Abstract
Powdery mildew is a fungal disease that threatens wheat production throughout the world. Breeding resistant cultivars is an effective way to reduce harm caused by powdery mildew. In this study, 35 wheat-Psathyrostachys huashanica-derived lines were developed by crossing common wheat and P. huashanica Keng (2n = 2x = 14, NsNs) using embryo culture. Resistance to powdery mildew in the derived lines was identified at the seedling and adult stages. Line H5-5-4-2 was selected with immunity to powdery mildew at both growth stages. The chromosome structure of this line was characterized by cytology, genomic in situ hybridization (GISH), and expressed sequence tag-sequence-tagged site (EST-STS) analysis. The chromosome configuration was 2n = 44 = 22II. Two P. huashanica chromosomes with strong hybridization signals were detected by GISH analysis. Among 83 EST-STS markers that covered all seven homologous groups in wheat, three pairs of STS markers, BE497584, BF202643, and BG262410, located in wheat homologous group 1 amplified clear specific bands related to P. huashanica. The results indicated that resistant line H5-5-4-2 was a wheat-P. huashanica 1Ns disomic addition line.
1. Introduction
Wheat (Triticum aestivum L.) is one of the most widely cultivated cereal crops worldwide and at least one third of the global population depends on wheat as the staple food []. Wheat production is threatened by many diseases and powdery mildew caused by Blumeria graminis f. sp. tritici (DC.) Speer (Bgt) is a particularly important foliar disease [,]. Fungicides are often used to control powdery mildew but their widespread application is inappropriate due to high cost, development of resistance in the pathogen, and environmental impacts []. Breeding resistant cultivars is also extremely important and this is the main method employed for effectively controlling powdery mildew in wheat []. However, pathogen populations undergo rapid mutation events and the coevolution with host resistance that cause resistance gene to become ineffective [,]. Therefore, it is necessary to discover more resistance resources that confer resistance to powdery mildew for application in wheat breeding []. Distant hybridization is an effective method for broadening the resistance spectrum by introducing novel resistance genes from wild relatives into the bread wheat gene pool [].
Psathyrostachys huashanica Keng (2n = 2x = 14, NsNs) is a perennial cross-pollination gramineae species discovered on a hillside near Huashan, in the Qingling Mountains of Shaanxi []. As a wild relative of common wheat, P. huashanica is a valuable source of resistance genes for breeding new cultivars, as well as being an endangered and protected species [,]. P. huashanica has attracted substantial attention from breeders owing to its agronomic characteristics, that include early maturity, short height, and drought and salinity tolerance, but especially its resistance to powdery mildew, stripe rust, wheat scab, and other diseases []. Thus, P. huashanica has genetic resources that enhance the tertiary gene pool for Triticum aestivum. [].
Plant breeders in China used P. huashanica × common wheat crosses as early as 1988 to obtain an F1 hybrid with a chromosome number of 28 and a heptaploid hybrid with 2n = 21II + 7I in a first backcross with 7182. A series of wheat-P. huashanica addition lines and substitution lines were then produced, as well as a new intergeneric amphiploid PHw-SA by treating the F1 hybrid from a cross between wheat cv Kaixianluohanmai and P. huashanica with colchicine [,]. Based on these studies, a small fragment wheat–P. huashanica translocation line K-13-835-3 was developed from a BC1F5 population of a cross between PHw-SA and wheat cv CN16, with attractive agronomic traits and high resistance to stripe rust was selected []. Zhang et al. [] conducted random amplified polymorphic DNA (RAPD) analysis using the genomic DNA from P. huashanica and common wheat, and characterized two novel sequence characterized amplified region (SCAR) markers called Psh-D15270 and Psh-F19558, that were present only in the Ns genome.
The use of resistance to powdery mildew found in P. huashanica to produce resistant wheat cultivars was mentioned rarely in previous studies. Therefore, we selected and analyzed the powdery mildew-resistant lines among 35 wheat-P. huashanica-derived lines obtained from the wheat cv 7182 × P. huashanica cross. The aims of the study were: (a) to evaluate the resistance to powdery mildew in 35 wheat-P. huashanica-derived lines; (b) to determine the chromosomal composition and genomic origin of the resistant materials based on cytogenetic observations and genomic in situ hybridization (GISH) analysis; and (c) to analyze the homoeologous relationships between exogenous chromosomes and wheat chromosomes by expressed sequence tag-sequence-tagged site (EST-STS) analysis.
2. Materials and Methods
2.1. Plant Materials
The genetic stocks used in this study comprised winter wheat cv 7182 (AABBDD, 2n = 2x = 42), P. huashanica (NsNs, 2n = 2x = 14), and 35 wheat-P. huashanica derived lines developed from cross 7182 × P. huashanica. The parental wheat cultivar 7182 and P. huashanica were employed as controls for assessing powdery mildew resistance and in the EST-STS analyses. Susceptible wheat cv Mingxian 169 was used as the control in powdery mildew tests, and the 35 lines were evaluated to test their resistance to powdery mildew. Genomic DNA from Chinese Spring was used as a blocker in GISH analyses. All of these plant materials are preserved at the Shaanxi Key Laboratory of Genetic Engineering for Plant Breeding, College of Agronomy, Northwest A&F University, Shaanxi, China.
