Low Molecular Weight Chitosan (LMWC)-based Polyplexes for pDNA Delivery: From Bench to Bedside
Abstract
:1. Introduction
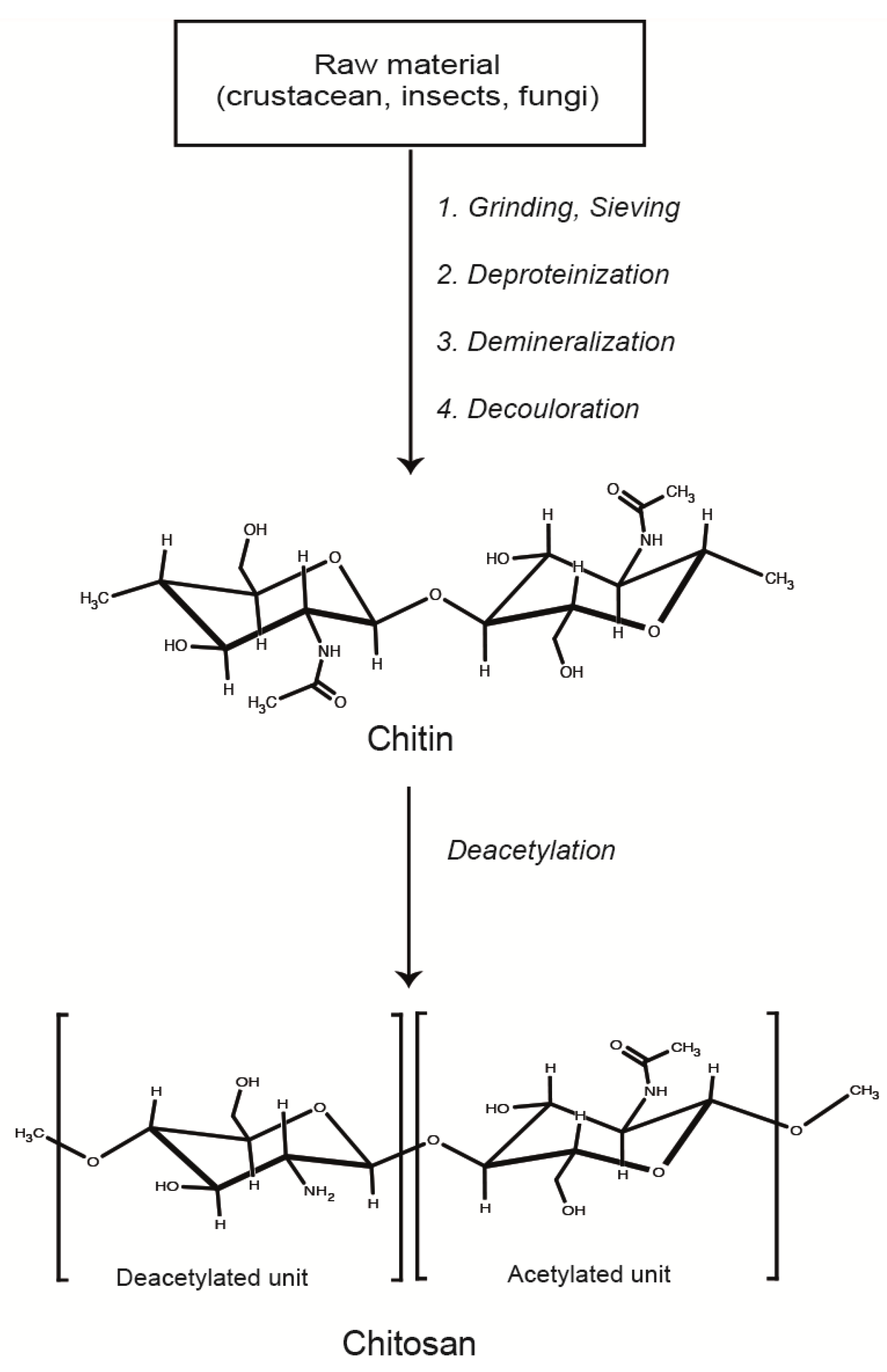
2. Formation and Characterization of LMWC-pDNA Polyplexes
2.1. LMWC-pDNA Polyplexes Formation Process
2.2. Physico-Chemical Characterization of LMWC-pDNA Polyplexes
| Characterization technique | Parameter |
|---|---|
| Acid-base titration | Buffering capacity |
| Agarose gel electrophoresis | Binding affinity Release capacity Protection against endonuclease Stability |
| Asymmetrical flow field-flow fractionation (AF4) coupled with light scattering | Size Stoichiometry |
| Atomic force microscopy | Size Morphology |
| Electronic microscopy | Size Morphology |
| Dynamic light scattering | Size Colloidal stability |
| EtBr displacement assay | Stability |
| Laser Doppler velocimetry | Zeta potential |
| Isothermal titration calorimetry | Binding affinity Stability |
| Nanoparticle tracking analysis | Size Mw Concentration |
| Orange II dye depletion assay (AF4 results confirmation) | Stoichiometry |
| Polyanion competition assay | Stability |
| Potentiometric titration | Buffering capacity |
| Static light scattering | Mw |
2.2.1. Size
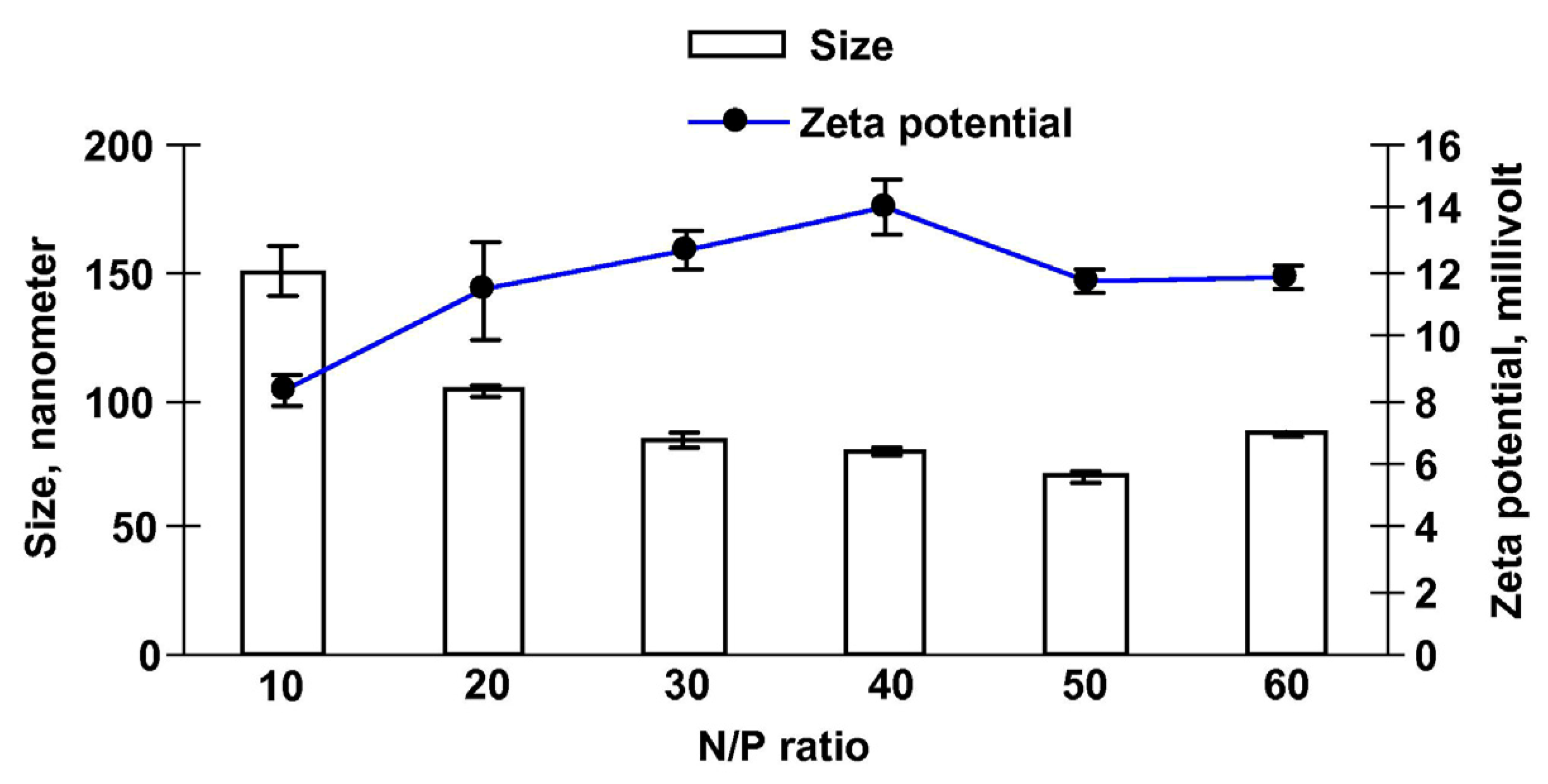
2.2.2. Zeta Potential
2.2.3. Morphology

2.2.4. Binding Affinity

2.2.5. Buffering Capacity

2.2.6. Colloidal Stability in Physiological Conditions
3. In vivo Evaluation of LMWC-pDNA Polyplexes Transfection Process

3.1. Cellular Binding and Uptake

