Nucleobase-Containing Polymers: Structure, Synthesis, and Applications
Abstract
:1. Introduction
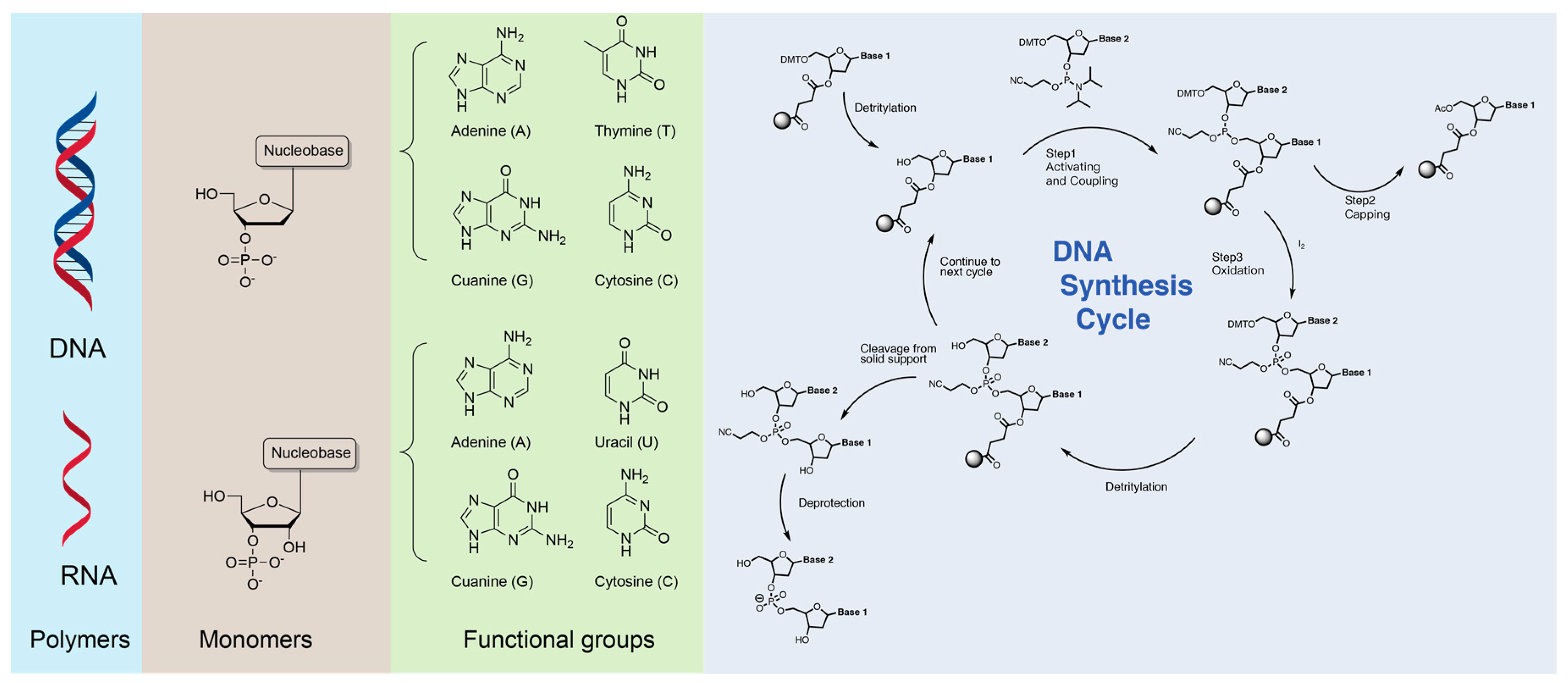
2. Nucleobase-Containing Polymers in Nature
3. Synthetic Nucleobase-Containing Polymers
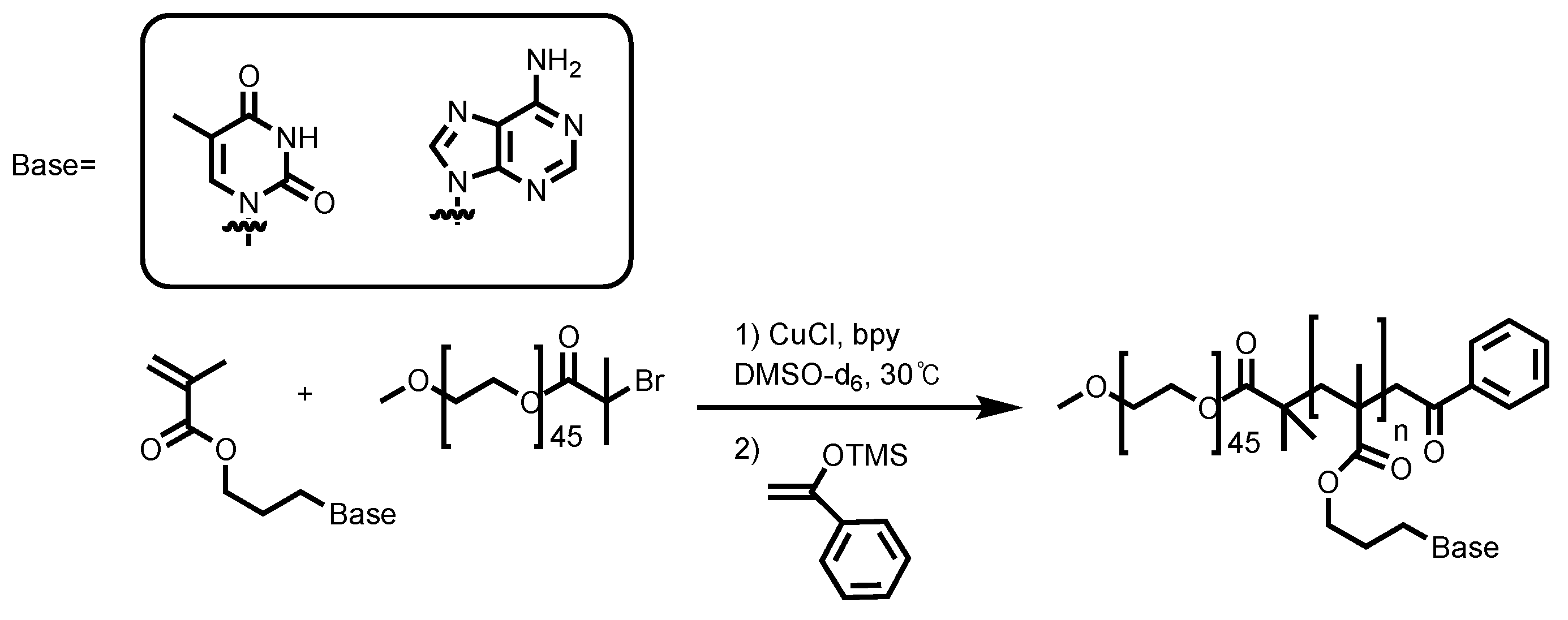
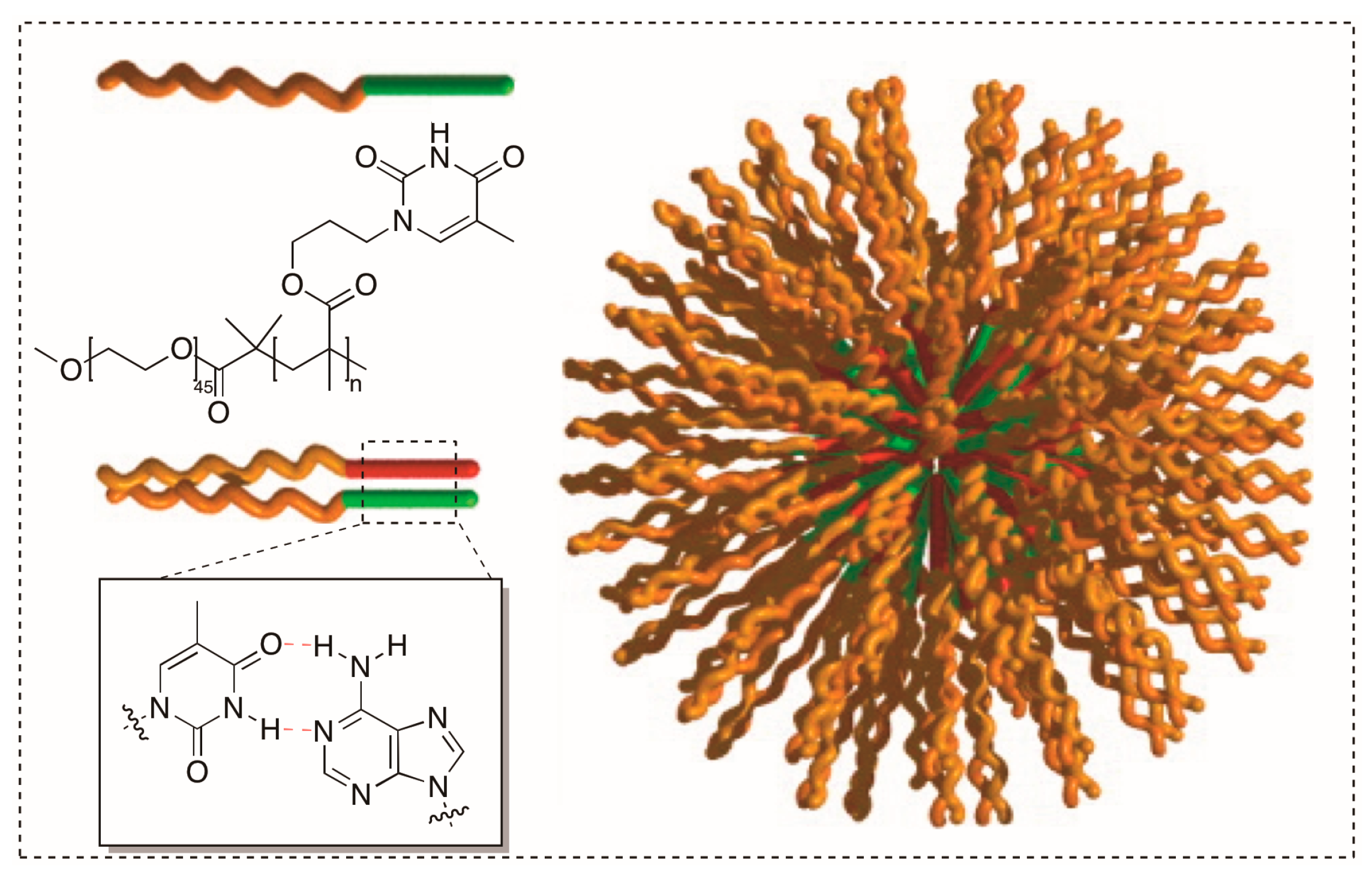
3.1. Synthesizing Nucleobase Polymers by ATRP
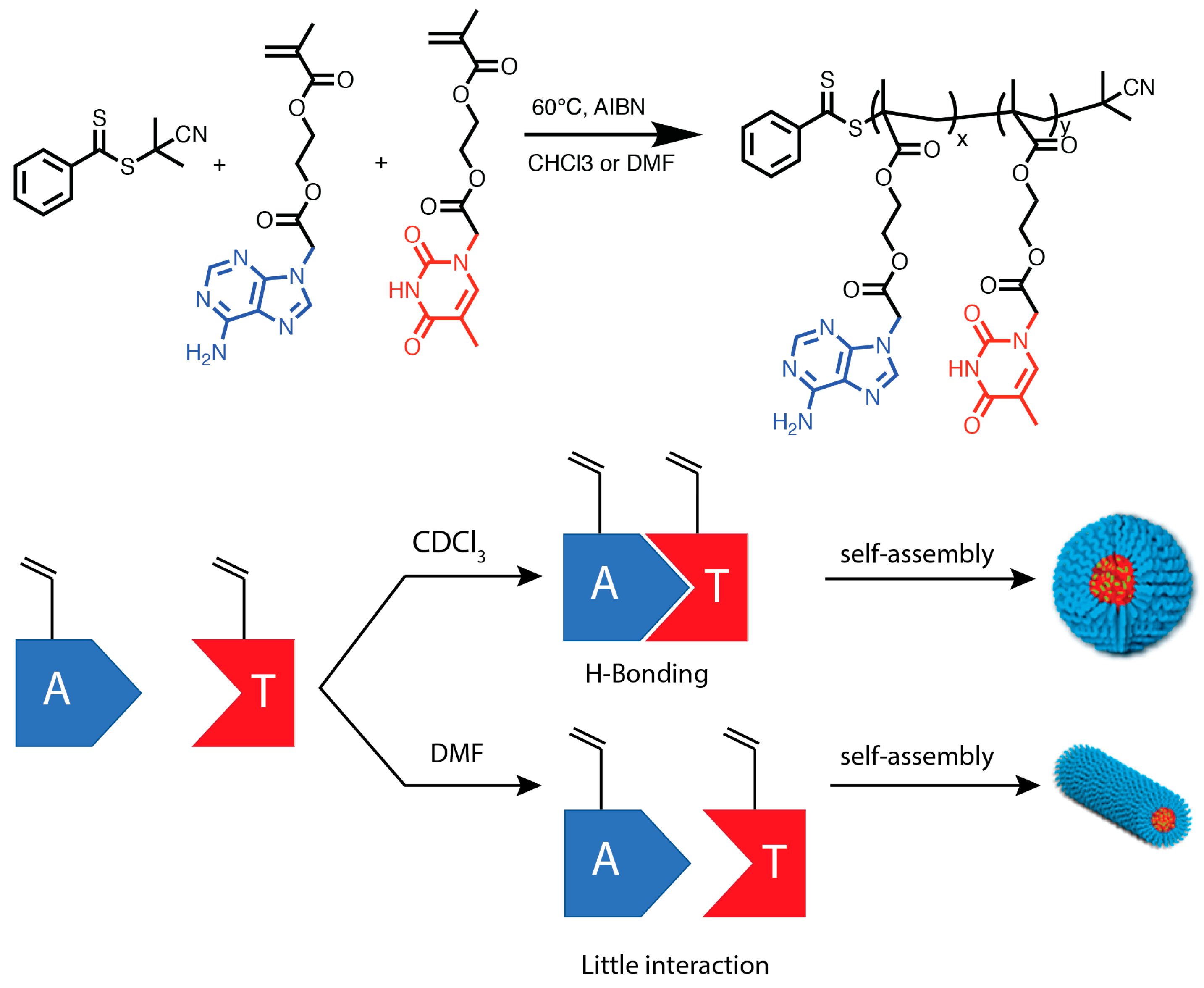
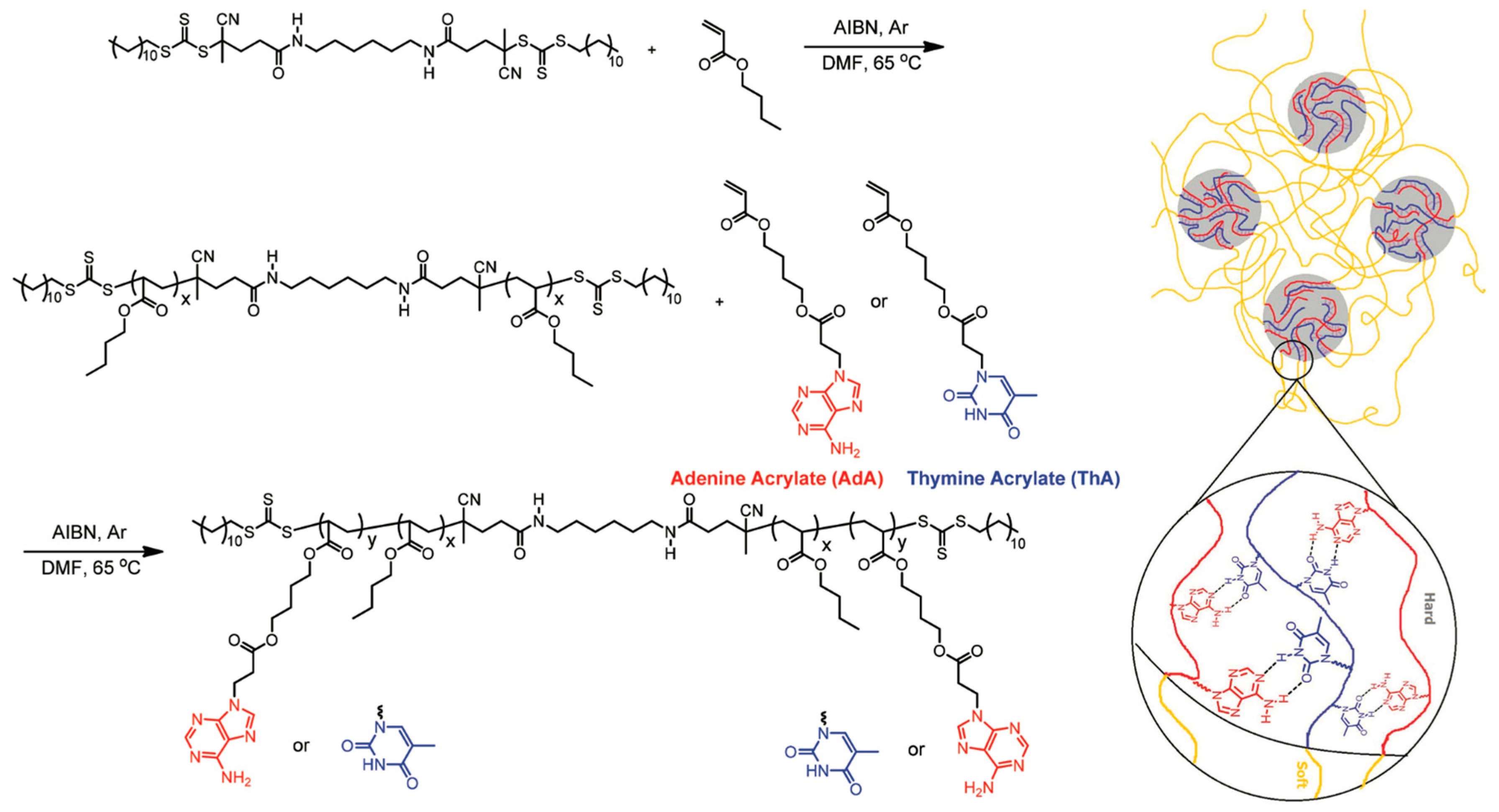
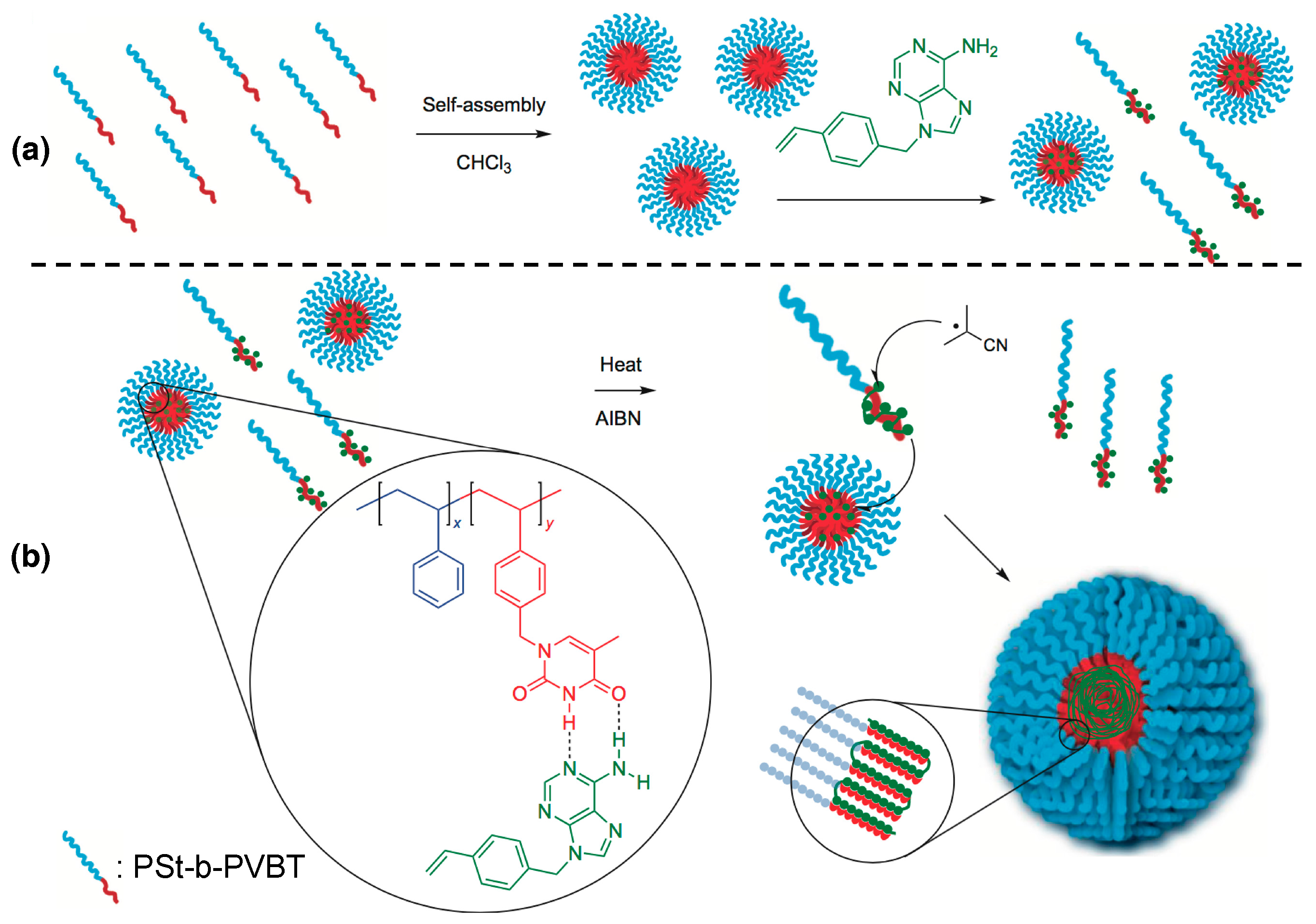
3.2. Synthesizing Nucleobase Polymers by RAFT
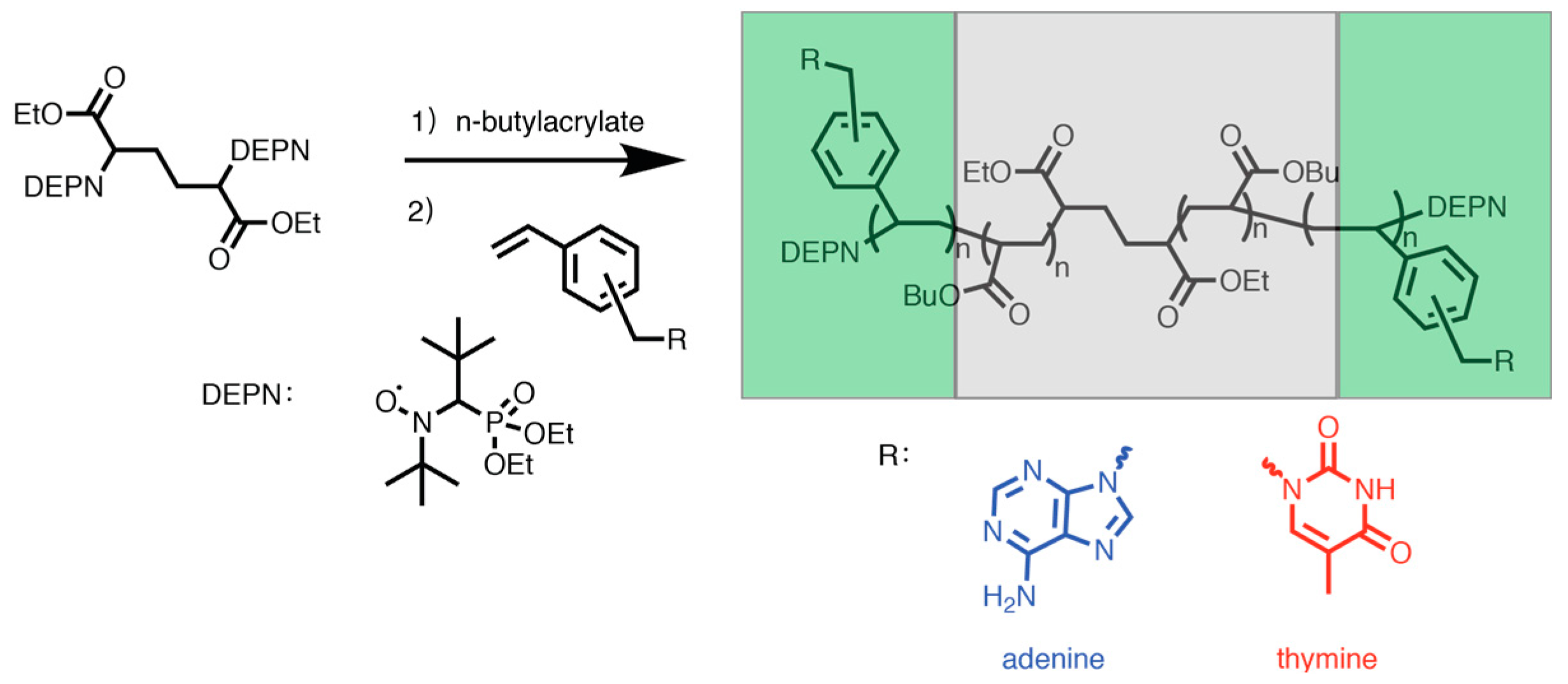
3.3. Synthesizing Nucleobase Polymers by NMP
3.4. Synthesizing Nucleobase Polymers by Conventional Radical Polymerization
3.5. Synthesizing Nucleobase Polymers by Click Chemistry
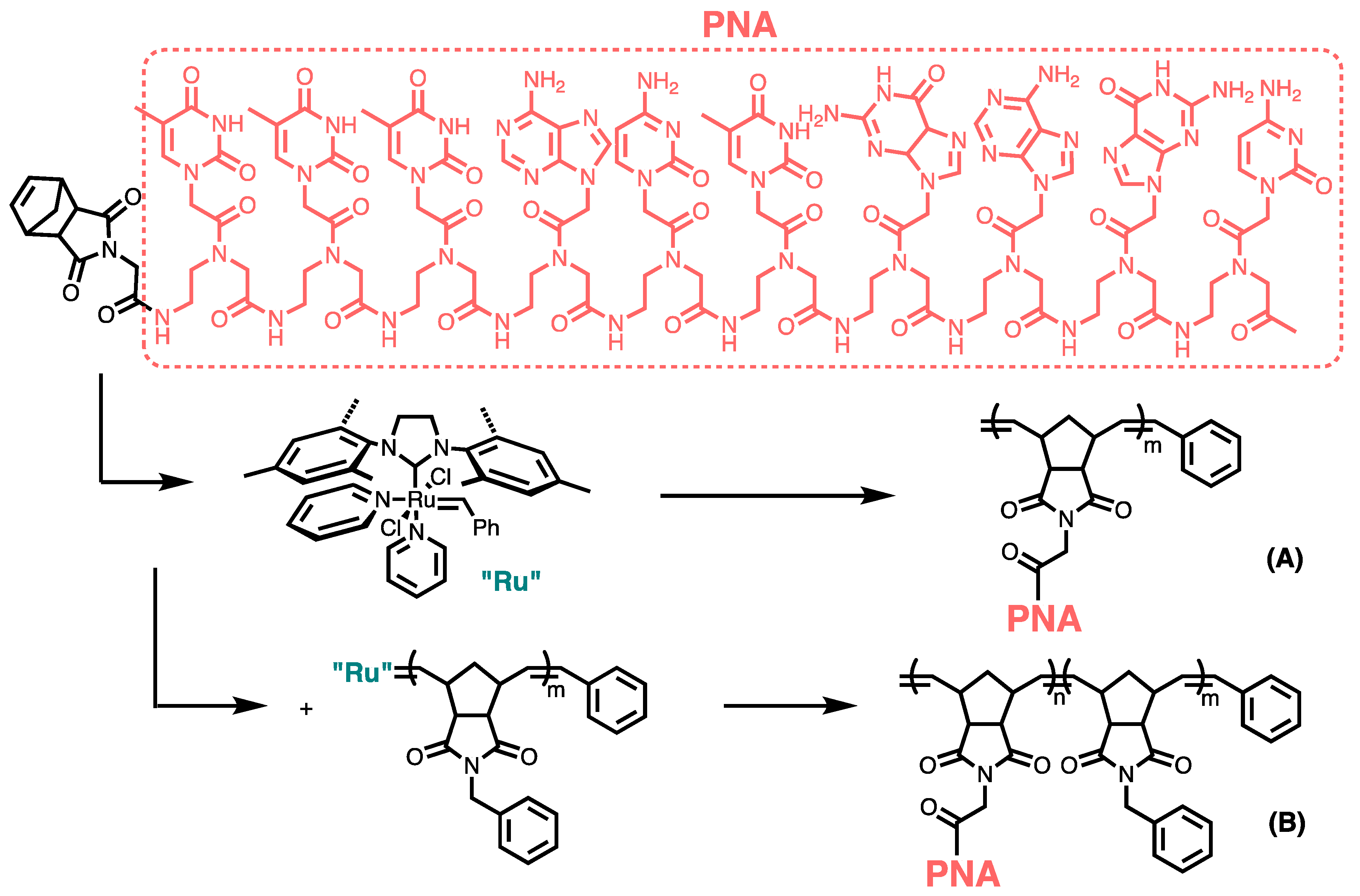
3.6. Synthesizing Nucleobase Polymers by ROMP
3.7. Synthesizing Nucleobase Polymers by Other Polymerizations
4. Applications of Nucleobase-Containing Polymers in Materials Science
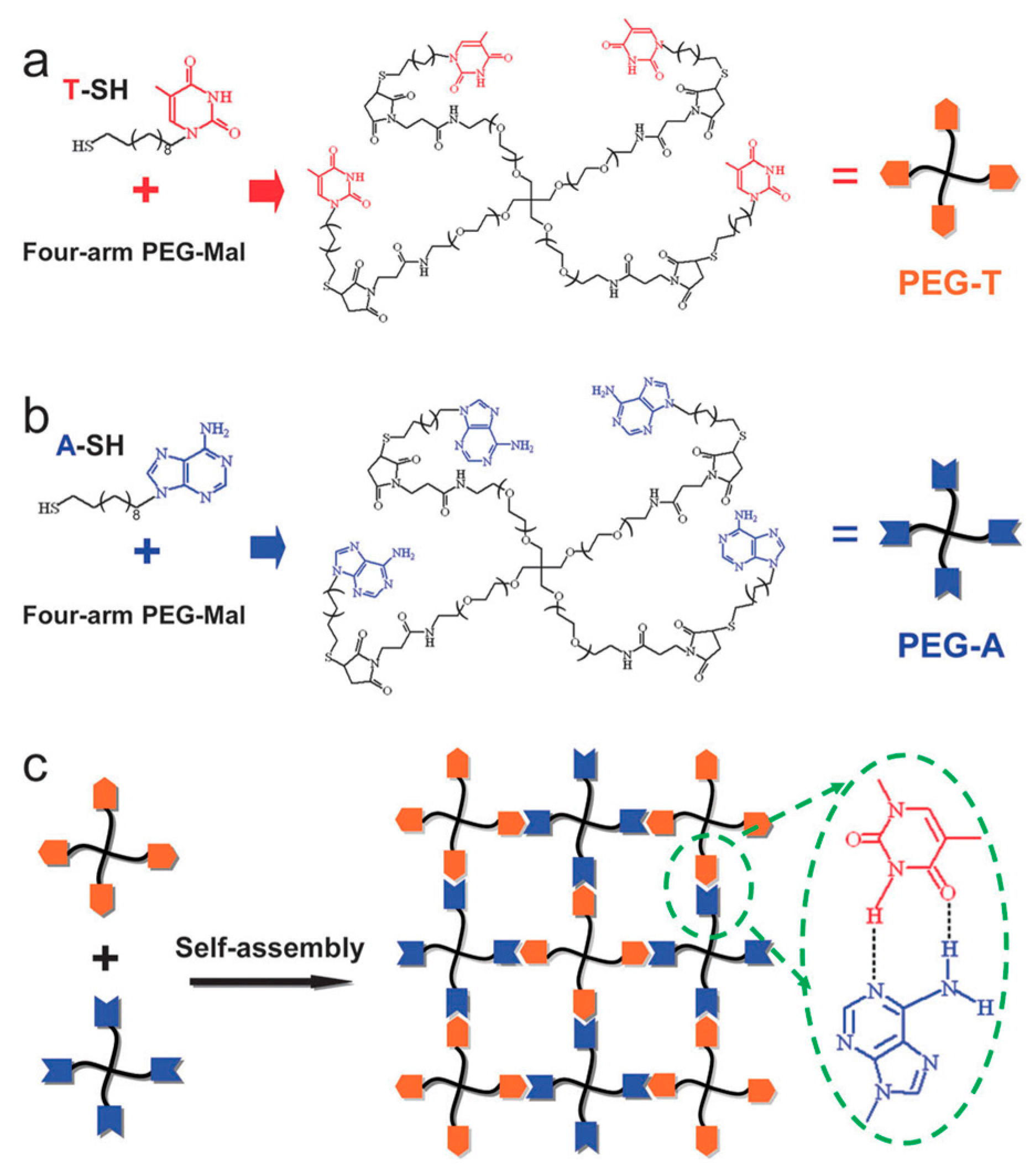
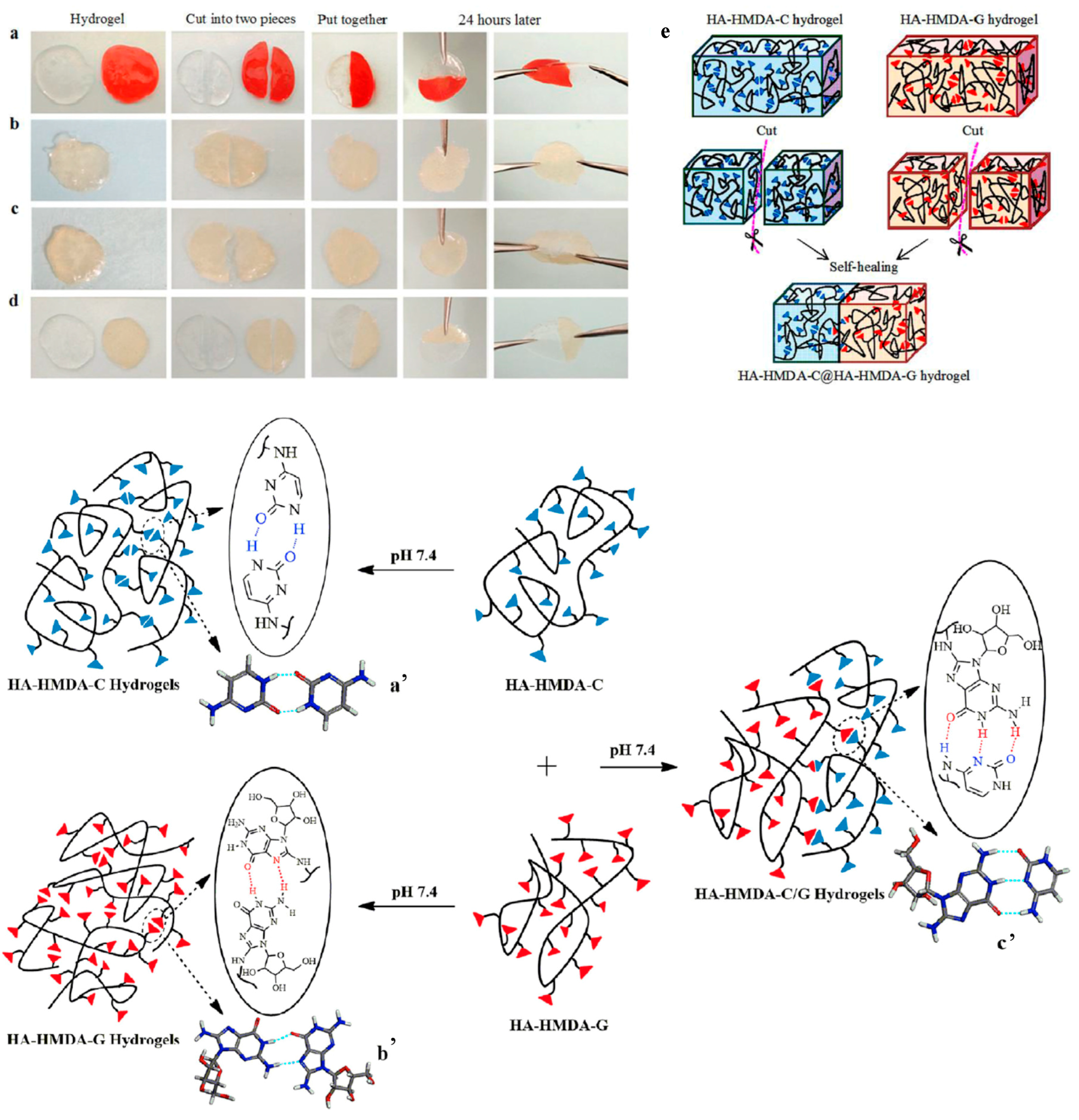
4.1. Hydrogels
4.2. DNA Delivery
4.3. Self-Healing Materials
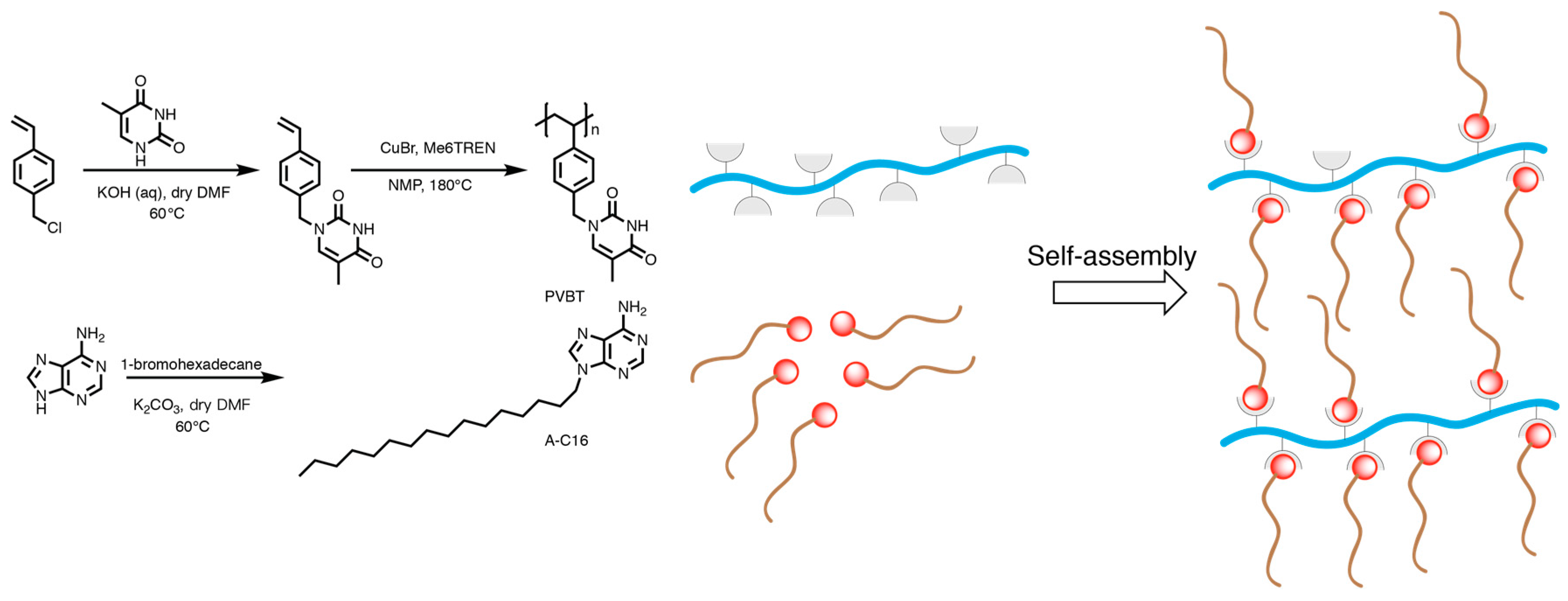
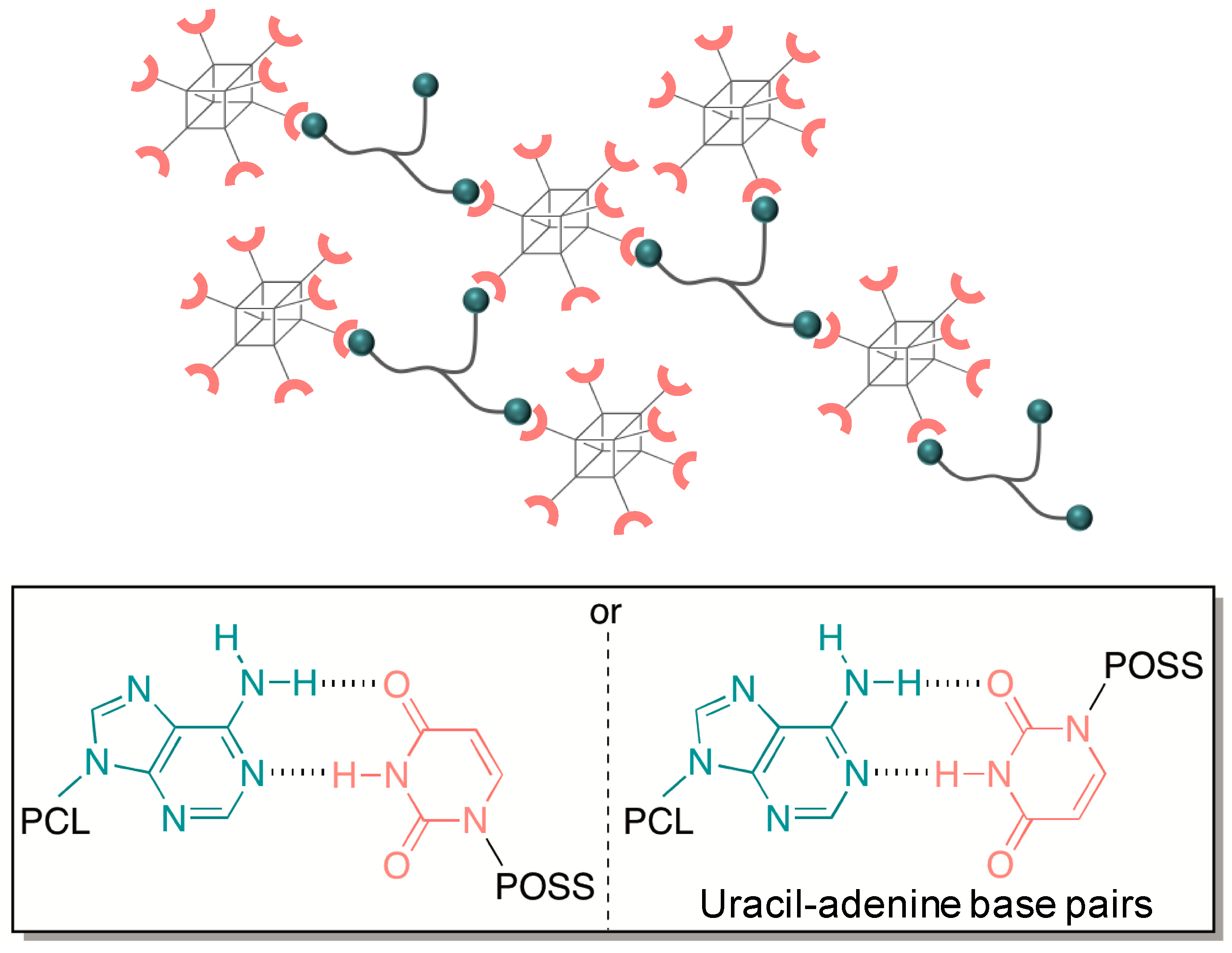
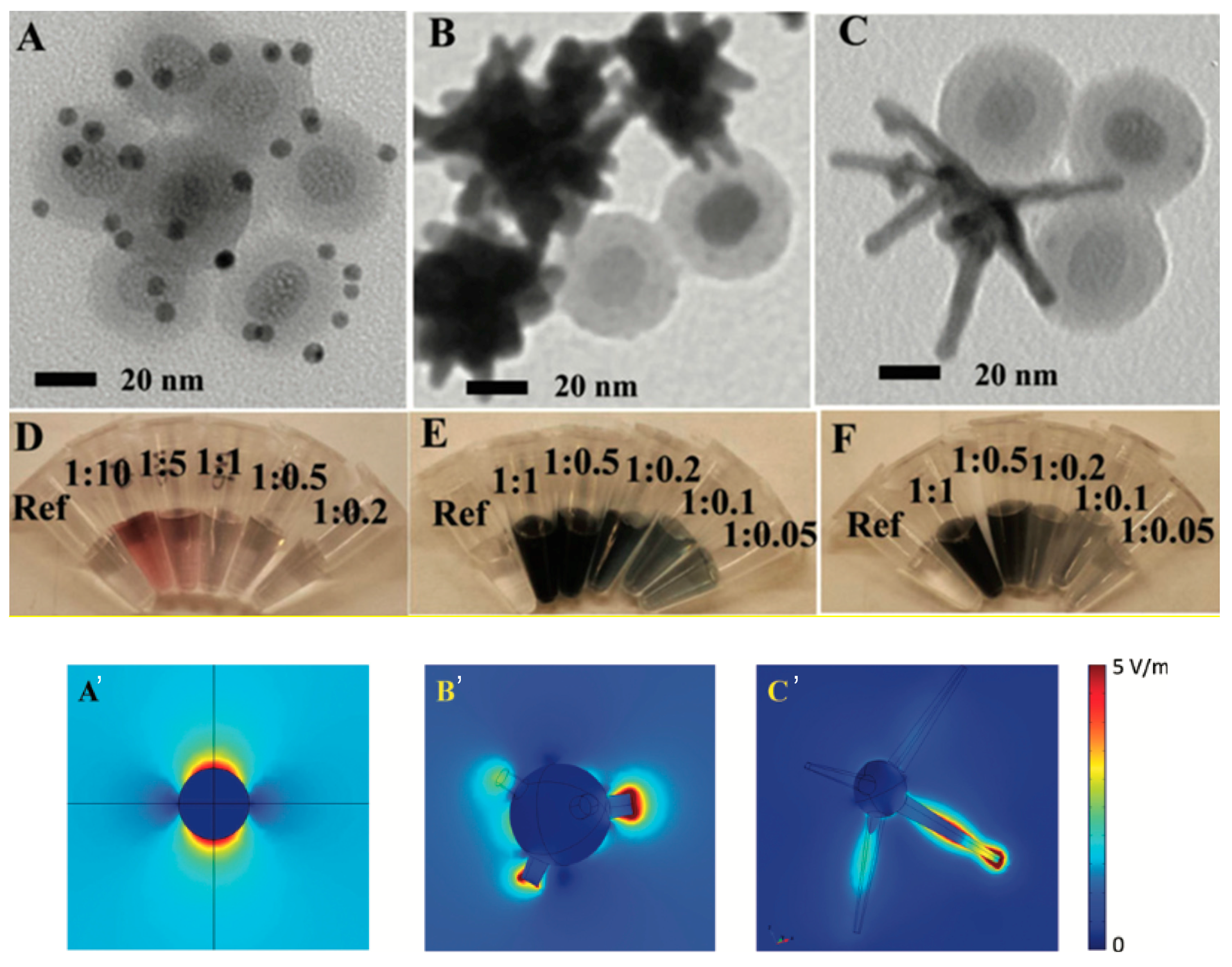
4.4. Self-Assembly of Nanoparticles
4.5. Metal–Nucleobase Coordination
5. Conclusions and Outlook
Author Contributions
Conflicts of Interest
References
- Nykypanchuk, D.; Maye, M.M.; van der Lelie, D.; Gang, O. DNA-guided crystallization of colloidal nanoparticles. Nature 2008, 451, 549–552. [Google Scholar] [CrossRef] [PubMed]
- Tkachenko, A.V. Morphological diversity of DNA-colloidal self-assembly. Phys. Rev. Lett. 2002, 89, 148303. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Harris, N.C.; Kiang, C.H. The reversible phase transition of DNA-linked colloidal gold assemblies. Phys. A Stat. Mach. Appl. 2005, 354, 1–9. [Google Scholar] [CrossRef]
- Leunissen, M.E.; Dreyfus, R.; Sha, R.J.; Wang, T.; Seeman, N.C.; Pine, D.J.; Chaikin, P.M. Towards self-replicating materials of DNA-functionalized colloids. Soft Matter 2009, 5, 2422–2430. [Google Scholar] [CrossRef]
- Rothemund, P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Hong, F.; Zhang, F.; Liu, Y.; Yan, H. DNA origami: Scaffolds for creating higher order structures. Chem. Rev. 2017, 117, 12584–12640. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.G.; Meyer, T.; Shih, W.M.; Bellot, G. Regulation at a distance of biomolecular interactions using a DNA origami nanoactuator. Nat. Commun. 2016, 7, 10935. [Google Scholar] [CrossRef] [PubMed]
- Marini, M.; Piantanida, L.; Musetti, R.; Bek, A.; Dong, M.D.; Besenbacher, F.; Lazzarino, M.; Firrao, G. A revertible, autonomous, self-assembled DNA-origami nanoactuator. Nano Lett. 2011, 11, 5449–5454. [Google Scholar] [CrossRef] [PubMed]
- Malashikhina, N.; Pavlov, V. DNA-decorated nanoparticles as nanosensors for rapid detection of ascorbic acid. Biosens. Bioelectron. 2012, 33, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Morales, J.M.; Skipwith, C.G.; Clark, H.A. Quadruplex integrated DNA (quid) nanosensors for monitoring dopamine. Sensors 2015, 15, 19912–19924. [Google Scholar] [CrossRef] [PubMed]
- Stougaard, M.; Ho, Y.P. DNA-based nanosensors for next-generation clinical diagnostics via detection of enzyme activity. Expert Rev. Mol. Diagn. 2014, 14, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Walsh, R.; Morales, J.M.; Skipwith, C.G.; Ruckh, T.T.; Clark, H.A. Enzyme-linked DNA dendrimer nanosensors for acetylcholine. Sci. Rep. 2015, 5, 14832. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Lu, D.Q.; Liang, H.; Xie, S.; Luo, C.; Hu, M.; Xu, L.; Zhang, X.; Tan, W. Fluorescence resonance energy transfer-based DNA tetrahedron nanotweezer for highly reliable detection of tumor-related mRNA in living cells. ACS Nano 2017, 11, 4060–4066. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Cheng, W.; Li, Y.; Xu, Y.; Li, X.; Yin, Y.; Ju, H.; Ding, S. Catalytic hairpin assembly actuated DNA nanotweezer for logic gate building and sensitive enzyme-free biosensing of microRNAs. Anal. Chem. 2016, 88, 7500–7506. [Google Scholar] [CrossRef] [PubMed]