2.2. Assessment of Powdery Mildew Resistance
The reactions to powdery mildew isolate Bgt E09 at the seedling stage were assessed as described by An et al. [], where wheat cultivar 7182, P. huashanica, Mingxian 169, and 35 derivative wheat cultivars/lines were tested. The plant materials were grown in plug trays in a greenhouse. Plants were inoculated with E09 at the two-leaf stage. When Mingxian 169 exhibited full disease symptoms, the tested lines were evaluated by an infection type (IT) scale from 0–4 (Table 1), where plants with IT = 0, 1, or 2 were considered resistant and these with IT = 3 or 4 were susceptible [,].

Table 1.
Classification standards for powdery mildew in the seedling stage in wheat.
The responses of adult plants to powdery mildew were determined in two replicates grown during 2018–2019 at the Yang Ling Wheat Experimental Station of Northwest A&F University. Resistance to powdery mildew in the field was assessed using a mixture of 30 different Bgt isolates as inocula. Each material was planted in two rows with a length of 1 m and row spacing of 25 cm, and Mingxian 169 were planted around the tested plants as a disease spreader. In the jointing stage, we artificially dusted the spores evenly over the leaves. When Mingxian 169 exhibited fully developed symptoms, reactions were evaluated and recorded on a scale from 0–9, where scores of 0–4 were considered resistant and 5–9 indicated susceptibility []. Moreover, the resistance of each material was also assigned based on the disease index (DI) calculated by 20 randomly selected plants. DI was calculated as [Σ(each disease level × number of diseased plants)/(highest disease level × total number of diseased plants)] × 100 []. The classification standards are shown in Table 2.

Table 2.
Evaluation criteria for resistance to powdery mildew in the wheat adult stage.
2.3. Cytogenetic Analysis
Seeds of the test materials were germinated on wet filter paper in dishes. Seedling root tips were cut and placed in ice water for 24 h and then transferred to ethanol–acetic acid (3:1) for 1 week. Pollen mother cells (PMCs) in metaphase I of meiosis were obtained from young panicles and fixed in anhydrous ethanol-chloroform-glacial acetic acid (6:3:1, v/v). Root tips and PMCs were squashed in 45% acetic acid after staining with 2% acetocarmine for at least 2 h [], and then used for cytological observations and GISH analysis. The prepared slides and observed with an Olympus BX60 microscope (Japan penguin) to assess chromosome structure and counts, and images were captured. The slides were frozen with liquid nitrogen, before removing the cover slips and storing at −20 °C.
2.4. Genomic In Situ Hybridization (GISH) Analysis
GISH was conducted to detect P. huashanica chromatin in the wheat-P. huashanica-derived lines. P. huashanica genomic DNA extracted from fresh leaves by an improved CTAB (Cetyl Trimethylammonium Ammonium Bromide) method [], was labeled with Dig-Nick-Translation Mix/digoxigenin (digoxigenin-11-dUTP, DIG; Roche, Germany) using the nick translation method. The hybridization solution contained 1 μL 10% (w/v) sodium dodecyl sulfate, 1 μL salmon sperm DNA (5 μg/μL), 3 μL probe DNA, 4 μL 20 × SSC solution, 8 μL 50% (w/v) dextran sulfate, and 20 μL deionized formamide, which was made up to a volume 40 μL with 3 μL double distilled H2O. Hybridization for GISH was performed by placing a drop of hybridization solution on a slide with the sample. The slides were then incubated at a temperature of 80 °C for 8 min and 37 °C for 16 h. Next, 60 μL 5% bovine serum albumin was dropped onto the slide, which was incubated at 37 °C for 20 min, before adding 50 μL of Anti-Dig-FITC (Fluorescein Isothiocyanate) for detecting and visualizing the labeled chromosomes. We observed the chromosomes using a fluorescence microscope (Olympus BX60) and photographed images using a Photometrics SenSys CCD camera (the USA).
2.5. Expressed Sequence Tag-Sequence-Tagged Site (EST-STS) Analysis
EST-STS markers were employed to determine the homoeologous relationships among the alien P. huashanica chromosomes in the wheat-P. huashanica lines. We extracted total genomic DNA from the test materials as well as cv 7182 and P. huashanica as described above [,]. In total, 83 EST-STS multiple-loci primer pairs (https://wheat.pw.usda.gov/SNP/new/pcr_primers.shtml) that covered seven wheat homoeologous groups were used for identifying P. huashanica chromosomes in line H5-5-4-2. The polymerase chain reaction (PCR) mixture with a volume of 20 μL contained 2 μL 10 × PCR Buffer, 2 μL (2.5 μmol/mL) primers, 2 μL (40–60 μg/μL) DNA template, 1.6 μL dNTPs (2.5 μmol/mL), 1.6 μL MgCl2 (2.5 mmol/mL), 0.1 μL (5 μmol/μL) Taq polymerase, and 10.7 μL double distilled H2O was compounded for conducting PCR amplification. The procedure comprised initial denaturation for 3 min at 94 °C, followed by 35 cycles of denaturation at 90 °C for 1 min, annealing at 55 °C for 50 s. and a final extension at 72 °C for 1 min. The PCR products were added to 2.5 μL loading buffer, fractionated by non-denatured polyacrylamide gel (8%) electrophoresis, run at 165 V and 120 mA for 2 h, and then visualized in an ultraviolet light box.
