompetitive allele-specific PCR |
| Bgt | B. graminis f. sp. tritici |
| PM | powdery mildew |
| SSR | simple sequence repeat |
References
- Brenchley, R.; Spannagl, M.; Pfeifer, M.; Barker, G.L.A.; D′Amore, R.; Allen, A.M.; McKenzie, N.; Kramer, M.; Kerhornou, A.; Bolser, D.; et al. Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 2012, 491, 705–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food security: The challenge of feeding 9 billion people. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atkinson, N.J.; Urwin, P.E. The interaction of plant biotic and abiotic stresses: From genes to the field. J. Exp. Bot. 2012, 63, 3523–3543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2006, 58, 221–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinto, R.S.; Reynolds, M.P.; Mathews, K.L.; McIntyre, C.L.; Olivares-Villegas, J.-J.; Chapman, S.C. Heat and drought adaptive QTL in a wheat population designed to minimize confounding agronomic effects. Theor. Appl. Genet. 2010, 121, 1001–1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ollier, M.; Talle, V.; Brisset, A.-L.; Le Bihan, Z.; Duerr, S.; Lemmens, M.; Goudemand, E.; Robert, O.; Hilbert, J.-L.; Buerstmayr, H. QTL mapping and successful introgression of the spring wheat-derived QTL Fhb1 for Fusarium head blight resistance in three European triticale populations. Theor. Appl. Genet. 2020, 133, 457–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graybosch, R.A. Mini Review: Uneasy unions: Quality effects of rye chromatin transfers to wheat. J. Cereal Sci. 2001, 33, 3–16. [Google Scholar] [CrossRef]
- Devos, K.M.; Atkinson, M.D.; Chinoy, C.N.; Francis, H.A.; Harcourt, R.L.; Koebner, R.M.; Liu, C.J.; Masojć, P.; Xie, D.X.; Gale, M.D. Chromosomal rearrangements in the rye genome relative to that of wheat. Theor. Appl. Genet. 1993, 85, 673–680. [Google Scholar] [CrossRef] [PubMed]
- Ren, T.; Li, Z.; Yan, B.; Tan, F.; Tang, Z.; Fu, S.; Yang, M.; Ren, Z. Targeted segment transfer from rye chromosome 2R to wheat chromosomes 2A, 2B, and 7B. Cytogenet. Genome Res. 2017, 151, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.D.; Qi, L.L.; Zhou, B.; Zhang, S.Z.; Liu, D.J. Development and molecular cytogenetic analysis of wheat-Haynaldia villosa 6VS/6AL translocation lines specifying resistance to powdery mildew. Theor. Appl. Genet. 1995, 91, 1125–1128. [Google Scholar] [CrossRef]
- Chhuneja, P.; Kaur, S.; Goel, R.K.; Aghaee-Sarbarzeh, M.; Prashar, M.; Dhaliwal, H.S. Transfer of leaf rust and stripe rust resistance from Aegilops umbellulata Zhuk. to bread wheat (Triticum aestivum L.). Genet. Resour. Crop Evol. 2008, 55, 849–859. [Google Scholar] [CrossRef]
- Qi, L.L.; Pumphrey, M.O.; Friebe, B.; Zhang, P.; Qian, C.; Bowden, R.L.; Rouse, M.N.; Jin, Y.; Gill, B.S. A novel Robertsonian translocation event leads to transfer of a stem rust resistance gene (Sr52) effective against race Ug99 from Dasypyrum villosum into bread wheat. Theor. Appl. Genet. 2011, 123, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Cao, M.; Chen, P.; Li, W.; Liu, D. Identification, mapping, and application of polymorphic DNA associated with resistance gene Pm21 of wheat. Genome 1996, 39, 191–197. [Google Scholar] [CrossRef]