- Lechardeur, D.; Verkman, A.S.; Lukacs, G.L. Intracellular routing of plasmid DNA during non-viral gene transfer. Adv. Drug Deliv. Rev. 2005, 57, 755–767. [Google Scholar] [CrossRef] [PubMed]
- Pack, D.W.; Hoffman, A.S.; Pun, S.; Stayton, P.S. Design and development of polymers for gene delivery. Nat. Rev. Drug Discov. 2005, 4, 581–593. [Google Scholar] [CrossRef] [PubMed]
- McHale, R.; O’Reilly, R.K. Nucleobase containing synthetic polymers: Advancing biomimicry via controlled synthesis and self-assembly. Macromolecules 2012, 45, 7665–7675. [Google Scholar] [CrossRef]
- Inaki, Y. Synthetic nucleic-acid analogs. Prog. Polym. Sci. 1992, 17, 515–570. [Google Scholar] [CrossRef]
- Smith, W.T. Nucleic acid models. Prog. Polym. Sci. 1996, 21, 209–253. [Google Scholar] [CrossRef]
- Watson, J.D.; Crick, F.H.C. Molecular structure of nucleic acids—A structure for deoxyribose nucleic acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef] [PubMed]
- Beaucage, S.L.; Iyer, R.P. Advances in the synthesis of oligonucleotides by the phosphoramidite approach. Tetrahedron 1992, 48, 2223–2311. [Google Scholar] [CrossRef]
- Reese, C.B. Oligo- and poly-nucleotides: 50 years of chemical synthesis. Org. Biomol. Chem. 2005, 3, 3851–3868. [Google Scholar] [CrossRef] [PubMed]
- Kosuri, S.; Church, G.M. Large-scale de novo DNA synthesis: Technologies and applications. Nat. Methods 2014, 11, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Navarro, L.A.; Chilkoti, A.; Zauscher, S. High-molecular-weight polynucleotides by transferase-catalyzed living chain-growth polycondensation. Angew. Chem. Int. Ed. 2017, 56, 6778–6782. [Google Scholar] [CrossRef] [PubMed]
- Spijker, H.; Van Delft, F.L.; Van Hest, J.C.M. Atom transfer radical polymerization of adenine, thymine, cytosine, and guanine nucleobase monomers. Macromolecules 2007, 40, 12–18. [Google Scholar] [CrossRef]
- Spijker, H.J.; Dirks, A.J.; van Hest, J.C.M. Synthesis and assembly behavior of nucleobase-functionalized block copolymers. J. Polym. Sci. Part A Polym. Chem. 2006, 44, 4242–4250. [Google Scholar] [CrossRef]
- Garcia, M.; Beecham, M.P.; Kempe, K.; Haddleton, D.M.; Khan, A.; Marsh, A. Water soluble triblock and pentablock poly(methacryloyl nucleosides) from copper-mediated living radical polymerisation using peg macroinitiators. Eur. Polym. J. 2015, 66, 444–451. [Google Scholar] [CrossRef]
- McHale, R.; Patterson, J.P.; Zetterlund, P.B.; O’Reilly, R.K. Biomimetic radical polymerization via cooperative assembly of segregating templates. Nat. Chem. 2012, 4, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Pitto-Barry, A.; Willcock, H.; Quan, W.-D.; Kirby, N.; Sanchez, A.M.; O’Reilly, R.K. Exploiting nucleobase-containing materials—From monomers to complex morphologies using raft dispersion polymerization. Polym. Chem. 2015, 6, 106–117. [Google Scholar] [CrossRef]
- Hua, Z.; Pitto-Barry, A.; Kang, Y.; Kirby, N.; Wilks, T.R.; O’Reilly, R.K. Micellar nanoparticles with tuneable morphologies through interactions between nucleobase-containing synthetic polymers in aqueous solution. Polym. Chem. 2016, 7, 4254–4262. [Google Scholar] [CrossRef]
- Zhang, K.; Aiba, M.; Fahs, G.B.; Hudson, A.G.; Chiang, W.D.; Moore, R.B.; Ueda, M.; Long, T.E. Nucleobase-functionalized acrylic ABA triblock copolymers and supramolecular blends. Polym. Chem. 2015, 6, 2434–2444. [Google Scholar] [CrossRef]
- Kang, Y.; Pitto-Barry, A.; Rolph, M.S.; Hua, Z.; Hands-Portman, I.; Kirby, N.; O’Reilly, R.K. Use of complementary nucleobase-containing synthetic polymers to prepare complex self-assembled morphologies in water. Polym. Chem. 2016, 7, 2836–2846. [Google Scholar] [CrossRef] [PubMed]