3. Results
3.1. Evaluations of Resistance to Powdery Mildew
The seedling reactions to powdery mildew by the 35 wheat-P. huashanica offspring as well as parents 7182 and P. huashanica are displayed in Table 3. Nine materials (25.71%) exhibited resistance to isolate E09 whereas the remainder were moderately or highly susceptible. H5-5-4-2 was selected for further study because of its immunity to powdery mildew. Wheat cv 7182 was moderately resistant, whereas P. huashanica and H5-5-4-2 were immune (Figure 1a).

Table 3.
Seedling reaction to powdery mildew in 35 wheat-P. huashanica-derived lines, their parents, and Mingxian 169 control.

Figure 1.
(a) Responses of H5-5-4-2, its parents 7182 and Psathyrostachys huashanica, and Mingxian 169 to powdery mildew (Bgt) isolate E09 in the seedling stage. (b) Reactions of the same plant materials to 30 mixed Bgt isolates at the adult stage. 1, Psathyrostachys huashanica; 2, H5-5-4-2; 3, 7182; 4, Mingxian 169.
The materials were assessed in the adult stage to determine their resistance to powdery mildew during wheat-growing seasons in 2018–2019. Line H5-5-4-2 and P. huashanica exhibited uniform immunity to the mixture of Bgt isolates. By contrast, wheat cultivar 7182 was moderately resistant to infection and Mingxian169 was highly susceptible (Figure 1b). The IT and DI scores for these four materials were as follows: P. huashanica, IT = 0, DI = 0; H5-5-4-2, IT = 0, DI = 0; line 7182, IT = 4, DI = 6.1; and Mingxian169, IT = 8, DI = 50.12. Therefore, the evaluation results indicated that H5-5-4-2 exhibited great resistance to powdery mildew in both the seedling and adult stages, where its resistance to powdery mildew was probably attributable to the introduction of two chromosomes from P. huashanica. Thus, a series of subsequent studies were conducted using this immune derivative line.
3.2. Cytogenetic Analysis of H5-5-4-2
Microscopic observations of H5-5-4-2 were conducted for root tip cells (RTCs) and PMCs during mitosis metaphase and meiosis metaphase I, respectively, to determine the chromosome number and pairing behaviors. The mitotic observations of H5-5-4-2 indicated that the RTCs had a chromosome number of 2n = 44 (Figure 2a). In addition, the chromosome configuration for the PMCs from H5-5-4-2 in metaphase I was 2n = 22II (Figure 2b).

Figure 2.
Mitotic and meiotic cytological observations of wheat-P. huashanica-derived line H5-5-4-2. (a) Chromosomes in the root tip somatic cells during mitotic metaphase, 2n = 44. (b) Chromosome behavior of pollen mother cells during metaphase I, 2n = 22 II.
3.3. GISH Analysis of H5-5-4-2
Mitotic and meiotic GISH analyses were conducted using the whole genomic DNA of P. huashanica as probe. GISH identification in mitosis demonstrated that H5-5-4-2 contained two added chromosomes with yellow-green hybridization signals, whereas the other 42 wheat chromosomes stained red (Figure 3a). In addition, one ring bivalent with a strong hybridization signal (bright yellow) was observed by GISH at meiotic metaphase I, (Figure 3b). These results demonstrated that two alien chromosomes were introduced into H5-5-4-2 from P. huashanica. Besides, the two alien chromosomes from P. huashanica could form a separate bivalent and undergo normal synapsis, pairing, and segregation in the wheat background. Thus, line H5-5-4-2 was a cytogenetically stable wheat-P. huashanica disomic addition line.
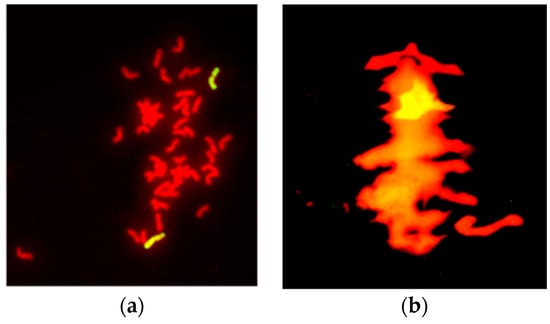
Figure 3.
Genomic in situ hybridization analysis was conducted with root tip cells (RTCs) and pollen mother cells (PMCs) from H5-5-4-2 by using genomic DNA from P. huashanica as a probe and genomes from 7182 as blocker. Fluorescent hybridization signals were exhibited by the alien chromosomes from P. huashanica whereas the wheat chromosomes were counterstained with propidium iodide (red). (a) RTCs during somatic metaphase showing two chromosomes with yellow-green hybridization signals from P. huashanica chromosomes. (b) PMCs in meiotic metaphase I showing a ring bivalent with a fluorescent hybridization signal (bright yellow).