- Ma, J.; Zhou, R.; Dong, Y.; Wang, L.; Wang, X.; Jia, J. Molecular mapping and detection of the yellow rust resistance gene Yr26 in wheat transferred from Triticum turgidum L. using microsatellite markers. Euphytica 2001, 120, 219–226. [Google Scholar] [CrossRef]
- Wang, R.; Wu, J.; Deng, X.; Liu, D.; Yan, Y. Drought-responsive protein identification in developing grains of a wheat–Haynaldia villosa 6VS/6AL translocation line. Crop Pasture Sci. 2019, 69, 1182–1196. [Google Scholar] [CrossRef]
- Rani, K.; Raghu, B.R.; Jha, S.K.; Agarwal, P.; Mallick, N.; Niranjana, M.; Sharma, J.B.; Singh, A.K.; Sharma, N.K.; Rajkumar, S.; et al. A novel leaf rust resistance gene introgressed from Aegilops markgrafii maps on chromosome arm 2AS of wheat. Theor. Appl. Genet. 2020, 133, 2685–2694. [Google Scholar] [CrossRef] [PubMed]
- Riley, R.; Chapman, V.; Johnson, R.O.Y. Introduction of yellow rust resistance of aegilops comosa into wheat by genetically induced homoeologous recombination. Nature 1968, 217, 383–384. [Google Scholar] [CrossRef]
- Schneider, A.; Molnár, I.; Molnár-Láng, M. Utilisation of Aegilops (goatgrass) species to widen the genetic diversity of cultivated wheat. Euphytica 2008, 163, 1–19. [Google Scholar] [CrossRef]
- Adonina, I.G.; Salina, E.A.; Pestsova, E.G.; Röder, M.S. Transferability of wheat microsatellites to diploid Aegilops species and determination of chromosomal localizations of microsatellites in the S genome. Genome 2005, 48, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Islam, A.K.M.R.; Shepherd, K.W.; Sparrow, D.H.B. Isolation and characterization of euplasmic wheat-barley chromosome addition lines. Heredity 1981, 46, 161–174. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Sun, S.; Ge, W.; Zhao, L.; Kong, L. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science 2020, 368, eaba5435. [Google Scholar] [CrossRef]
- Riley, R.; Chapman, V. The comparison of wheat-rye and wheat- Aegilops amphidiploids. J. Agric. Sci. 1957, 49, 246–250. [Google Scholar] [CrossRef]
- Nemeth, C.; Yang, C.-Y.; Kasprzak, P.; Hubbart, S.; Scholefield, D.; Mehra, S.; Skipper, E.; King, I.; King, J.; Jenkins, G. Generation of amphidiploids from hybrids of wheat and related species from the genera Aegilops, Secale, Thinopyrum, and Triticum as a source of genetic variation for wheat improvement. Genome 2015, 58, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, T.; Taketa, S.; Murai, K. Differential effects of cultivated and wild barley 5H chromosomes on heading characters in wheat-barley chromosome addition lines. Hereditas 2002, 136, 195–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rawat, N.; Tiwari, V.K.; Neelam, K.; Randhawa, G.S.; Chhuneja, P.; Singh, K.; Dhaliwal, H.S. Development and characterization of Triticum aestivum–Aegilops kotschyi amphiploids with high grain iron and zinc contents. Plant Genet. Resour. 2009, 7, 271–280. [Google Scholar] [CrossRef]
- Jiang, J.; Friebe, B.; Gill, B.S. Recent advances in alien gene transfer in wheat. Euphytica 1993, 73, 199–212. [Google Scholar] [CrossRef]
- Djanaguiraman, M.; Prasad, P.V.V.; Kumari, J.; Sehgal, S.K.; Friebe, B.; Djalovic, I.; Chen, Y.; Siddique, K.H.M.; Gill, B.S. Alien chromosome segment from Aegilops speltoides and Dasypyrum villosum increases drought tolerance in wheat via profuse and deep root system. BMC Plant Biol. 2019, 19, 242. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Tang, H.; Ding, P.; Mu, Y.; Habib, A.; Liu, Y.; Jiang, Q.; Chen, G.; Kang, H.; Wei, Y.; et al. Effects of the 1BL/1RS translocation on 24 traits in a recombinant inbred line population. Cereal Res. Commun. 2020, 48, 225–232. [Google Scholar] [CrossRef]