- Mather, B.D.; Baker, M.B.; Beyer, F.L.; Berg, M.A.G.; Green, M.D.; Long, T.E. Supramolecular triblock copolymers containing complementary nucleobase molecular recognition. Macromolecules 2007, 40, 6834–6845. [Google Scholar] [CrossRef]
- Wu, Y.-S.; Wu, Y.-C.; Kuo, S.-W. Thymine- and adenine-functionalized polystyrene form self-assembled structures through multiple complementary hydrogen bonds. Polymers 2014, 6, 1827–1845. [Google Scholar] [CrossRef]
- Lutz, J.; Pfeifer, S.; Chanana, M.; Thunemann, A.F.; Bienert, R. H-bonding-directed self-assembly of synthetic copolymers containing nucleobases: Organization and colloidal fusion in a noncompetitive solvent. Langmuir 2006, 22, 7411–7415. [Google Scholar] [CrossRef] [PubMed]
- Nakahata, M.; Takashima, Y.; Hashidzume, A.; Harada, A. Macroscopic self-assembly based on complementary interactions between nucleobase pairs. Chemistry 2015, 21, 2770–2774. [Google Scholar] [CrossRef] [PubMed]
- Lutz, J.-F.; Thünemann, A.F.; Nehring, R. Preparation by controlled radical polymerization and self-assembly via base-recognition of synthetic polymers bearing complementary nucleobases. J. Polym. Sci. Part A Polym. Chem. 2005, 43, 4805–4818. [Google Scholar] [CrossRef]
- Xi, W.; Pattanayak, S.; Wang, C.; Fairbanks, B.; Gong, T.; Wagner, J.; Kloxin, C.J.; Bowman, C.N. Clickable nucleic acids: Sequence-controlled periodic copolymer/oligomer synthesis by orthogonal thiol-x reactions. Angew. Chem. Int. Ed. 2015, 54, 14462–14467. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Zheng, Y.; Zhao, B.; Li, S.; Zhang, Y.; Gao, C. Sequentially hetero-functional, topological polymers by step-growth thiol-yne approach. Sci. Rep. 2014, 4, 4387. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Fairbanks, B.; He, L.; Liu, T.; Shah, P.; Cha, J.N.; Stansbury, J.W.; Bowman, C.N. Water-soluble clickable nucleic acid (CNA) polymer synthesis by functionalizing the pendant hydroxyl. Chem. Commun. 2017, 53, 10156–10159. [Google Scholar] [CrossRef] [PubMed]
- Lo, P.K.; Sleiman, H.F. Nucleobase-templated polymerization: Copying the chain length and polydispersity of living polymers into conjugated polymers. J. Am. Chem. Soc. 2009, 131, 4182–4183. [Google Scholar] [CrossRef] [PubMed]
- James, C.R.; Rush, A.M.; Insley, T.; Vukovic, L.; Adamiak, L.; Kral, P.; Gianneschi, N.C. Poly(oligonucleotide). J. Am. Chem. Soc. 2014, 136, 11216–11219. [Google Scholar] [CrossRef] [PubMed]
- Tsao, Y.T.; Wooley, K.L. Synthetic, functional thymidine-derived polydeoxyribonucleotide analogues from a six-membered cyclic phosphoester. J. Am. Chem. Soc. 2017, 139, 5467–5473. [Google Scholar] [CrossRef] [PubMed]
- Bauerle, P.; Emge, A. Specific recognition of nucleobase-functionalized polythiophenes. Adv. Mater. 1998, 10, 324–330. [Google Scholar] [CrossRef]
- Wang, J.; Matyjaszewski, K. Controlled/“living” radical polymerization. Atom transfer radical polymerization in the presence of transition-metal complexes. J. Am. Chem. Soc. 1995, 117, 5614–5615. [Google Scholar] [CrossRef]
- Kato, M.; Kamigaito, M.; Sawamoto, M.; Higashimura, T. Polymerization of methyl-methacrylate with the carbon-tetrachloride dichlorotris(triphenylphosphine)ruthenium(II) methylaluminum bis(2,6-di-tert-butylphenoxide) initiating system—Possibility of living radical polymerization. Macromolecules 1995, 28, 1721–1723. [Google Scholar] [CrossRef]
- Cheng, C.C.; Huang, C.F.; Yen, Y.C.; Chang, F.C. A “plug and play” polymer through biocomplementary hydrogen bonding. J. Polym. Sci. Part A Polym. Chem. 2008, 46, 6416–6424. [Google Scholar] [CrossRef]
- Chiefari, J.; Chong, Y.K.; Ercole, F.; Krstina, J.; Jeffery, J.; Le, T.P.T.; Mayadunne, R.T.A.; Meijs, G.F.; Moad, C.L.; Moad, G. Living free-radical polymerization by reversible addition-fragmentation chain transfer: The raft process. Macromolecules 1998, 31, 5559–5562. [Google Scholar] [CrossRef]