3.4. EST-STS Analysis of H5-5-4-2
In this study, 83 pairs of EST-STS primers distributed throughout all over homoeologous groups were screened for polymorphisms in 7182 and P. huashanica. These polymorphic primers were used to amplify DNA samples from the disomic addition line H5-5-4-2 and its parents 7182 and P. huashanica. Three EST-STS primers, BE497584, BF202643, and BG262410 (Table 4) that mapped to homoeologous group I (1AL, 1AS, 1BL, 1BS, 1DL, and 1DS) amplified clear P. huashanica-specific bands in line H5-5-4-2 but none in the wheat parent 7182 (Figure 4). However, amplification products of other primers produced no specific bands. We inferred that the chromosomes from P. huashanica belonged to homoeologous group I and that the added chromosome pair in H5-5-4-2 could be designated 1Ns.

Table 4.
EST-STS markers from homoeologous group I used to analyze the linkage relationship with the 1Ns chromosome of P. huashanica.
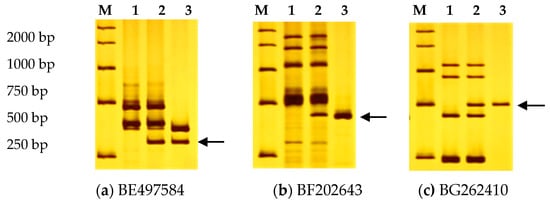
Figure 4.
Identification of the homoeologous group of the alien chromosome pair in H5-5-4-2 and wheat using expressed sequence tag-sequence-tagged site (EST-STS) markers. Lane M, DNA ladder (DL2000); lane 1, 7182; lane 2, H5-5-4-2; lane 3, P. huashanica. Arrows indicate the specific amplification product from P. huashanica.
4. Discussion
Distant hybridization is a major focused research in many laboratories worldwide. It allows genes from related species to be transferred into wheat to improve quality and productivity []. Researchers in China began conducting crosses between wheat with Thinopyrum intermedium and Thinopyrum elongatum in the 1950s, and a number of wheat cultivars were developed []. A wheat–rye 1BL·1RS translocation line was obtained by transferring many resistance genes (e.g., Yr9, Lr26, Pm8, and Sr31) from rye into a wheat background, and it is an outstanding example of genetic improvement in wheat by distant hybridization [,]. Recently, the exploitation of available genes in the wild relatives of wheat has diversified greatly. In particular, An et al. [] obtained a new wheat–rye addition line called WR35 via crossing Xiaoyan 6 with the rye cultivar German White, where the addition line exhibited resistance to powdery mildew and stripe rust, and a high kernel number per spike. Pan et al. [] also found that the 1P chromosome from Agropyron cristatum enhanced the spike length and tiller number in wheat, and a wheat-A. cristatum 1P addition line was obtained using molecular and phenotypic identification techniques.
Crosses between wheat and P. huashanica are relatively recent, and P. huashanica is significant among numerous species related to wheat because of its excellent agronomic characteristics. Several wheat-P. huashanica progeny lines were generated in previous studies, including disomic addition lines [], disomic substitution lines [], and small fragment translocation lines []. In our laboratory, 35 derived lines were developed by crossing common wheat 7182 with P. huashanica via embryo rescue culture and backcrossing, and these intermediate materials with different agronomic traits provide a suitable foundation for exploiting genes from P. huashanica.
Powdery mildew severely hinders yield and quality improvements in wheat and the loss of effective genetic resistance to powdery mildew due to the high variability of pathogens and the uniformity of resistance sources is leading to a crisis in wheat production [,,]. Among the 92 powdery mildew resistance genes (Pm1~Pm65) that have been officially named, many are derived from wild relatives [,,]. Thus, many studies have aimed to identify new resistance genes in the relatives of wheat [,]. In previous studies, the resistance genes Pm7, Pm8, Pm17, and Pm20 were identified in rye, and Pm8 was utilized widely, although the effectiveness of its resistance has now been lost [,,]. Pm21, which is located on the short arm of chromosome 6V in Haynaldia villosa (2n = 14, VV) and it is an excellent gene that confers broad-spectrum resistance to powdery mildew in wheat []. Zhan et al. [] showed that alien chromosome fragments possessed genetic loci with resistance to powdery mildew and stripe rust in wheat–Thinopyrum intermedium translocation lines. In addition, Li et al. [] detected resistance genes in the 2P chromosome in Agropyron cristatum.
The powdery mildew resistance genes in P. huashanica have not been identified in recent studies, and wheat-P. huashanica-derived lines with resistance have rarely been reported, so it is significant to conduct further research into powdery mildew resistance in P. huashanica. In the present study, we examined the seeding responses of 35 wheat-P. huashanica-derived lines to powdery mildew and showed that line H5-5-4-2 was immune (Table 3, Figure 1a). Moreover, the resistance evaluation of H5-5-4-2 in the adult stage was immunity (Figure 1b). Our results suggested that this resistance originated from P. huashanica. We further speculated that this resistance may be a quantitative trait controlled by multiple genes as the derived lines displayed different evaluation of immunity, high resistance, moderate resistance, moderate susceptibility and high susceptibility. These findings provide a great precondition to explore resistance genes in P. huashanica and the development of novel resistance germplasm resources.