- He, H.; Zhu, S.; Zhao, R.; Jiang, Z.; Ji, Y.; Ji, J.; Qiu, D.; Li, H.; Bie, T. Pm21, encoding a typical CC-NBS-LRR protein, confers broad-spectrum resistance to wheat powdery mildew disease. Mol. Plant 2018, 11, 879–882. [Google Scholar] [CrossRef] [Green Version]
- He, H.; Du, H.; Liu, R.; Liu, T.; Yang, L.; Gong, S.; Tang, Z.; Du, H.; Liu, C.; Han, R.; et al. Characterization of a new gene for resistance to wheat powdery mildew on chromosome 1RL of wild rye Secale sylvestre. Theor. Appl. Genet. 2021, 1–7. [Google Scholar] [CrossRef]
- McIntosh RA, D.J.; Rogers, W.J.; Morris, C.; Xia, X.C. Catalogue of Gene Symbols for Wheat: 2017 Supplement (KOMUGI Wheat Genetic Resource Database). 2017. Available online: https://shigen.nig.ac.jp/wheat/komugi/genes/symbolClassList.jsp (accessed on 1 November 2020).
- Li, Y.; Shi, X.; Hu, J.; Wu, P.; Li, H. Identification of a Recessive Gene PmQ Conferring Resistance to Powdery Mildew in Wheat Landrace Qingxinmai Using BSR-Seq Analysis. Plant Dis. 2019, 104, 743–751. [Google Scholar] [CrossRef]
- Friebe, B.; Heun, M.; Tuleen, N.; Zeller, F.J.; Gill, B.S. Cytogenetically Monitored Transfer of Powdery Mildew Resistance from Rye into Wheat. Crop Sci. 1994, 34, 621–625. [Google Scholar] [CrossRef]
- Singh, S.P.; Hurni, S.; Ruinelli, M.; Brunner, S.; Sanchez-Martin, J.; Krukowski, P.; Peditto, D.; Buchmann, G.; Zbinden, H.; Keller, B. Evolutionary divergence of the rye Pm17 and Pm8 resistance genes reveals ancient diversity. Plant Mol. Biol. 2018, 98, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Hao, M.; Liu, M.; Luo, J.; Fan, C.; Yi, Y.; Zhang, L.; Yuan, Z.; Ning, S.; Zheng, Y.; Liu, D. Introgression of Powdery Mildew Resistance Gene Pm56 on Rye Chromosome Arm 6RS Into Wheat. Front. Plant Sci. 2018, 9, 1040. [Google Scholar] [CrossRef]
- Zhang, R.; Sun, B.; Chen, J.; Cao, A.; Xing, L.; Feng, Y.; Lan, C.; Chen, P. Pm55, a developmental-stage and tissue-specific powdery mildew resistance gene introgressed from Dasypyrum villosum into common wheat. Theor. Appl. Genet. 2016, 129, 1975–1984. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Fan, Y.; Kong, L.; Wang, Z.; Wu, J. Pm62, an adult-plant powdery mildew resistance gene introgressed from Dasypyrum villosum chromosome arm 2VL into wheat. Theor. Appl. Genet. 2018, 131, 2613–2620. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Xiong, C.; Mu, H.; Yao, R.; Cao, A. Pm67, a new powdery mildew resistance gene transferred from Dasypyrum villosum chromosome 1V to common wheat (Triticum aestivum L.). Crop J. 2020. [Google Scholar] [CrossRef]
- Li, H.; Xin, Z.Y.; Ma, Y.Z.; Xu, H.J.; Jia, X. Development and identification of wheat–Haynaldia villosa T6DL.6VS chromosome translocation lines conferring resistance to powdery mildew. Plant Breed. 2005, 124, 203–205. [Google Scholar] [CrossRef]
- Gao, S.G. Identification and analysis of resistance of NAU 92R wheat lines. Acta Agron. Sin. 1999, 25, 389–391. (In Chinese) [Google Scholar]