3.2. Endolysosomal Escape and Polyplex Dissociation
3.3. Tools for the Study of Uptake Pathways

3.4. Nuclear Import
3.5. Factors Involved in the Transfection Efficiency of LMWC based Polyplexes
4. Chemical Modifications of Ch to Overcome Transfection Barriers
5. Freeze-Drying of Polyplexes
6. In vitro Applications of LMWC based Vectors for pDNA Delivery
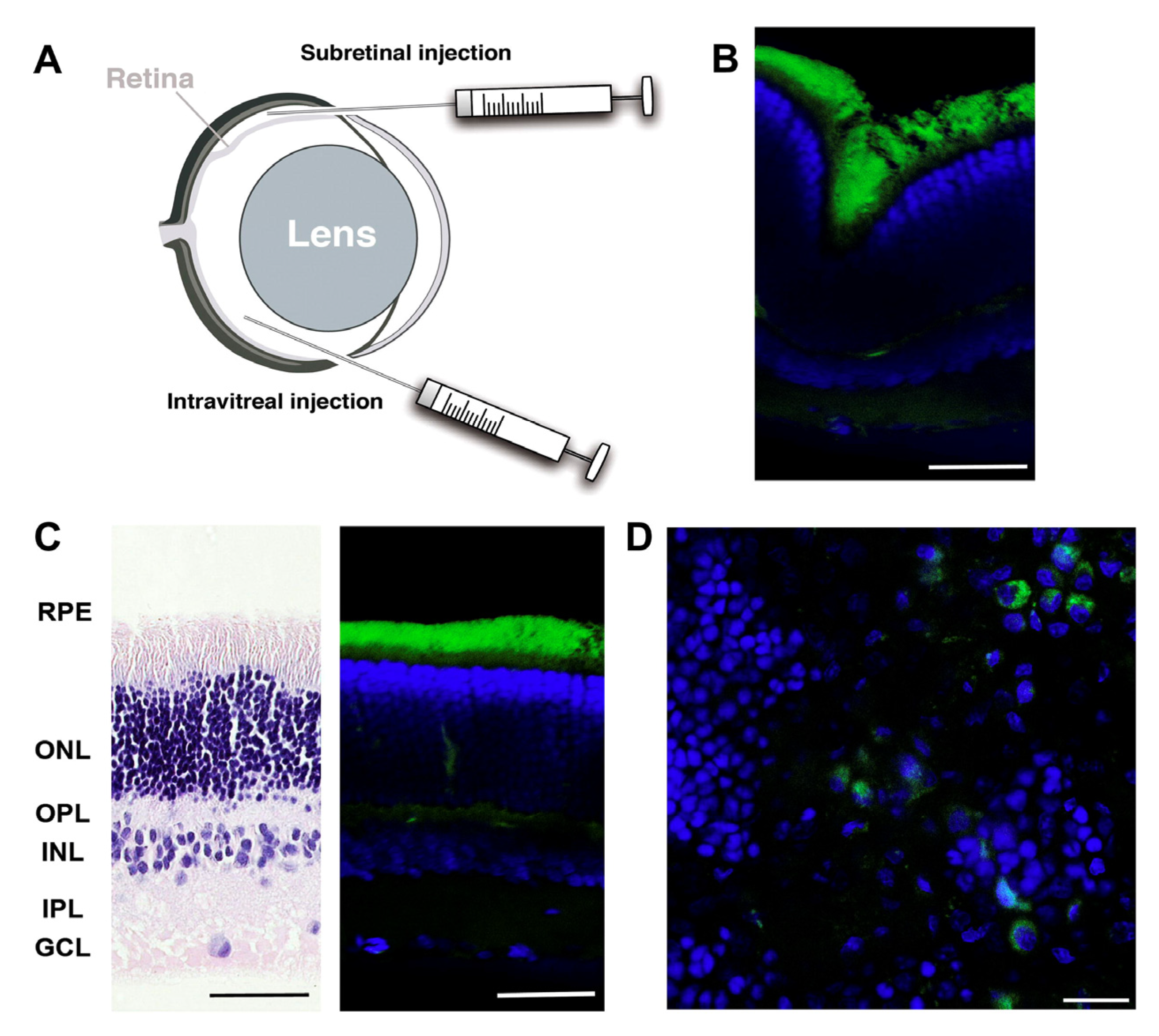
6.1. Ocular Delivery
6.2. Lung Delivery
6.3. Other Delivery Routes
| Route of Administration | Animal Model | Chitosan | pDNA | Comments | Objective | Reference |
|---|---|---|---|---|---|---|
| Corneal injection | Sprague-Dawley rats | Novavect O15 (5.7 kDa, DDA 99%); Novafect O25 (7.3 kDa, DDA 99%) | gWiz-Luc (luciferase encoding reporter gene) 1.5 μg; gWiz-GFP (GFP encoding reporter gene) 1.5 μg | – | Treatment of acquired and inherited corneal disorders | [27] |
| Corneal injection | Sprague-Dawley rats | Novafect O15 (5.7 kDa, DDA 99%) | gWiz,-Luc, pCpG-Luc, pEPI-CMV, pEPI-UbC (encoding for luciferase) 1.5 μg; gWiz-GFP, pCpG-GFP (encoding for GFP) 1.5 μg | – | Treatment of acquired and inherited corneal disorders | [33] |
| Subretinal, intravitreous injection | Sprague-Dawley rats | Novafect O15 (5.7 kDa, DDA 99%) | pCMS-EGFP reporter gene (100 ng) | – | Treatment of retinal disorders | [28] |
| Subretinal, intravitreous injection | Sprague-Dawley rats | Novafect O25 (7.3 kDa, DDA 99%) | pCMS-EGFP reporter gene (100 ng) | – | Treatment of retinal disorders | [32] |