The resistance performance of wheat varieties may differ during various growth periods. Comprehensive assessments of disease resistance need to be conducted in both the seedling and adult stage. Varieties with adult resistance mostly exhibit sustained quantitative resistance (horizontal resistance). If a variety exhibits resistance in both the seedling and adult stages, then it will show strong resistance throughout the whole growth period []. In the present study, we found that derivative line H5-5-4-2 not only exhibited high resistance to a mixture of races in the adult stage, but it was also immune to Bgt isolate E09 in the seedling stage. Thus, we speculated that the resistance genes derived from P. huashanica may be the major genes, which may allow this line to exhibit strong broad-spectrum resistance to powdery mildew, thereby providing a resistance gene resource for breeding wheat cultivars with resistance to powdery mildew.
Mitotic and meiotic cytological observations as well as GISH were performed in order to determine the origin of resistance gene in the immune derivative line H5-5-4-2, where the results demonstrated that H5-5-4-2 is a disomic addition line. GISH can simply determine the presence of alien chromosomes but molecular markers are valuable analysis tools for identifying the homoeology of alien chromosomes. Thus, a wheat-Aegilops geniculata 7Mg addition line was identified using EST-STS markers by Wang et al. []. In addition, Wang et al. [] identified a wheat-Leymus racemosus translocation line T5AS-7LrL·7LrS by using EST-STS markers. In the present study, three pairs of primers belonging to homoeologous group I amplified Ns genome-specific bands in H5-5-4-2, thereby indicating the presence of a pair of 1Ns chromosomes derived from P. huashanica, and thus the powdery mildew-resistant derivative line H5-5-4-2 is a wheat-P. huashanica 1Ns disomic addition line.
Wheat disomic addition lines had substantial value in various applications as basic materials for investigating the genetic relationships between wheat and its relatives and gene mapping, they can also be employed as bridge materials for producing substitution lines and translocation lines []. A complete set of 1Ns–7Ns wheat-P. huashanica disomic addition lines was developed by Du et al. []. In addition, in a previous study of wheat-P. huashanica 1Ns disomic addition lines, Zhao et al. [] found that the addition of the 1Ns chromosome from P. huashanica in H9021-28-5 had the effects of improving agronomic traits such as the grain weight, resistance to stripe rust, flour-processing properties, and the contents of some microelements. Moreover, Du et al. [] showed that the 1Ns disomic addition line 12-3 exhibited high resistance to leaf rust, and it possessed large and awnless panicles. However, previous studies have not investigated powdery mildew resistance in wheat–P. huashanica 1Ns disomic addition lines.
Therefore, our successful identification of the wheat-P. huashanica 1Ns chromosome disomic addition line H5-5-4-2 with resistance to powdery mildew may facilitate the breeding of varieties with resistance to powdery mildew, as well as enriching the wheat germplasm resource pool and laying a material foundation for fully utilizing the excellent traits of P. huashanica in wheat resistance breeding. Future studies may focus on the resistance of 1Ns chromosome in P. huashanica with referencing our study to promote the investigation and development of more wheat-P. huashanica 1Ns alien addition lines.
Author Contributions
J.W. (Jun Wu) and X.C. conceived and designed the study; J.H., Y.L. and Q.Z. performed the study; J.L. and Q.Y. contributed new methods or models; C.H. and J.W. (Jinglin Wang) analyzed the date; J.H. and Y.L. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Key Research and Development Project of Shaanxi Province, grant number 2019ZDLNY04-05.
Acknowledgments
We sincerely thank the College of Agronomy, Northwest A&F University, for providing the laboratory of this study. We also truly appreciate Dr. Duncan E. Jackson for his language editing and checking of scientific consistency.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Yang, M.J.; Huang, K.Y.; Han, Q.D. Research progresses on wheat powdery mildew and its resistance. Mol. Plant Breed. 2016, 14, 1244–1254. [Google Scholar] [CrossRef]