- Liu, C.; Wang, K.; Wang, M.J.; Li, S.J.; Lin, Z.S. Genetic behavior of Triticum aestivum–Dasypyrum villosum translocation chromosomes T6V#4S·6DL and T6V#2S·6AL carrying powdery mildew resistance. J. Integr. Agric. 2017, 16, 2136–2144. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.L.; Wang, M.J.; Zhang, Y.; Chu, C.P.; Zhang, X.S. Development and application of functional markers specific to powdery mildew resistance on chromosome arm 6VS from different origins of Haynaldia villosa. Acta Agron. Sin. 2013, 38, 1827–1832. (In Chinese) [Google Scholar] [CrossRef]
- Liu, C.; Qi, L.; Liu, W.; Zhao, W.; Wilson, J.; Friebe, B.; Gill, B.S. Development of a set of compensating Triticum aestivum—Dasypyrum villosum Robertsonian translocation lines. Genome 2011, 54, 836–844. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.L.; Wang, S.L.; Chen, P.D.; Liu, D.J.; Gill, B.S. Identification and physical mapping of three Haynaldia villosa chromosome-6V deletion lines. Theor. Appl. Genet. 1998, 97, 1042–1046. [Google Scholar] [CrossRef]
- Zhao, R.; Liu, B.; Jiang, Z.; Chen, T.; Wang, L.; Ji, Y.; Hu, Z.; He, H.; Bie, T. Comparative analysis of genetic effects of wheat-Dasypyrum villosum translocations T6V#2S·6AL and T6V#4S·6DL. Plant Breed. 2019, 138, 503–512. [Google Scholar] [CrossRef]
- Li, S.; Jia, Z.; Wang, K.; Du, L.; Li, H.; Lin, Z.; Ye, X. Screening and functional characterization of candidate resistance genes to powdery mildew from Dasypyrum villosum#4 in a wheat line Pm97033. Theor. Appl. Genet. 2020, 133, 3067–3083. [Google Scholar] [CrossRef]
- Bie, T.; Zhao, R.; Zhu, S.; Chen, S.; Cen, B.; Zhang, B.; Gao, D.; Jiang, Z.; Chen, T.; Wang, L.; et al. Development and characterization of marker MBH1 simultaneously tagging genes Pm21 and PmV conferring resistance to powdery mildew in wheat. Mol. Breed. 2015, 35, 189. [Google Scholar] [CrossRef]
- Li, S.; Wang, J.; Wang, K.; Chen, J.; Wang, K.; Du, L.; Ni, Z.; Lin, Z.; Ye, X. Development of PCR markers specific to Dasypyrum villosum genome based on transcriptome data and their application in breeding Triticum aestivum-D. villosum#4 alien chromosome lines. BMC Genom. 2019, 20, 289. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.; Dong, Z.; Zhao, L.; Ren, Y.; Zhang, N.; Chen, F. The wheat 660K SNP array demonstrates great potential for marker-assisted selection in polyploid wheat. Plant Biotechnol. J. 2020, 18, 1354–1360. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Xie, Y.; Li, Y.; Wu, S.; Wang, C. High-resolution mapping of the novel early leaf senescence gene Els2 in common wheat. Plants 2020, 9, 698. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, Q.; Liu, S.; Huang, S.; Mu, J.; Zeng, Q.; Huang, L.; Han, D.; Kang, Z. Saturation mapping of a major effect QTL for stripe rust resistance on wheat chromosome 2B in cultivar Napo 63 wsing SNP genotyping arrays. Front. Plant Sci. 2017, 8, 653. [Google Scholar] [CrossRef] [Green Version]