- Kang, Y.; Lu, A.; Ellington, A.; Jewett, M.C.; O’Reilly, R.K. Effect of complementary nucleobase interactions on the copolymer composition of raft copolymerizations. ACS Macro Lett. 2013, 2, 581–586. [Google Scholar] [CrossRef]
- Kang, Y.; Pitto-Barry, A.; Maitland, A.; O’Reilly, R.K. Raft dispersion polymerization: A method to tune the morphology of thymine-containing self-assemblies. Polym. Chem. 2015, 6, 4984–4992. [Google Scholar] [CrossRef]
- Fong, D.; Hua, Z.; Wilks, T.R.; O’Reilly, R.K.; Adronov, A. Dispersion of single-walled carbon nanotubes using nucleobase-containing poly(acrylamide) polymers. J. Polym. Sci. Part A Polym. Chem. 2017, 55, 2611–2617. [Google Scholar] [CrossRef]
- Moad, G.; Rizzardo, E. Alkoxyamine-initiated living radical polymerization: Factors affecting alkoxyamine homolysis rates. Macromolecules 1995, 28, 8722–8728. [Google Scholar] [CrossRef]
- Nicolas, J.; Guillaneuf, Y.; Lefay, C.; Bertin, D.; Gigmes, D.; Charleux, B. Nitroxide-mediated polymerization. Prog. Polym. Sci. 2013, 38, 63–235. [Google Scholar] [CrossRef]
- Kolb, H.C.; Finn, M.G.; Sharpless, K.B. Click chemistry: Diverse chemical function from a few good reactions. Angew. Chem. 2001, 40, 2004–2021. [Google Scholar] [CrossRef]
- Binder, W.H.; Sachsenhofer, R. ‘Click’ chemistry in polymer and material science: An update. Macromol. Rapid. Commum. 2008, 29, 952–981. [Google Scholar] [CrossRef]
- Xi, W.X.; Scott, T.F.; Kloxin, C.J.; Bowman, C.N. Click chemistry in materials science. Adv. Funct. Mater. 2014, 24, 2572–2590. [Google Scholar] [CrossRef]
- Zhang, T.; Zheng, Z.H.; Cheng, X.; Ding, X.B.; Peng, Y.X. “Click” chemistry in materials science. Prog. Chem. 2008, 20, 1090–1101. [Google Scholar]
- Chernykh, A.; Agag, T.; Ishida, H. Synthesis of linear polymers containing benzoxazine moieties in the main chain with high molecular design versatility via click reaction. Polymer 2009, 50, 382–390. [Google Scholar] [CrossRef]
- Helms, B.; Mynar, J.L.; Hawker, C.J.; Frechet, J.M.J. Dendronized linear polymers via “click chemistry”. J. Am. Chem. Soc. 2004, 126, 15020–15021. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.A.; Finn, M.G.; Koberstein, J.T.; Turro, N.J. Construction of linear polymers, dendrimers, networks, and other polymeric architectures by copper-catalyzed azide-alkyne cycloaddition “click” chemistry. Macromol. Rapid Commum. 2008, 29, 1052–1072. [Google Scholar] [CrossRef]
- Kolli, B.; Mishra, S.P.; Joshi, M.P.; Mohan, S.R.; Dhami, T.S.; Samui, A.B. Synthesis and characterization of linear polymers for non-linear optics through click chemistry route. Adv. Mater. Res. 2012, 584, 8–12. [Google Scholar] [CrossRef]
- Polaske, N.W.; McGrath, D.V.; McElhanon, J.R. Thermally reversible dendronized linear AB step-polymers via “click” chemistry. Macromolecules 2011, 44, 3203–3210. [Google Scholar] [CrossRef]
- Zhang, W.A.; Muller, A.H.E. A “click chemistry” approach to linear and star-shaped telechelic poss-containing hybrid polymers. Macromolecules 2010, 43, 3148–3152. [Google Scholar] [CrossRef]
- Chen, H.; Jia, J.Q.; Duan, X.; Yang, Z.; Kong, J. Reduction-cleavable hyperbranched polymers with limited intramolecular cyclization via click chemistry. J. Polym. Sci. Part A Polym. Chem. 2015, 53, 2374–2380. [Google Scholar] [CrossRef]
- Han, Y.; Gao, C. Miktoarms hyperbranched polymer brushes: One-step fast synthesis by parallel click chemistry and hierarchical self-assembly. Sci. China Chem. 2010, 53, 2461–2471. [Google Scholar] [CrossRef]
- Li, S.P.; Han, J.; Gao, C. High-density and hetero-functional group engineering of segmented hyperbranched polymers via click chemistry. Polym. Chem. 2013, 4, 1774–1787. [Google Scholar] [CrossRef]
- Semsarilar, M.; Ladmiral, V.; Perrier, S. Highly branched and hyperbranched glycopolymers via reversible addition-fragmentation chain transfer polymerization and click chemistry. Macromolecules 2010, 43, 1438–1443. [Google Scholar] [CrossRef]