- Dean, R.; Van Kan, J.; Pretorius, Z.A.; E Hammond-Kosack, K.; Di Pietro, A.; Spanu, P.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Morgounov, A.; Tufan, H.A.; Sharma, R.; Akin, B.; Bagci, A.; Braun, H.-J.; Kaya, Y.; Keser, M.; Payne, T.S.; Sonder, K.; et al. Global incidence of wheat rusts and powdery mildew during 1969–2010 and durability of resistance of winter wheat variety Bezostaya 1. Eur. J. Plant Pathol. 2011, 132, 323–340. [Google Scholar] [CrossRef]
- Khong, N.G.; Randoux, B.; Tayeh, C.; Coutte, F.; Bourdon, N.; Tisserant, B.; Laruelle, F.; Jacques, P.; Reignault, P. Induction of resistance in wheat against powdery mildew by bacterial cyclic lipopeptides. Commun. Agric. Appl. Boil. Sci. 2012, 77, 39–51. [Google Scholar]
- Tan, C.; Li, G.; Cowger, C.; Carver, B.F.; Xu, X. Characterization of Pm59, a novel powdery mildew resistance gene in Afghanistan wheat landrace PI 181356. Theor. Appl. Genet. 2018, 131, 1145–1152. [Google Scholar] [CrossRef]
- Ma, P.; Xu, H.; Xu, Y.; Li, L.; Qie, Y.; Luo, Q.; Zhang, X.; Li, X.; Zhou, Y.; An, D. Molecular mapping of a new powdery mildew resistance gene Pm2b in Chinese breeding line KM2939. Theor. Appl. Genet. 2015, 128, 613–622. [Google Scholar] [CrossRef]
- McDonald, B.; Linde, C. The population genetics of plant pathogens and breeding strategies for durable resistance. Euphytica 2002, 124, 163–180. [Google Scholar] [CrossRef]
- Hou, L.; Zhang, X.; Li, X.; Jia, J.; Yang, H.; Zhan, H.; Qiao, L.; Guo, H.; Chang, Z. Mapping of Powdery Mildew Resistance Gene pmCH89 in a Putative Wheat-Thinopyrum intermedium Introgression Line. Int. J. Mol. Sci. 2015, 16, 17231–17244. [Google Scholar] [CrossRef]
- Lin, Z.-S.; Cui, Z.-F.; Zeng, X.-Y.; Ma, Y.-Z.; Zhang, Z.-Y.; Nakamura, T.; Ishikawa, G.; Nakamura, K.; Yoshida, H.; Xin, Z.-Y. Analysis of wheat-Thinopyrum intermedium derivatives with BYDV-resistance. Euphytica 2007, 158, 109–118. [Google Scholar] [CrossRef]
- Wang, X.-J.; Chen, X.-H.; Pang, Y.-H.; Jing, F.; Zhang, J.; Hu, S.-Y.; Zan, K.; Wu, J.; Yang, Q.-H.; Zhao, J.-X. Molecular Cytogenetics Identification of a wheat Psathyrostachys huashanica Substitution Line DH2322. Acta Agron. Sin. 2015, 41, 207. [Google Scholar] [CrossRef]
- Baden, C. A taxonomic revision of Psathyrostachys (Poaceae). Nord. J. Bot. 1991, 11, 3–26. [Google Scholar] [CrossRef]
- Du, W.; Wang, J.; Lu, M.; Sun, S.; Chen, X.; Zhao, J.; Yang, Q.; Wu, J. Molecular cytogenetic identification of a wheat–Psathyrostachys huashanica Keng 5Ns disomic addition line with stripe rust resistance. Mol. Breed. 2013, 31, 879–888. [Google Scholar] [CrossRef]
- Han, Y.C.; Wang, C.Y.; Chen, C.H.; Tian, Z.R.; Ji, W.Q. Molecular cytogenetic study on wheat-Psathyrostachys huashanica 1Ns disomic addition line. J. Triticeae Crop. 2015, 35, 1044–1049. [Google Scholar] [CrossRef]
- Ma, D.-F.; Hou, L.; Sun, C.; Zhang, X.; Yin, J.-L.; Guo, Q.-Y.; Zhu, Y.-X. Molecular mapping of stripe rust resistance gene YrH9017 in wheat-Psathyrostachys huashanica introgression line H9017-14-16-5-3. J. Integr. Agric. 2019, 18, 108–114. [Google Scholar] [CrossRef]
- Chen, S.Y.; Hou, W.S.; Zhang, A.J.; Fu, J.; Yang, Q.H. Breeding and cytogenetic study of Triticum aestivum-Psathyrostachys huashanica alien addition lines. Acta Genet. Sin. 1996, 23, 447–452. [Google Scholar]
- Kang, H.Y.; Wang, Y.; Sun, G.L.; Zhang, H.Q.; Fan, X.; Zhou, Y.H. Production and characterization of an amphiploid between common wheat and Psathyrostachys huashanica Keng ex Kuo. Plant Breed. 2009, 128, 36–40. [Google Scholar] [CrossRef]
- Kang, H.-Y.; Zhang, Z.-J.; Xu, L.-L.; Qi, W.-L.; Tang, Y.; Wang, H.; Zhu, W.; Li, D.-Y.; Zeng, J.; Wang, Y.; et al. Characterization of wheat–Psathyrostachys huashanica small segment translocation line with enhanced kernels per spike and stripe rust resistance. Genome 2016, 59, 221–229. [Google Scholar] [CrossRef]
- Zhang, J.; Jiang, Y.; Guo, Y.L.; Li, Y.H.; Wang, Y.; Pu, X. Cloning of Ns genome-specific sequence of Psathyrostachys huashanica and construction of molecular markers. J. Agric. Biotechnol. 2017, 25, 1391–1399. [Google Scholar] [CrossRef]