- Xin, F.; Zhu, T.; Wei, S.; Han, Y.; Zhao, Y.; Zhang, D.; Ma, L.; Ding, Q. QTL mapping of kernel traits and validation of a major QTL for kernel length-width ratio using SNP and bulked segregant analysis in wheat. Sci Rep. UK 2020, 10, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, D.; Wen, W.; Fu, L.; Li, F.; Li, J.; Xie, L.; Xia, X.; Ni, Z.; He, Z.; Cao, S. Genetic dissection of a major QTL for kernel weight spanning the Rht-B1 locus in bread wheat. Theor. Appl. Genet. 2019, 132, 3191–3200. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Liang, F.; Xu, H.; Zhang, X.; Zhai, H.; Sun, Q.; Ni, Z. Identification of QTL for grain size and shape on the D genome of natural and synthetic allohexaploid wheats with near-identical AABB genomes. Front. Plant Sci. 2017, 8, 1705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, P.; Liang, X.; Zhao, H.; Feng, B.; Xu, E.; Wang, L.; Hu, Y. Identification of the quantitative trait loci controlling spike-related traits in hexaploid wheat (Triticum aestivum L.). Planta 2019, 250, 1967–1981. [Google Scholar] [CrossRef] [PubMed]
- Guan, P.; Di, N.; Mu, Q.; Shen, X.; Wang, Y.; Wang, X.; Yu, K.; Song, W.; Chen, Y.; Xin, M.; et al. Use of near-isogenic lines to precisely map and validate a major QTL for grain weight on chromosome 4AL in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 2019, 132, 2367–2379. [Google Scholar] [CrossRef] [PubMed]
- Guan, P.; Shen, X.; Mu, Q.; Wang, Y.; Wang, X.; Chen, Y.; Zhao, Y.; Chen, X.; Zhao, A.; Mao, W.; et al. Dissection and validation of a QTL cluster linked to Rht-B1 locus controlling grain weight in common wheat (Triticum aestivum L.) using near-isogenic lines. Theor. Appl. Genet. 2020, 133, 2639–2653. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Guo, H.; Zhou, C.; Guo, X.; Xie, Y.; Zhao, L.; Gu, J.; Zhao, S.; Ding, Y.; Liu, L. A combined association mapping and t-test analysis of SNP loci and candidate genes involving in resistance to low nitrogen traits by a wheat mutant population. PLoS ONE 2019, 14, e0211492. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wen, W.; Liu, J.; Zhang, Y.; Cao, S.; He, Z.; Rasheed, A.; Jin, H.; Zhang, C.; Yan, J.; et al. Genetic architecture of grain yield in bread wheat based on genome-wide association studies. BMC Plant Biol. 2019, 19, 168. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Yuan, F.; Zhang, H.; Zhang, Z.; Zhao, J.; Yang, Q.; Wu, J.; Chen, X. Characterization of the wheat-Psathyrostachys huashania Keng 2Ns/2D substitution line H139: A novel germplasm with enhanced resistance to wheat take-all. Front. Plant Sci. 2020, 11, 233. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, Q.; Xu, L.; Mu, J.; Wu, J.; Zeng, Q.; Yu, S.; Huang, L.; Han, D.; Kang, Z. Rapid identification of a major effect QTL conferring adult plant resistance to stripe rust in wheat cultivar Yaco“S”. Euphytica 2017, 213, 124. [Google Scholar] [CrossRef]
- Wu, J.; Wang, X.; Chen, N.; Yu, R.; Yu, S.; Wang, Q.; Huang, S.; Wang, H.; Singh, R.P.; Bhavani, S.; et al. Association analysis identifies new loci for resistance to chinese Yr26-virulent races of the stripe rust pathogen in a diverse panel of wheat germplasm. Plant Dis. 2020, 104, 1751–1762. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Zhang, J.; Che, Y.; Liu, W.; Lu, Y.; Yang, X.; Li, X.; Jia, J.; Liu, X.; Li, L. Construction of Agropyron Gaertn. genetic linkage maps using a wheat 660K SNP array reveals a homoeologous relationship with the wheat genome. Plant Biotechnol. J. 2018, 16, 818–827. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, Q.S. Chinese Wheat Improvement and Pedigree Analysis; China Agriculture Press: Beijing, China, 2003. (In Chinese) [Google Scholar]