- Medel, S.; Bosch, P.; de la Torre, C.; Ramirez, P. Click chemistry to fluorescent hyperbranched polymers. 1-synthesis, characterization and spectroscopic properties. Eur. Polym. J. 2014, 59, 290–301. [Google Scholar] [CrossRef]
- Pandey, S.; Mishra, S.P.; Kolli, B.; Kanai, T.; Samui, A.B. Hyperbranched photo responsive and liquid crystalline azo-siloxane polymers synthesized by click chemistry. J. Polym. Sci. Part A Polym. Chem. 2012, 50, 2659–2668. [Google Scholar] [CrossRef]
- Hoogenboom, R. Thiol-yne chemistry: A powerful tool for creating highly functional materials. Angew. Chem. Int. Ed. 2010, 49, 3415–3417. [Google Scholar] [CrossRef] [PubMed]
- Popescu, D.L.; Tsarevsky, N.V. Multibrominated hyperbranched polymers: Synthesis and further functionalizations by arget atrp or click chemistry. Macromol. Rapid. Commum. 2012, 33, 869–875. [Google Scholar] [CrossRef] [PubMed]
- Saha, A.; Ramakrishnan, S. Site-specific functionalization of hyperbranched polymers using “click” chemistry. Macromolecules 2009, 42, 4028–4037. [Google Scholar] [CrossRef]
- Xu, J.T.; Tao, L.; Boyer, C.; Lowe, A.B.; Davis, T.P. Combining thio-bromo “click” chemistry and raft polymerization: A powerful tool for preparing functionalized multiblock and hyperbranched polymers. Macromolecules 2010, 43, 20–24. [Google Scholar] [CrossRef]
- Arseneault, M.; Wafer, C.; Morin, J.F. Recent advances in click chemistry applied to dendrimer synthesis. Molecules 2015, 20, 9263–9294. [Google Scholar] [CrossRef] [PubMed]
- Deraedt, C.; Pinaud, N.; Astruc, D. Recyclable catalytic dendrimer nanoreactor for part-per-million Cu(I) catalysis of “click” chemistry in water. J. Am. Chem. Soc. 2014, 136, 12092–12098. [Google Scholar] [CrossRef] [PubMed]
- Franc, G.; Kakkar, A. Dendrimer design using Cu(I)-catalyzed alkyne-azide “click-chemistry”. Chem. Commun. 2008, 5267–5276. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Kukowska-Latallo, J.F.; Tang, S.; Zong, H.; Johnson, K.B.; Desai, A.; Gordon, C.L.; Leroueil, P.R.; Baker, J.R., Jr. The facile synthesis of multifunctional pamam dendrimer conjugates through copper-free click chemistry. Bioorg. Med. Chem. Lett. 2012, 22, 3152–3156. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, Z.; Wu, L.; Zhang, Y.; Yu, M.; Wei, L. Constructing metal nanoparticle multilayers with polyphenylene dendrimer/gold nanoparticles via “click” chemistry. Langmuir 2013, 29, 3943–3949. [Google Scholar] [CrossRef] [PubMed]
- Meesaragandla, B.; Mahalingam, V. Synthesis of upconverting hydrogel nanocomposites using thiol-ene click chemistry: Template for the formation of dendrimer-like gold nanoparticle assemblies. Chemistry 2015, 21, 16811–16817. [Google Scholar] [CrossRef] [PubMed]
- Qin, A.J.; Lam, J.W.Y.; Tang, B.Z. Click polymerization: Progresses, challenges, and opportunities. Macromolecules 2010, 43, 8693–8702. [Google Scholar] [CrossRef]
- Jiang, B.; Pu, H.T.; Pan, H.Y.; Chang, Z.H.; Jin, M.; Wan, D.C. Proton conducting membranes based on semi-interpenetrating polymer network of fluorine-containing polyimide and perfluorosulfonic acid polymer via click chemistry. Electrochim. Acta 2014, 132, 457–464. [Google Scholar] [CrossRef]
- Koyuncu, F.B.; Davis, A.R.; Carter, K.R. Emissive conjugated polymer networks with tunable band-gaps via thiol-ene click chemistry. Chem. Mater. 2012, 24, 4410–4416. [Google Scholar] [CrossRef]
- Pan, H.Y.; Pu, H.T.; Jin, M.; Wan, D.C.; Modestov, A.D. Semi-interpenetrating polymer networks based-on end-group crosslinked fluorine-containing polyimide via click chemistry. Electrochim. Acta 2013, 89, 577–584. [Google Scholar] [CrossRef]
- Retailleau, M.; Pierrel, J.; Ibrahim, A.; Croutxe-Barghorn, C.; Allonas, X. Sequenced click chemistry and photopolymerization: A new approach toward semi-interpenetrating polymer networks. Polym. Adv. Technol. 2017, 28, 491–495. [Google Scholar] [CrossRef]