- An, D.; Zheng, Q.; Zhou, Y.; Ma, P.; Lv, Z.; Li, L.; Li, B.; Luo, Q.; Xu, H.; Xu, Y. Molecular cytogenetic characterization of a new wheat–rye 4R chromosome translocation line resistant to powdery mildew. Chromosom. Res. 2013, 21, 419–432. [Google Scholar] [CrossRef]
- Yu, X.; Ren, S.; Zhao, L.; Guo, J.; Bao, Y.; Ma, Y.; Wang, H.; Ohm, H.W.; Yu, D.; Li, H.; et al. Molecular mapping of a novel wheat powdery mildew resistance gene Ml92145E8-9 and its application in wheat breeding by marker-assisted selection. Crop. J. 2018, 6, 621–627. [Google Scholar] [CrossRef]
- Zeng, F.-S.; Yang, L.-J.; Gong, S.-J.; Shi, W.-Q.; Zhang, X.-J.; Wang, H.; Xiang, L.-B.; Xue, M.-F.; Yu, D.-Z. Virulence and Diversity of Blumeria graminis f. sp. tritici Populations in China. J. Integr. Agric. 2014, 13, 2424–2437. [Google Scholar] [CrossRef]
- Sheng, B.Q.; Duan, X.Y. Improvement of scale 0-9 method for scoring adult plant resistance to powdery mildew of wheat. J. Beijing Agric. Sci. 1991, 1, 38–39. [Google Scholar]
- Yan, Z.Y.; Fan, J.R.; Liu, W.; Zhou, Y.L. Models of disease index estimation of wheat powdery mildew based on the concentrations of Blumeria graminis f. sp. tritici conidia in the fields. Acta Phytopathol. Sin. 2017, 47, 253–261. [Google Scholar] [CrossRef]
- Tang, X.; Shi, D.; Xu, J.; Li, Y.; Li, W.; Ren, Z.; Fu, T. Molecular cytogenetic characteristics of a translocation line between common wheat and Thinopyrum intermedium with resistance to powdery mildew. Euphytica 2014, 197, 201–210. [Google Scholar] [CrossRef]
- Cota-Sánchez, J.H.; Remarchuk, K.; Ubayasena, K. Ready-to-use DNA extracted with a CTAB method adapted for herbarium specimens and mucilaginous plant tissue. Plant Mol. Boil. Rep. 2006, 24, 161–167. [Google Scholar] [CrossRef]
- Lee, M.H.; Nicholson, P. Isolation of genomic DNA from plant tissues. Nat. Biotechnol. 1997, 15, 805–806. [Google Scholar] [CrossRef]
- An, D.; Xu, H.; Xu, Y.-F. Enhancement of wheat distant hybridization germplasm. Chin. J. Eco-Agric. 2012, 19, 1011–1019. [Google Scholar] [CrossRef]
- Dong, Y.C. Wheat Breeding through Distant Crossing. In Proceedings of the International Symposium on Genetics and Breeding of Wheat, Zhengzhou, Henan, China, 9–11 May 2001. [Google Scholar]
- Howell, T.; Hale, I.; Jankuloski, L.; Bonafede, M.; Gilbert, M.; Dubcovsky, J. Mapping a region within the 1RS.1BL translocation in common wheat affecting grain yield and canopy water status. Theor. Appl. Genet. 2014, 127, 2695–2709. [Google Scholar] [CrossRef]
- Ren, T.-H.; Chen, F.; Yan, B.-J.; Zhang, H.-Q.; Ren, Z.-L. Genetic diversity of wheat–rye 1BL.1RS translocation lines derived from different wheat and rye sources. Euphytica 2011, 183, 133–146. [Google Scholar] [CrossRef]
- An, D.; Ma, P.; Zheng, Q.; Fu, S.; Li, L.; Han, F.; Han, G.; Wang, J.; Xu, Y.; Jin, Y.; et al. Development and molecular cytogenetic identification of a new wheat-rye 4R chromosome disomic addition line with resistances to powdery mildew, stripe rust and sharp eyespot. Theor. Appl. Genet. 2018, 132, 257–272. [Google Scholar] [CrossRef]
- Pan, C.; Li, Q.; Lu, Y.; Zhang, J.; Yang, X.; Li, X.; Li, L.; Liu, W. Chromosomal localization of genes conferring desirable agronomic traits from Agropyron cristatum chromosome 1P. PLoS ONE 2017, 12, e0175265. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Wang, J.; Lu, M.; Sun, S.; Chen, X.; Zhao, J.; Yang, Q.; Wu, J. Characterization of a wheat-Psathyrostachys huashanica Keng 4Ns disomic addition line for enhanced tiller numbers and stripe rust resistance. Planta 2013, 239, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yao, X.; Yang, Z.; Cheng, X.; Yuan, F.; Liu, Y.; Wu, J.; Yang, Q.; Zhao, J.; Chen, X. Molecular cytogenetic characterization of a novel wheat–Psathyrostachys huashanica Keng 5Ns (5D) disomic substitution line with stripe rust resistance. Mol. Breed. 2019, 39, 109. [Google Scholar] [CrossRef]
- Cowger, C.; Miranda, L.; Griffey, C.; Hall, M.; Murphy, J.P.; Maxwell, J. Wheat powdery mildew. In Disease Resistance in Wheat; CABI Publishing: Wallingford, UK, 2012; pp. 84–119. [Google Scholar]
- He, F.; Bao, Y.; Qi, X.; Ma, Y.; Li, X.; Wang, H. Molecular cytogenetic identification of a wheat–Thinopyrum ponticum translocation line resistant to powdery mildew. J. Genet. 2017, 96, 165–169. [Google Scholar] [CrossRef]