- Si Quanmin, Z.X.; Sheng, B.Q.; Duan, X.Y. Identification of physiologic race of Erysiphe graminis f. sp. Tritici. Sci. Agric. Sin. 1987, 20, 64–70. (In Chinese) [Google Scholar]
- Su, H.; Liu, Y.; Liu, C.; Shi, Q.; Huang, Y.; Han, F. Centromere satellite repeats have undergone rapid changes in polyploid wheat subgenomes. Plant Cell 2019, 31, 2035–2051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, A.; Xing, L.; Wang, X.; Yang, X.; Wang, W.; Sun, Y.; Qian, C.; Ni, J.; Chen, Y.; Liu, D. Serine/threonine kinase gene Stpk-V, a key member of powdery mildew resistance gene Pm21, confers powdery mildew resistance in wheat. Proc. Natl. Acad. Sci. USA 2011, 108, 7727–7732. [Google Scholar] [CrossRef] [Green Version]
- Lin, Z.S.; Zhang, Y.L.; Wang, M.J.; Li, J.R.; Wang, K.; Chen, X.; Xu, Q.F.; Zhang, X.S.; Ye, X.G. Isolation and molecular analysis of genes Stpk-V2 and Stpk-V3 homologous to powdery mildew resistance gene Stpk-V in a Dasypyrum villosum accession and its derivatives. J. Appl. Genet. 2013, 54, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Xing, L.; Hu, P.; Liu, J.; Witek, K.; Zhou, S.; Xu, J.; Zhou, W.; Gao, L.; Huang, Z.; Zhang, R.; et al. Pm21 from Haynaldia villosa encodes a CC-NBS-LRR protein conferring powdery mildew resistance in wheat. Mol. Plant 2018, 11, 874–878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, H.; Ji, J.; Li, H.; Tong, J.; Feng, Y.; Wang, X.; Han, R.; Bie, T.; Liu, C.; Zhu, S. Genetic diversity and evolutionary analyses reveal the powdery mildew resistance gene Pm21 undergoing diversifying selection. Front. Genet. 2020, 11, 489. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, V.; Gahlaut, V.; Mathur, S.; Agarwal, P.; Khandelwal, M.; Khurana, P.; Tyagi, A.; Balyan, H.; Gupta, P. Identification of novel SNP in promoter sequence of TaGW2-6A associated with grain weight and other agronomic traits in wheat (Triticum aestivum L.). PLoS ONE 2015, 10, e0129400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, Z.; Hao, C.; Wang, L.; Dong, Y.; Zhang, X. Identification and development of a functional marker of TaGW2 associated with grain weight in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 2011, 122, 211–223. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Bai, Z.; Li, X.; Wang, P.; Wu, Q.; Yang, L.; Li, L.; Li, X. SNP identification and allelic-specific PCR markers development for TaGW2, a gene linked to wheat kernel weight. Theor. Appl. Genet. 2012, 125, 1057–1068. [Google Scholar] [CrossRef] [PubMed]

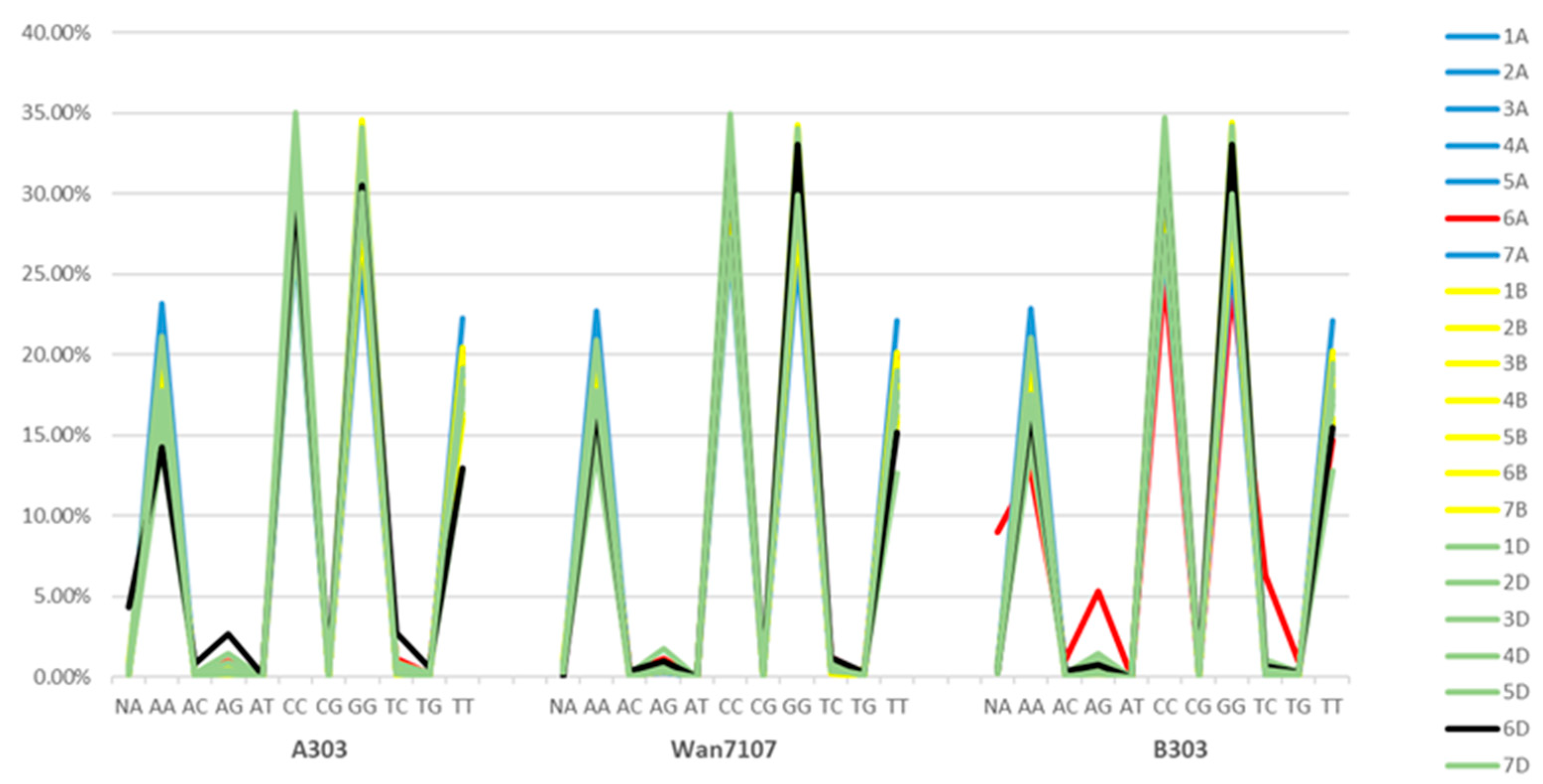


| Wan7107 | NA (%) | AA (%) | AC (%) | AG (%) | AT (%) | CC (%) | CG (%) | GG (%) | TC (%) | TG (%) | TT (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1A | 0.66 | 19.21 | 0.14 | 0.39 | 0.04 | 30.44 | 0.06 | 29.33 | 0.53 | 0.07 | 19.13 |
| 2A | 0.46 | 17.99 | 0.09 | 0.41 | 0.00 | 32.08 | 0.00 | 30.61 | 0.41 | 0.03 | 17.91 |
| 3A | 0.57 | 18.78 | 0.16 | 0.55 | 0.02 | 30.38 | 0.08 | 30.12 | 0.35 | 0.12 | 18.85 |
| 4A | 0.44 | 19.35 | 0.07 | 0.39 | 0.00 | 29.87 | 0.13 | 30.71 | 0.33 | 0.05 | 18.68 |
| 5A | 0.53 | 18.07 | 0.11 | 0.32 | 0.04 | 31.73 | 0.11 | 30.11 | 0.46 | 0.15 | 18.34 |
| 6A | 0.59 | 18.74 | 0.30 | 1.14 | 0.00 | 29.67 | 0.34 | 27.89 | 1.19 | 0.25 | 19.88 |
| 7A | 0.59 | 22.78 | 0.10 | 0.64 | 0.04 | 26.81 | 0.13 | 26.16 | 0.51 | 0.08 | 22.16 |
| 1B | 0.39 | 19.23 | 0.12 | 0.73 | 0.05 | 29.76 | 0.02 | 29.59 | 0.71 | 0.05 | 19.35 |