| Topic administration | Rabbits | Ultrapure Ch hydrochloride salt (113 kDa) | pEGFP reporter gene (25 μg, 50 μg, 100 μg) | Ch was mixed with HA salt, and NPs were prepared by ionotropic gelification | Treatment of ocular diseases | [89] |
| Aerosol | Balb/c mice | Ch Chitoclear (126 kDa, DDA 98%) | pGL3-control plasmid encoding luciferase | Electrostatially formed polyplexes were conjugated with FAP-B | Lung targeting | [90] |
| Intratracheal administration | Balb/c mice | UPC, Protasan UPG 210 | gWiz-Luc, pCMV-Luc (luciferase encoding reporter genes) 5 μg, 10 μg, 25 μg | Fully deacetylated Ch was depolymerized to obtain oligomers with number average DPn 25 and 18 | Lung targeting | [21] |
| Intratracheal administration | Balb/c mice | Fully de- N-acetylated Ch (3.6–7 kDa) | gWiz-Luc, pCMV-Luc (luciferase encoding reporter genes) | Ch oligomers were substituted with trisaccharides, obtaining oligormes with 7%, 23%, 40% of substituted amines | Lung targeting | [50] |
| Intranasal administration | Sprague-Dawley rats | Ch Mw: 5, 173 kDa | pEGFP-C3 encoding GFP;pDNA encoding CETP-C | – | Immunotherapeutic DNA vaccine for atherosclerosis treatment | [93] |
| Intranasal administration | C57BL/6 mice | Ch Mw: 115 kDa, DDA 95% | pGRP (0.5 mg) | Ch was conjugated with D-mannose | Production of anti-GRP IgG and inhibition of tumor growth | [94] |