- Xu, X.-D.; Feng, J.; Fan, J.; Liu, Z.; Li, Q.; Zhou, Y.; Ma, Z.-H. Identification of the resistance gene to powdery mildew in Chinese wheat landrace Baiyouyantiao. J. Integr. Agric. 2018, 17, 37–45. [Google Scholar] [CrossRef]
- Li, G.; Cowger, C.; Wang, X.; Carver, B.F.; Xu, X. Characterization of Pm65, a new powdery mildew resistance gene on chromosome 2AL of a facultative wheat cultivar. Theor. Appl. Genet. 2019, 132, 2625–2632. [Google Scholar] [CrossRef]
- Tan, C.; Li, G.; Cowger, C.; Carver, B.F.; Xu, X. Characterization of Pm63, a powdery mildew resistance gene in Iranian landrace PI 628024. Theor. Appl. Genet. 2018, 132, 1137–1144. [Google Scholar] [CrossRef]
- Zhang, D.; Zhu, K.; Dong, L.; Liang, Y.; Li, G.; Fang, T.; Guo, G.; Wu, Q.; Xie, J.; Chen, Y.; et al. Wheat powdery mildew resistance gene Pm64 derived from wild emmer (Triticum turgidum var. dicoccoides) is tightly linked in repulsion with stripe rust resistance gene Yr5. Crop. J. 2019, 7, 761–770. [Google Scholar] [CrossRef]
- Grewal, S.; Yang, C.; Hubbart-Edwards, S.; Scholefield, D.; Ashling, S.; Burridge, A.; King, I.P.; King, J. Characterisation of Thinopyrum bessarabicum chromosomes through genome-wide introgressions into wheat. Theor. Appl. Genet. 2017, 131, 389–406. [Google Scholar] [CrossRef]
- Rahmatov, M.; Rouse, M.N.; Nirmala, J.; Danilova, T.; Friebe, B.; Steffenson, B.J.; Johansson, E. A new 2DS·2RL Robertsonian translocation transfers stem rust resistance gene Sr59 into wheat. Theor. Appl. Genet. 2016, 129, 1383–1392. [Google Scholar] [CrossRef]
- An, D.; Zheng, Q.; Luo, Q.; Ma, P.; Zhang, H.; Li, L.; Han, F.; Xu, H.; Xu, Y.; Zhang, X.; et al. Molecular Cytogenetic Identification of a New Wheat-Rye 6R Chromosome Disomic Addition Line with Powdery Mildew Resistance. PLoS ONE 2015, 10, e0134534. [Google Scholar] [CrossRef] [PubMed]
- Friebe, B.; Heun, M.; Tuleen, N.; Zeller, F.J.; Gill, B.S. Cytogenetically Monitored Transfer of Powdery Mildew Resistance from Rye into Wheat. Crop. Sci. 1994, 34, 621–625. [Google Scholar] [CrossRef]
- Zhan, H.; Zhang, X.; Li, G.; Pan, Z.; Hu, J.; Li, X.; Qiao, L.; Jia, J.; Guo, H.; Chang, Z.; et al. Molecular Characterization of a New Wheat-Thinopyrum intermedium Translocation Line with Resistance to Powdery Mildew and Stripe Rust. Int. J. Mol. Sci. 2015, 16, 2162–2173. [Google Scholar] [CrossRef] [PubMed]
- Xing, L.; Hu, P.; Liu, J.; Witek, K.; Zhou, S.; Xu, J.; Zhou, W.; Gao, L.; Huang, Z.; Zhang, R.; et al. Pm21 from Haynaldia villosa Encodes a CC-NBS-LRR Protein Conferring Powdery Mildew Resistance in Wheat. Mol. Plant 2018, 11, 874–878. [Google Scholar] [CrossRef]
- Li, Q.; Lu, Y.; Pan, C.; Zhang, J.; Liu, W.; Yang, X.; Li, X.; Xi, Y.; Li, L. Characterization of a Novel Wheat-Agropyron cristatum 2P Disomic Addition Line with Powdery Mildew Resistance. Crop. Sci. 2016, 56, 2390–2400. [Google Scholar] [CrossRef]
- Wang, Z.L.; Li, M.X.; Xi, Y.J.; Liu, S.D. Appraisal of Resistance to Powdery Mildew and Molecular Marker Analysis of Pm4b of Wheat Varieties (Lines). Acta Agric. Boreal.-Sin. 2014, 29, 56–61. [Google Scholar] [CrossRef]
- Wang, Y.; Quan, W.; Peng, N.; Wang, C.; Yang, X.; Liu, X.; Zhang, H.; Chen, C.; Ji, W. Molecular cytogenetic identification of a wheat–Aegilops geniculata Roth 7Mg disomic addition line with powdery mildew resistance. Mol. Breed. 2016, 36. [Google Scholar] [CrossRef]
- Wang, L.-S.; Zhang, Y.-L.; Nan, G.-H. Molecular and Cytogenetic Identification of Triticum aestivum-Leymus racemosus Translocation Line T5AS-7LrL·7LrS. Acta Agron. Sin. 2018, 44, 1442–1447. [Google Scholar] [CrossRef]
- Du, W.; Wang, J.; Pang, Y.; Wu, J.; Zhao, J.; Liu, S.; Yang, Q.; Chen, X. Development and application of PCR markers specific to the 1Ns chromosome of Psathyrostachys huashanica Keng with leaf rust resistance. Euphytica 2014, 200, 207–220. [Google Scholar] [CrossRef]
- Zhao, J.X.; Wu, J.; Cheng, X.N.; Dong, J.; Chen, X.H.; Liu, S.H.; Du, W.L.; Pang, Y.Y.; Yang, Q.H.; Ji, W.Q. Agronomic and quality traits of a wheat-Psathyrostachys huashanica 1Ns disomic addition line. Acta Agron. Sin. 2010, 36, 1610–1614. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).