| 2B | 1.11 | 18.55 | 0.24 | 1.02 | 0.02 | 30.18 | 0.16 | 29.37 | 0.87 | 0.34 | 18.15 |
| 3B | 0.68 | 19.49 | 0.12 | 0.51 | 0.02 | 29.91 | 0.17 | 28.39 | 0.50 | 0.07 | 20.12 |
| 4B | 0.43 | 15.74 | 0.09 | 0.37 | 0.03 | 32.37 | 0.09 | 34.30 | 0.22 | 0.09 | 16.26 |
| 5B | 0.49 | 19.43 | 0.10 | 0.49 | 0.00 | 29.75 | 0.04 | 29.45 | 0.48 | 0.16 | 19.61 |
| 6B | 0.49 | 15.98 | 0.19 | 0.50 | 0.02 | 33.42 | 0.06 | 32.87 | 0.44 | 0.13 | 15.90 |
| 7B | 0.48 | 17.97 | 0.13 | 0.67 | 0.08 | 31.36 | 0.19 | 30.42 | 0.56 | 0.13 | 18.00 |
| 1D | 0.50 | 17.06 | 0.17 | 0.99 | 0.06 | 32.36 | 0.28 | 30.49 | 0.88 | 0.33 | 16.90 |
| 2D | 0.39 | 14.11 | 0.26 | 1.74 | 0.00 | 34.99 | 0.58 | 34.02 | 0.97 | 0.32 | 12.63 |
| 3D | 0.81 | 16.48 | 0.16 | 0.57 | 0.00 | 31.66 | 0.32 | 31.18 | 0.89 | 0.32 | 17.61 |
| 4D | 0.71 | 20.93 | 0.00 | 0.63 | 0.08 | 27.25 | 0.16 | 30.41 | 0.71 | 0.08 | 19.04 |
| 5D | 0.41 | 15.85 | 0.06 | 0.53 | 0.06 | 31.47 | 0.00 | 33.29 | 0.41 | 0.23 | 17.67 |
| 6D | 0.00 | 16.69 | 0.38 | 0.99 | 0.00 | 31.87 | 0.30 | 33.08 | 1.21 | 0.30 | 15.17 |
| 7D | 0.29 | 17.76 | 0.10 | 0.34 | 0.00 | 31.95 | 0.00 | 29.95 | 0.54 | 0.15 | 18.93 |
| Average | 0.53 | 18.10 | 0.15 | 0.66 | 0.03 | 30.92 | 0.15 | 30.56 | 0.63 | 0.17 | 18.11 |
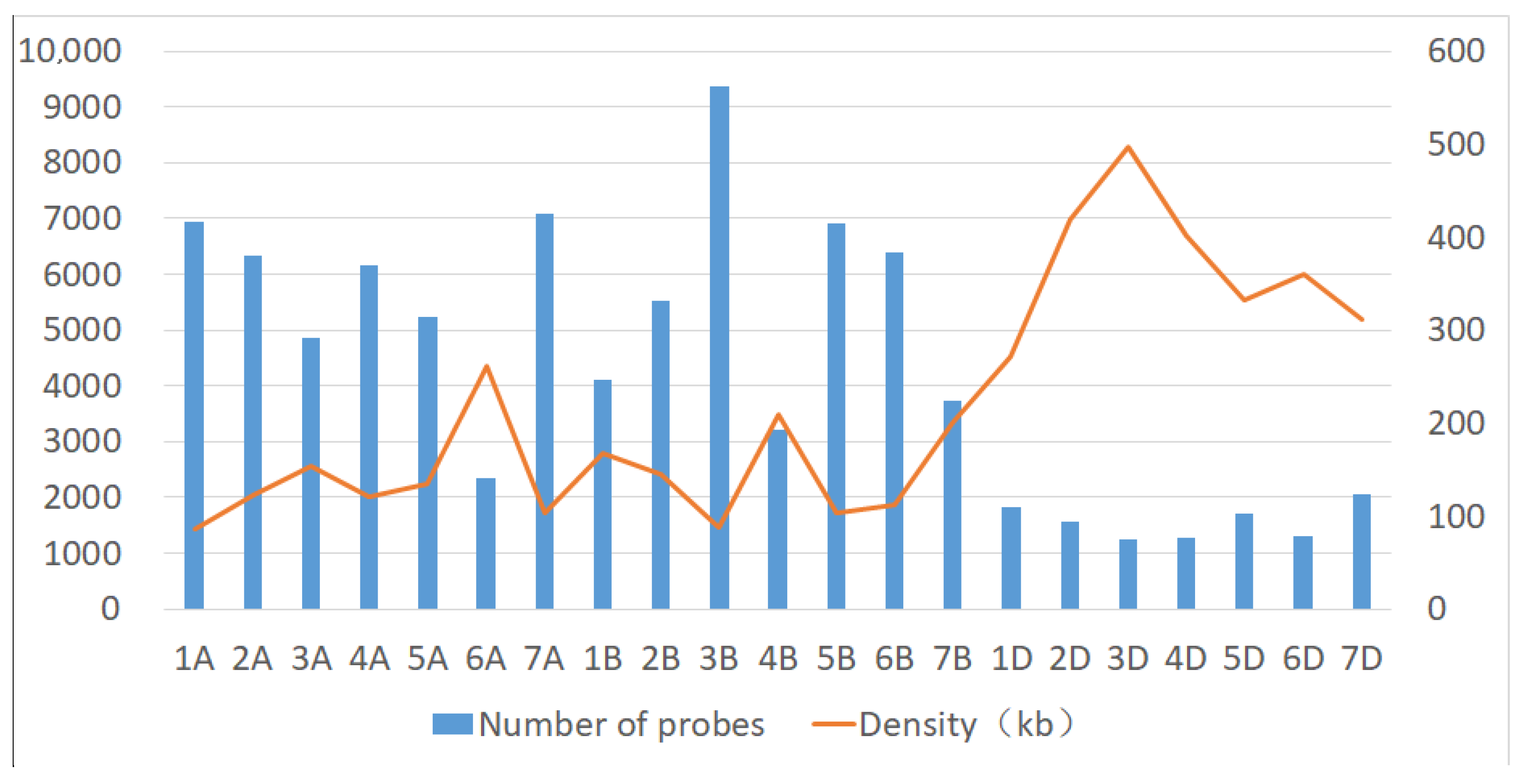
| Chromosome | Physical Range (Mb) | Number of Probes | Density (kb) | A303 vs. Wan7107(SNP) | B303 vs. Wan7107(SNP) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Number | Density (kb) | Proportion (%) | Number | Density (kb) | Proportion (%) | ||||
| 1A | 593.07 | 6949 | 85.35 | 34 | 17,443.24 | 0.49 | 91 | 6517.25 | 1.31 |
| 2A | 780.26 | 6327 | 123.32 | 49 | 15,923.67 | 0.77 | 72 | 10,836.94 | 1.14 |
| 3A | 750.55 | 4871 | 154.09 | 30 | 25,018.33 | 0.62 | 59 | 12,721.19 | 1.21 |
| 4A | 743.86 | 6151 | 120.93 | 31 | 23,995.48 | 0.5 | 50 | 14,877.20 | 0.81 |
| 5A | 709.42 | 5234 | 135.54 | 36 | 19,706.11 | 0.69 | 93 | 7628.17 | 1.78 |
| 6A | 616.46 | 2359 | 261.32 | 25 | 24,658.40 | 1.06 | 648 | 951.33 | 27.47 |
| 7A | 736.44 | 7072 | 104.13 | 40 | 18,411.00 | 0.57 | 107 | 6882.62 | 1.51 |
| 1B | 688.58 | 4113 | 167.42 | 24 | 28,690.83 | 0.58 | 46 | 14,969.13 | 1.12 |
| 2B | 801.18 | 5516 | 145.25 | 43 | 18,632.09 | 0.78 | 77 | 10,404.94 | 1.4 |
| 3B | 830.14 | 9382 | 88.48 | 40 | 20,753.50 | 0.43 | 92 | 9023.26 | 0.98 |
| 4B | 673.24 | 3222 | 208.95 | 6 | 112,206.67 | 0.19 | 30 | 22,441.33 | 0.93 |
| 5B | 712.96 | 6911 | 103.16 | 33 | 21,604.85 | 0.48 | 81 | 8801.98 | 1.17 |
| 6B | 720.39 | 6382 | 112.88 | 35 | 20,582.57 | 0.55 | 81 | 8893.70 | 1.27 |
| 7B | 750.54 | 3734 | 201 | 12 | 62,545.00 | 0.32 | 35 | 21,444.00 | 0.94 |
| 1D | 495.15 | 1817 | 272.51 | 6 | 82,525.00 | 0.33 | 17 | 29,126.47 | 0.94 |
| 2D | 651.43 | 1552 | 419.74 | 7 | 93,061.43 | 0.45 | 15 | 43,428.67 | 0.97 |
| 3D | 614.95 | 1238 | 496.73 | 2 | 307,475.00 | 0.16 | 13 | 47,303.85 | 1.05 |
| 4D | 508.64 | 1266 | 401.77 | 14 | 36,331.43 | 1.11 | 15 | 33,909.33 | 1.18 |
| 5D | 565.47 | 1703 | 332.04 | 11 | 51,406.36 | 0.65 | 21 | 26,927.14 | 1.23 |
| 6D | 473.51 | 1318 | 359.26 | 147 | 3221.16 | 11.15 | 28 | 16,911.07 | 2.12 |
| 7D | 638.31 | 2050 | 311.37 | 9 | 70,923.33 | 0.44 | 15 | 42,554.00 | 0.73 |
| Isolate of Bgt | Funo | A303 | B303 | Nannong9918 |
|---|---|---|---|---|
| E01 | 4 | 0; | 0; | 0; |