- Yao, F.; Xu, L.Q.; Fu, G.D.; Lin, B.P. Sliding-graft interpenetrating polymer networks from simultaneous “click chemistry” and atom transfer radical polymerization. Macromolecules 2010, 43, 9761–9770. [Google Scholar] [CrossRef]
- Hoyle, C.E.; Bowman, C.N. Thiol-ene click chemistry. Angew. Chem. Int. Ed. 2010, 49, 1540–1573. [Google Scholar] [CrossRef] [PubMed]
- Lowe, A.B. Thiol-ene “click” reactions and recent applications in polymer and materials synthesis. Polym. Chem. 2010, 1, 17–36. [Google Scholar] [CrossRef]
- Lowe, A.B. Thiol-ene “click” reactions and recent applications in polymer and materials synthesis: A first update. Polym. Chem. 2014, 5, 4820–4870. [Google Scholar] [CrossRef]
- Hoyle, C.E.; Lowe, A.B.; Bowman, C.N. Thiol-click chemistry: A multifaceted toolbox for small molecule and polymer synthesis. Chem. Soc. Rev. 2010, 39, 1355–1387. [Google Scholar] [CrossRef] [PubMed]
- Harguindey, A.; Domaille, D.W.; Fairbanks, B.D.; Wagner, J.; Bowman, C.N.; Cha, J.N. Synthesis and assembly of click-nucleic-acid-containing PEG-PLGA nanoparticles for DNA delivery. Adv. Mater. 2017, 29, 1700743. [Google Scholar] [CrossRef] [PubMed]
- Mu, X.; Sowan, N.; Tumbic, J.A.; Bowman, C.N.; Mather, P.T.; Qi, H.J. Photo-induced bending in a light-activated polymer laminated composite. Soft Matter 2015, 11, 2673–2682. [Google Scholar] [CrossRef] [PubMed]
- Grubbs, R.H.; Tumas, W. Polymer synthesis and organotransition metal chemistry. Science 1989, 243, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Mutch, A.; Leconte, M.; Lefebvre, F.; Basset, J.M. Effect of alcohols and epoxides on the rate of romp of norbornene by a ruthenium trichloride catalyst. J. Mol. Catal. A Chem. 1998, 133, 191–199. [Google Scholar] [CrossRef]
- Bazzi, H.S.; Sleiman, H.F. Adenine-containing block copolymers via ring-opening metathesis polymerization: Synthesis and self-assembly into rod morphologies. Macromolecules 2002, 35, 9617–9620. [Google Scholar] [CrossRef]
- Nielsen, P.E. Sequence-selective DNA recognition by synthetic ligands. Bioconjug. Chem. 1991, 2, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wittung, P.; Nielsen, P.E.; Buchardt, O.; Egholm, M.; Norden, B. DNA-like double helix formed by peptide nucleic-acid. Nature 1994, 368, 561–563. [Google Scholar] [CrossRef] [PubMed]
- Egholm, M.; Buchardt, O.; Christensen, L.; Behrens, C.; Freier, S.M.; Driver, D.A.; Berg, R.H.; Kim, S.K.; Norden, B.; Nielsen, P.E. PNA hybridizes to complementary oligonucleotides obeying the watson-crick hydrogen-bonding rules. Nature 1993, 365, 566–568. [Google Scholar] [CrossRef] [PubMed]
- Kaihatsu, K.; Janowski, B.A.; Corey, D.R. Recognition of chromosomal DNA by PNAs. Chem. Biol. 2004, 11, 749–758. [Google Scholar] [CrossRef] [PubMed]
- Gregory, G.L.; Hierons, E.M.; Kociok-Köhn, G.; Sharma, R.I.; Buchard, A. CO2-driven stereochemical inversion of sugars to create thymidine-based polycarbonates by ring-opening polymerisation. Polym. Chem. 2017, 8, 1714–1721. [Google Scholar] [CrossRef]
- Lutolf, M.P.; Hubbell, J.A. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat. Biotechnol. 2005, 23, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Caliari, S.R.; Burdick, J.A. A practical guide to hydrogels for cell culture. Nat. Methods 2016, 13, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Guvendiren, M.; Lu, H.D.; Burdick, J.A. Shear-thinning hydrogels for biomedical applications. Soft Matter 2012, 8, 260–272. [Google Scholar] [CrossRef]
- Tan, H.P.; Xiao, C.; Sun, J.C.; Xiong, D.S.; Hu, X.H. Biological self-assembly of injectable hydrogel as cell scaffold via specific nucleobase pairing. Chem. Commun. 2012, 48, 10289–10291. [Google Scholar] [CrossRef] [PubMed]
- Zeng, P.; Xu, Y.; Zeng, C.H.; Ren, H.; Peng, M.L. Chitosan-modified poly(d,l-lactide-co-glycolide) nanospheres for plasmid DNA delivery and HBV gene-silencing. Int. J. Pharm. 2011, 415, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Spelios, M.; Kearns, M.; Savva, M. From gene delivery to gene silencing: Plasmid DNA-transfecting cationic lipid 1,3-dimyristoylamidopropane-2-[bis(2-dimethylaminoethane)] carbamate efficiently promotes small interfering RNA-induced RNA interference. Biochemistry 2010, 49, 5753–5759. [Google Scholar] [CrossRef] [PubMed]