| Intramuscular and subcutaneous administration | Balb/c mice | Depolymerized Ch 92–10 and 80–80 (Mw-DDA) | pVax1-4sFGF-2 and pVax1-PDGF-BB (encoding for FGF-2 and PDGF recombinant proteins) | – | Enhancing the repair of cartilage lesions or enhancing bone defect fill | [95] |
| Intramuscular and subcutaneous administration | Zucker Diabetic Fatty rats | Depolymerized Ch 92–10 and 80–80 (Mw-DDA) | pVax1-GLP1 encoding for the recombinant GLP-1 (165 μg) | – | Type 2 diabetes treatment | [96] |
| Intratumoral administration | C.B-17/Icr-scid-bg mice | Ch Mw 15.5 kDa, DDA 75%–85% | pAcEGFP1-C1 and Luc reporter plasmids encoding GFP and luciferase (100 μg) | – | Cancer treatment | [97] |
| Intravenous administration | Mice | – | pUC 19 encoding β-galactosidase reporter gene | Hydrophobically modified LMWC | – | [98] |
| Intravenous administration | Balb/c mice | Depolimerized Ch (7 kDa and 10 kDa) | pGL3 luciferase reporting gene (25 μg) | Ch was conjugated with bPEI and further with tuftsin | – | [99] |
7. Future Prospects
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Dewey, R.A.; Morrissey, G.; Cowsill, C.M.; Stone, D.; Bolognani, F.; Dodd, N.J.; Southgate, T.D.; Klatzmann, D.; Lassmann, H.; Castro, M.G.; et al. Chronic brain inflammation and persistent herpes simplex virus 1 thymidine kinase expression in survivors of syngeneic glioma treated by adenovirus-mediated gene therapy: Implications for clinical trials. Nat. Med. 1999, 5, 1256–1263. [Google Scholar] [CrossRef]
- Fox, J.L. Gene-therapy death prompts broad civil lawsuit. Nat. Biotechnol. 2000, 18. [Google Scholar] [CrossRef]