| E05 | 4 | 0; | 0 | 0; |
| E06 | 4 | 0; | 0 | 0; |
| E07 | 4 | 0; | 0 | 0 |
| E09 | 4 | 0 | 0; | 0; |
| E11 | 4 | 0; | 0; | 0; |
| E13 | 4 | 0 | 0; | 0; |
| E15 | 3 | 0; | 2+0; | 0; |
| E16 | 4 | 0 | 0 | 0; |
| E17 | 4 | 0; | 0 | 0; |
| E18 | 4 | 0; | 0; | 0 |
| E20 | 4 | 1 | 0; | 0; |
| E21 | 4 | 0 | 0; | 0; |
| E23-(1) | 4 | 0 | 0; | 0; |
| E23-(2) | 4 | 0 | 0; | 0; |
| E26 | 3 | 0; | 0; | 0 |
| E30-(1) | 4 | 0 | 0 | 0 |
| E30-(2) | 4 | 0; | 0; | 0; |
| E31 | 4 | 0; | 0 | 0; |
| E32 | 4 | 0; | 0; | 0; |
| E49 | 4 | 0; | 0; | 0 |
| E50 | 3 | 0 | 0 | 0; |
| E60 | 4 | 0 | 0 | 0; |
| E69 | 4 | 0; | 0 | 0; |
| Materials | Individuals | PH(cm) | EL(cm) | SN | GNPS | EN | SFN | TGW(g) |
|---|---|---|---|---|---|---|---|---|
| A303 | 30 | 78.55 ± 3.52 | 7.75 ± 0.96 | 19.63 ± 1.03 | 54.87 ± 6.61 | 4.33 ± 1.58 | 0.17 ± 0.38 | 39.67 ± 1.07 |
| p value | ** | ** | ns | ns | ns | ns | ** | |
| B303-1 | 30 | 75.52 ± 3.86 | 7.45 ± 0.86 | 19.20 ± 1.63 | 50.67 ± 7.09 | 4.63 ± 1.43 | 0.63 ± 0.76 | 39.07 ± 0.40 |
| p value | ** | ns | * | * | ns | * | ** | |
| B303-2 | 30 | 75.13 ± 4.67 | 7.11 ± 0.91 | 18.67 ± 1.44 | 52.90 ± 7.54 | 4.80 ± 1.39 | 0.43 ±0.62 | 40.56 ± 0.16 |
| p value | ** | ns | ** | ns | ns | ns | ** | |
| Wan7107 | 30 | 71.60 ± 2.85 | 7.15 ± 0.66 | 20.10 ± 1.52 | 55.67 ± 6.48 | 4.63 ± 1.49 | 0.23 ± 0.43 | 33.78 ± 0.83 |
| Pm97033/ Wan7107 | Individuals | PH | EL | SN | GNPS | FN | EN | SFN | TGW | PM Reaction |
|---|---|---|---|---|---|---|---|---|---|---|
| BC2F3 | 56 | 82.98 ± 6.25 | 6.08 ± 0.51 | 21.19 ± 1.57 | 33.32 ± 8.22 | 43.30 ± 5.79 | 1.96 ± 0.87 | 3.91 ± 1.52 | 44.82 ± 0.18 | R |
| 34 | 78.41 ± 6.15 | 5.75 ± 0.64 | 22.00 ± 1.41 | 26.82 ± 7.25 | 40.47 ± 6.45 | 2.26 ± 0.86 | 6.12 ± 1.71 | 40.48 ± 0.16 | S | |
| p value | ** | ** | * | ** | * | ns | ** | ** | ||
| BC2F4-1 | 22 | 82.50 ± 5.34 | 6.41 ± 0.52 | 21.05 ± 1.13 | 34.9 ± 5.84 | 47.41 ± 6.58 | 2.23 ± 1.15 | 3.5 ± 1.33 | 44.03 ± 0.03 | R |
| 23 | 81.83 ± 3.02 | 6.22 ± 0.44 | 21.95 ± 1.11 | 29.71 ± 6.08 | 43.52 ± 5.52 | 2.48 ± 0.73 | 4.86 ± 1.28 | 40.16 ± 0.20 | S | |
| p value | ns | ns | * | * | * | ns | ** | ** | ||
| BC2F4-2 | 22 | 81.35 ± 6.20 | 6.49 ± 0.61 | 20.23 ± 1.34 | 34.86 ± 9.93 | 45.27 ± 6.36 | 1.90 ± 1.15 | 3.77 ± 1.87 | 43.27 ± 0.73 | R |
| 25 | 78.64 ± 5.90 | 6.26 ± 0.47 | 22.40 ± 1.00 | 31.62 ± 6.10 | 46.20 ± 3.91 | 2.00 ± 0.64 | 4.80 ± 1.52 | 36.66 ± 0.16 | S | |
| p value | ns | ns | ** | ns | ns | ns | * | ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, B.; Ma, X.; Ye, X.; Zhou, Y.; Ma, Y.; Lin, Z. Genotypic and Phenotypic Characterization of Two Triticum aestivum L.—Dasypyrum villosum Translocations Lines in the Same Wheat Genetic Background. Agronomy 2021, 11, 399. https://doi.org/10.3390/agronomy11020399
Wang B, Ma X, Ye X, Zhou Y, Ma Y, Lin Z. Genotypic and Phenotypic Characterization of Two Triticum aestivum L.—Dasypyrum villosum Translocations Lines in the Same Wheat Genetic Background. Agronomy. 2021; 11(2):399. https://doi.org/10.3390/agronomy11020399
Chicago/Turabian StyleWang, Baicui, Xiaolan Ma, Xingguo Ye, Yilin Zhou, Youzhi Ma, and Zhishan Lin. 2021. "Genotypic and Phenotypic Characterization of Two Triticum aestivum L.—Dasypyrum villosum Translocations Lines in the Same Wheat Genetic Background" Agronomy 11, no. 2: 399. https://doi.org/10.3390/agronomy11020399