- Ewert, K.K.; Zidovska, A.; Ahmad, A.; Bouxsein, N.F.; Evans, H.M.; McAllister, C.S.; Samuel, C.E.; Safinya, C.R. Cationic liposome-nucleic acid complexes for gene delivery and silencing: Pathways and mechanisms for plasmid DNA and siRNA. Top. Curr. Chem. 2010, 296, 191–226. [Google Scholar] [PubMed]
- Beale, G.; Hollins, A.J.; Benboubetra, M.; Sohail, M.; Fox, S.P.; Benter, I.; Akhtar, S. Gene silencing nucleic acids designed by scanning arrays: Anti-EGFR activity of siRNA, ribozyme and DNA enzymes targeting a single hybridization-accessible region using the same delivery system. J. Drug Target. 2003, 11, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Oupicky, D.; Konak, C.; Ulbrich, K.; Wolfert, M.A.; Seymour, L.W. DNA delivery systems based on complexes of DNA with synthetic polycations and their copolymers. J. Control. Release 2000, 65, 149–171. [Google Scholar] [CrossRef]
- Luo, D.; Saltzman, W.M. Synthetic DNA delivery systems. Nat. Biotechnol. 2000, 18, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Hager, M.D.; Greil, P.; Leyens, C.; Der Zwaag, S.V.; Schubert, U.S. Self-healing materials. Adv. Mater. 2007, 22, 5424–5430. [Google Scholar] [CrossRef] [PubMed]
- Wool, R.P. Self-healing materials: A review. Soft Matter 2008, 4, 400–418. [Google Scholar] [CrossRef]
- Neal, J.A.; Mozhdehi, D.; Guan, Z. Enhancing mechanical performance of a covalent self-healing material by sacrificial noncovalent bonds. J. Am. Chem. Soc. 2015, 137, 4846–4850. [Google Scholar] [CrossRef] [PubMed]
- Tuncaboylu, D.C.; Sari, M.; Oppermann, W.; Okay, O. Tough and self-healing hydrogels formed via hydrophobic interactions. Macromolecules 2011, 44, 4997–5005. [Google Scholar] [CrossRef]
- Jacob, R.S.; Ghosh, D.; Singh, P.K.; Basu, S.K.; Jha, N.N.; Das, S.; Sukul, P.K.; Patil, S.B.; Sathaye, S.; Kumar, A. Self healing hydrogels composed of amyloid nano fibrils for cell culture and stem cell differentiation. Biomaterials 2015, 54, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Heo, Y.; Malakooti, M.H.; Sodano, H.A. Self-healing polymers and composites for extreme environments. J. Mater. Chem. 2016, 4, 17403–17411. [Google Scholar] [CrossRef]
- Cheng, C.; Chang, F.; Dai, S.A.; Lin, Y.; Lee, D. Bio-complementary supramolecular polymers with effective self-healing functionality. RSC Adv. 2015, 5, 90466–90472. [Google Scholar] [CrossRef]
- Ye, X.; Li, X.; Shen, Y.; Chang, G.; Yang, J.; Gu, Z. Self-healing pH-sensitive cytosine- and guanosine-modified hyaluronic acid hydrogels via hydrogen bonding. Polymer 2017, 108, 348–360. [Google Scholar] [CrossRef]
- Grzelczak, M.; Vermant, J.; Furst, E.M.; Lizmarzan, L.M. Directed self-assembly of nanoparticles. ACS Nano 2010, 4, 3591–3605. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Li, G.; Stansbury, J.W.; Zhu, X.; Wang, X.; Nie, J. A smart antibacterial surface made by photopolymerization. ACS Appl. Mater. Interfaces 2016, 8, 28047–28054. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Stansbury, J.W.; Ai, X.; Hu, R.; Tang, H.; Maitlo, I.; Nie, J. Nanostructure superhydrophobic surface prepared by photopolymerization. Chem. Lett. 2017, 46, 371–373. [Google Scholar] [CrossRef]
- Samanta, D.; Hosseininassab, N.; Zare, R.N. Electroresponsive nanoparticles for drug delivery on demand. Nanoscale 2016, 8, 9310–9317. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Chen, J.; Chen, X. Nanoparticles for gene delivery. Small 2013, 9, 2034–2044. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Mao, C.; Cho, S.; Ma, K.; Xi, W.; Bowman, C.N.; Park, W.; Cha, J.N. Experimental and theoretical photoluminescence studies in nucleic acid assembled gold-upconverting nanoparticle clusters. Nanoscale 2015, 7, 17254–17260. [Google Scholar] [CrossRef] [PubMed]
- Keene, F.R.; Smith, J.A.; Collins, J.G. Metal complexes as structure-selective binding agents for nucleic acids. Coord. Chem. Rev. 2009, 253, 2021–2035. [Google Scholar] [CrossRef]
- Ward, W.L.; Plakos, K.; Derose, V.J. Nucleic acid catalysis: Metals, nucleobases, and other cofactors. Chem. Rev. 2014, 114, 4318–4342. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.M. Potential metal-binding domains in nucleic acid binding proteins. Science 1986, 232, 485–487. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Mclaughlin, C.K.; Aldaye, F.A.; Hamblin, G.D.; Rys, A.Z.; Rouiller, I.; Sleiman, H.F. Metal-nucleic acid cages. Nat. Chem. 2009, 1, 390–396. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Chao, J.; Xiao, S.; Seeman, N.C. A proximity-based programmable DNA nanoscale assembly line. Nature 2010, 465, 202–205. [Google Scholar] [CrossRef] [PubMed]
- Seeman, N.C.; Lukeman, P.S. Nucleic acid nanostructures: Bottom-up control of geometry on the nanoscale. Rep. Prog. Phys. 2005, 68, 237–270. [Google Scholar] [CrossRef] [PubMed]
- Watson, R.M.; Skorik, Y.A.; Patra, G.K.; Achim, C. Influence of metal coordination on the mismatch tolerance of ligand-modified PNA duplexes. J. Am. Chem. Soc. 2005, 127, 14628–14639. [Google Scholar] [CrossRef] [PubMed]
- Beobide, G.; Castillo, O.; Cepeda, J.; Luque, A.; Perezyanez, S.; Roman, P.; Thomasgipson, J. Metal-carboxylato-nucleobase systems: From supramolecular assemblies to 3D porous materials. Coord. Chem. Rev. 2013, 257, 2716–2736. [Google Scholar] [CrossRef]
- Amoochoa, P.; Zamora, F. Coordination polymers with nucleobases: From structural aspects to potential applications. Coord. Chem. Rev. 2014, 276, 34–58. [Google Scholar] [CrossRef]
- Olea, D.; Alexandre, S.S.; Amoochoa, P.; Guijarro, A.; De Jesus, F.; Soler, J.M.; De Pablo, P.J.; Zamora, F.; Gomezherrero, J. From coordination polymer macrocrystals to nanometric individual chains. Adv. Mater. 2005, 17, 1761–1765. [Google Scholar] [CrossRef]
- García-Terán, J.P.; Castillo, O.; Luque, A.; García-Couceiro, U.; Román, P.; Lezama, L. An unusual 3D coordination polymer based on bridging interactions of the nucleobase adenine. Inorg. Chem. 2004, 43, 4549–4551. [Google Scholar] [CrossRef] [PubMed]

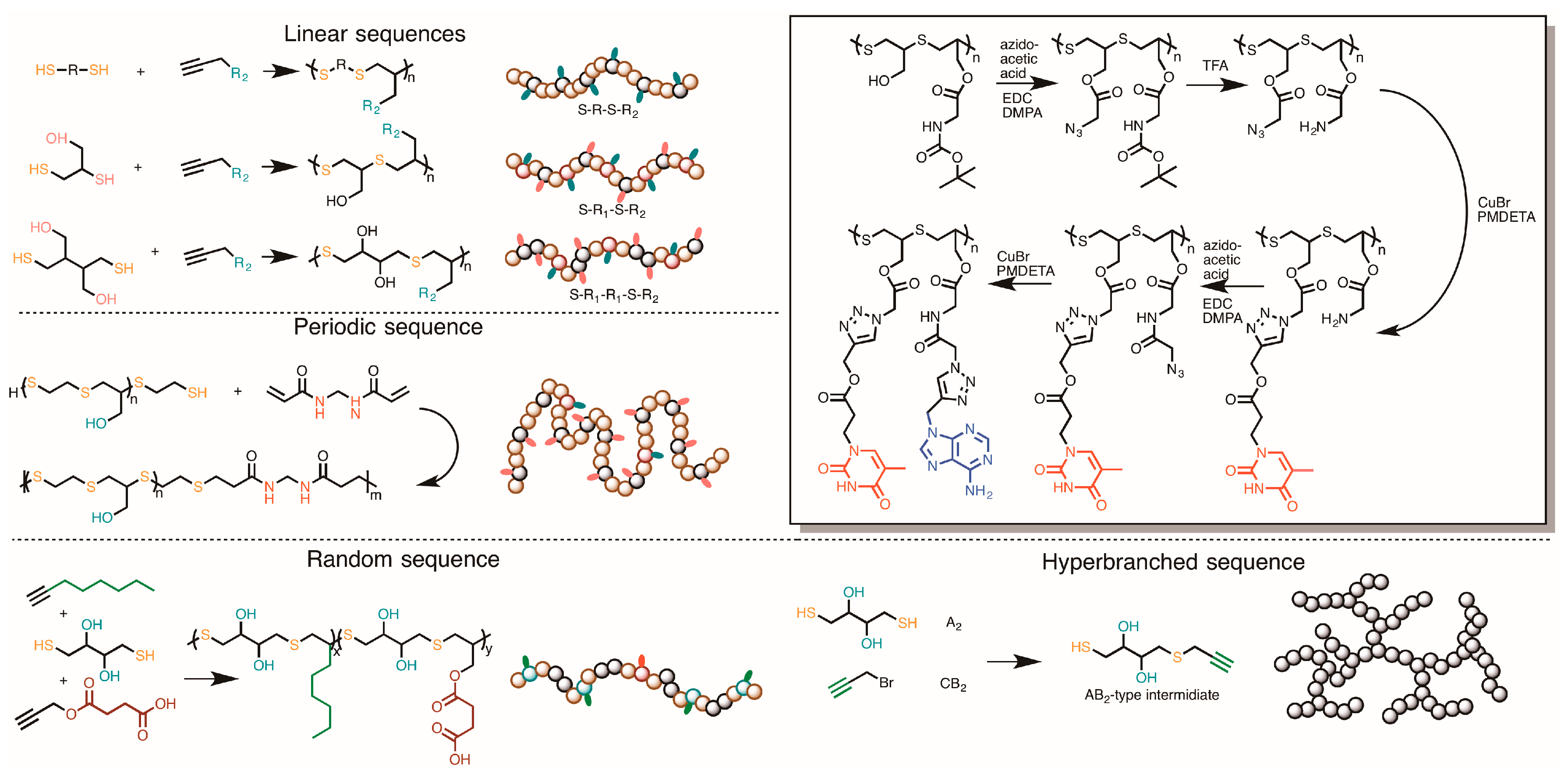
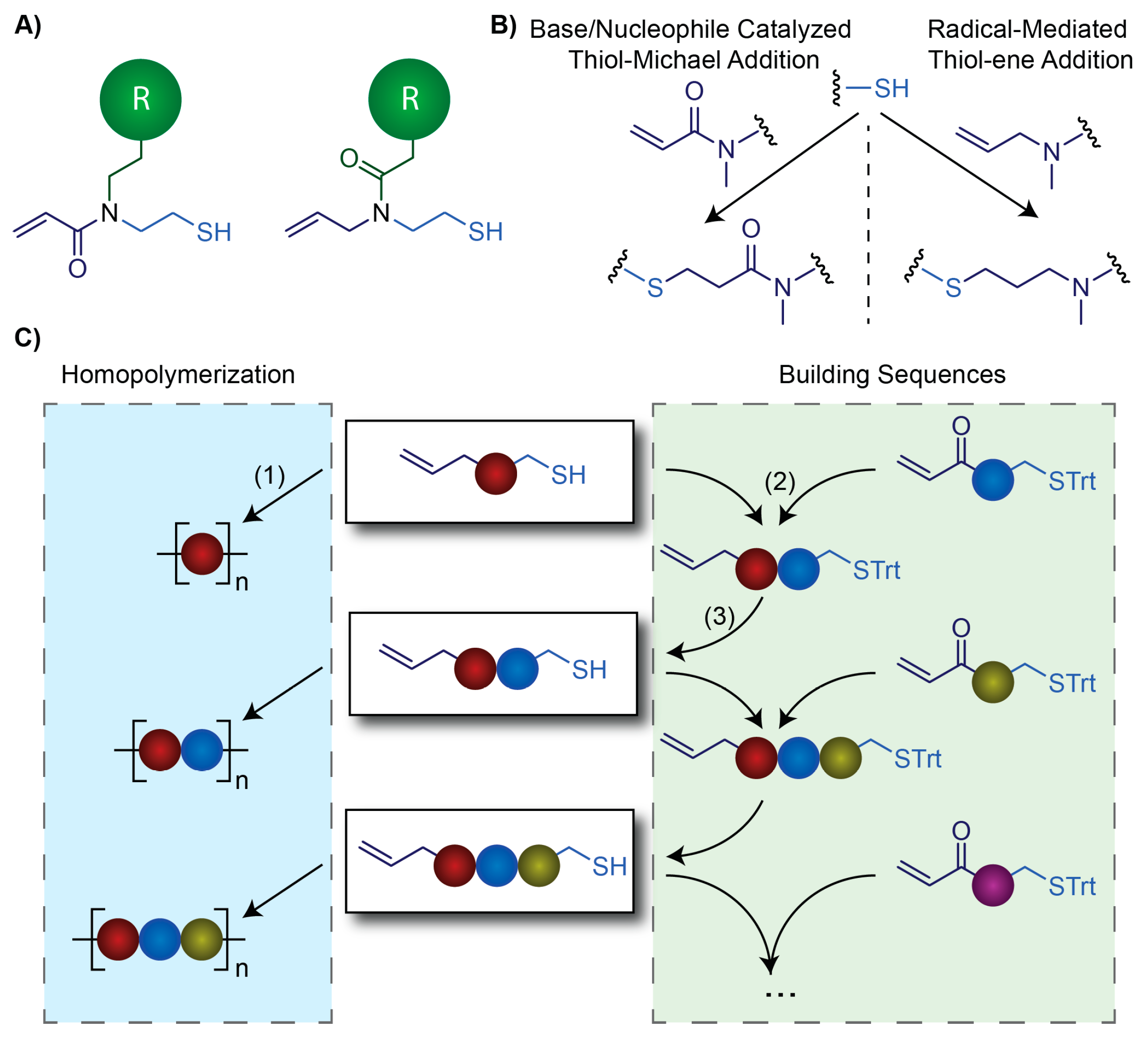
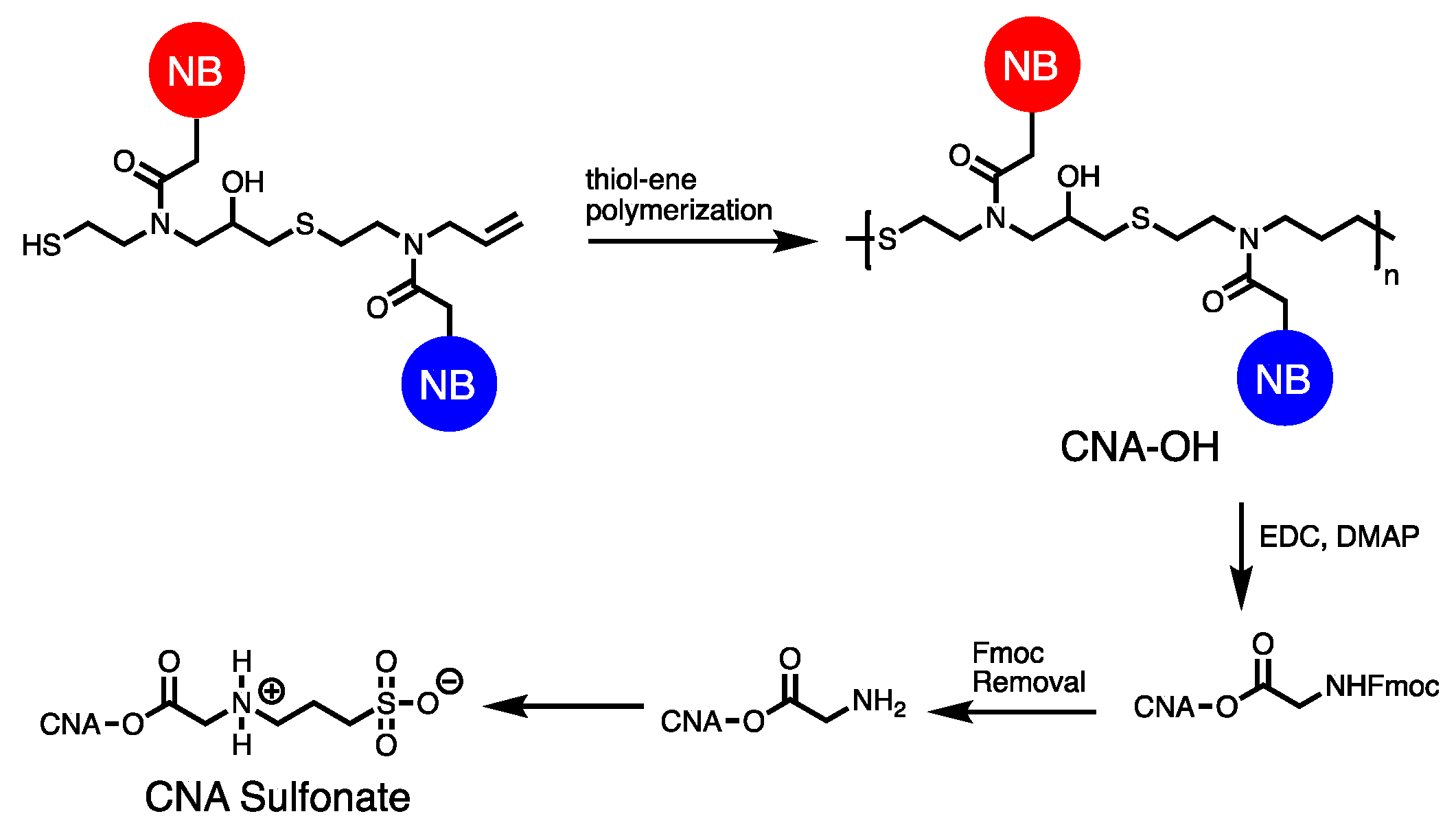

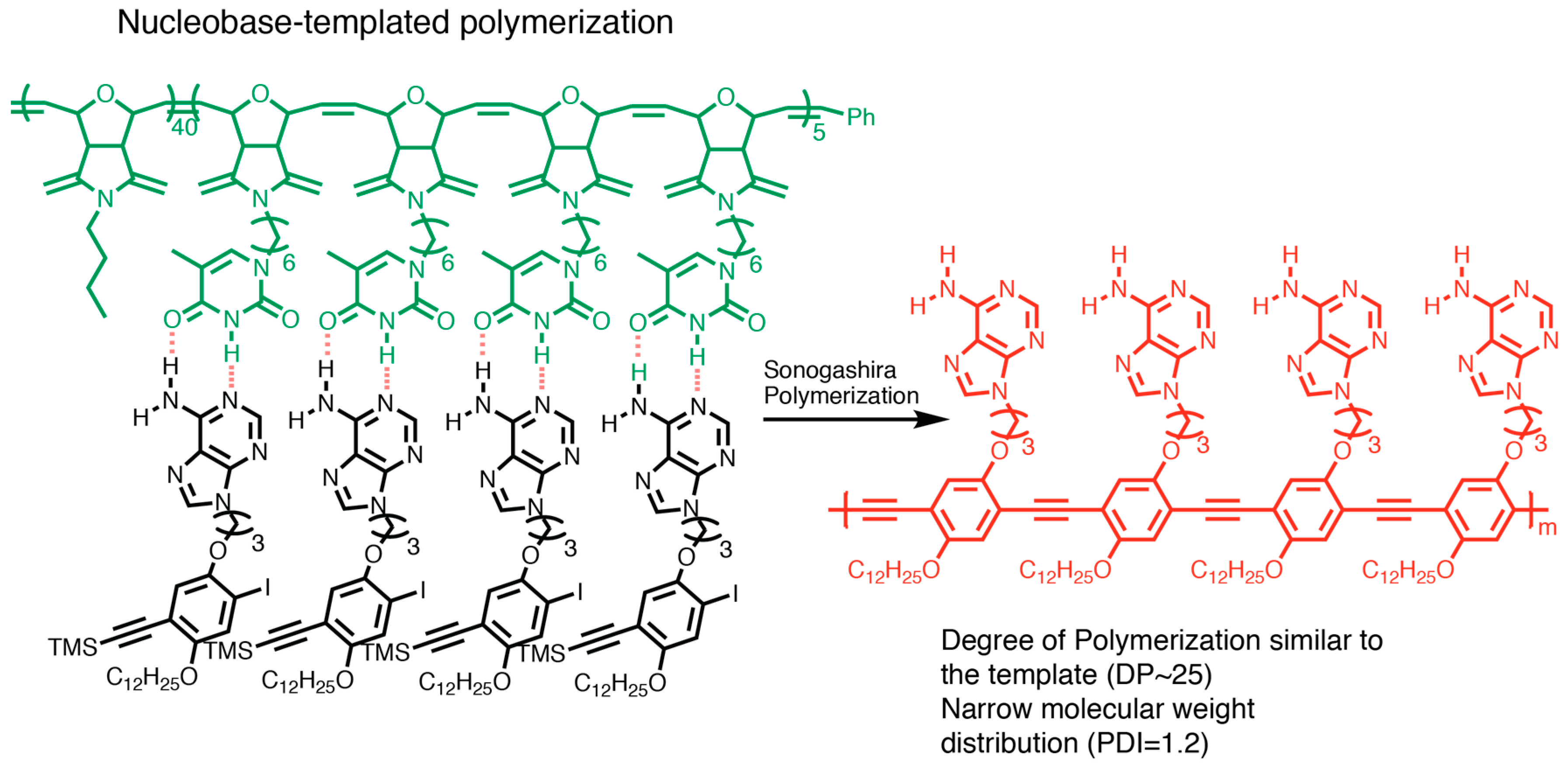



| Polymerization method | NB-Monomer | Length | References |
|---|---|---|---|
| ATRP | Styrenes and derivatives | 10~100 bases | [25,26,27] |
| Acrylates | |||
| Methacrylates | |||
| (Meth)acrylamides | |||
| Miscellaneous monomers | |||
| RAFT | Styrenes and derivatives | 10~100 bases | [28,29,30,31,32] |
| Acrylates | |||
| Methacrylates | |||
| (meth)acrylamides | |||
| NMP | Styrenes and derivatives | 20~100 bases | [33,34] |
| Acrylates | |||
| Conventional radical polymerization | Common monomers | 80~200 bases | [35,36,37] |
| Click chemistry | Azides | 10~50 bases | [38,39,40] |
| Alkynes | |||
| Thiols | |||
| Vinyls | |||
| Acrylamides | |||
| Dienes | |||
| Ring opening polymerization | Cyclic monomers (e.g., cyclic olefins, acetals, lactams, lactones) | 10~40 bases | [41,42,43] |
| Other polymerization | Thiophenes and derivatives | N.A. | [44] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, H.; Xi, W. Nucleobase-Containing Polymers: Structure, Synthesis, and Applications. Polymers 2017, 9, 666. https://doi.org/10.3390/polym9120666
Yang H, Xi W. Nucleobase-Containing Polymers: Structure, Synthesis, and Applications. Polymers. 2017; 9(12):666. https://doi.org/10.3390/polym9120666
Chicago/Turabian StyleYang, Haitao, and Weixian Xi. 2017. "Nucleobase-Containing Polymers: Structure, Synthesis, and Applications" Polymers 9, no. 12: 666. https://doi.org/10.3390/polym9120666