- Li, S.D.; Huang, L. Gene therapy progress and prospects: Non-viral gene therapy by systemic delivery. Gene Ther. 2006, 13, 1313–1319. [Google Scholar] [CrossRef]
- Ibraheem, D.; Elaissari, A.; Fessi, H. Gene therapy and dna delivery systems. Int. J. Pharm. 2014, 459, 70–83. [Google Scholar] [CrossRef]
- Lv, H.; Zhang, S.; Wang, B.; Cui, S.; Yan, J. Toxicity of cationic lipids and cationic polymers in gene delivery. J. Control. Release 2006, 114, 100–109. [Google Scholar] [CrossRef]
- Tong, H.; Qin, S.; Fernandes, J.; Li, L.; Dai, K.; Zhang, X. Progress and prospects of chitosan and its derivatives as non-viral gene vectors in gene therapy. Curr. Gene Ther. 2009, 496–502. [Google Scholar]
- Mao, S.; Sun, W.; Kissel, T. Chitosan-based formulations for delivery of DNA and siRNA. Adv. Drug Deliv. Rev. 2010, 62, 12–27. [Google Scholar] [CrossRef]
- Garcia-Fuentes, M.; Alonso, M.J. Chitosan-based drug nanocarriers: Where do we stand? J. Control. Release 2012, 161, 496–504. [Google Scholar] [CrossRef]
- Raftery, R.; O'Brien, F.; Cryan, S. Chitosan for gene delivery and orthopedic tissue engineering applications. Molecules 2013, 18, 5611–5647. [Google Scholar] [CrossRef]
- Buschmann, M.D.; Merzouki, A.; Lavertu, M.; Thibault, M.; Jean, M.; Darras, V. Chitosans for delivery of nucleic acids. Adv. Drug Deliv. Rev. 2013, 65, 1234–1270. [Google Scholar] [CrossRef]
- Kean, T.; Thanou, M. Biodegradation, biodistribution and toxicity of chitosan. Adv. Drug Deliv. Rev. 2010, 62, 3–11. [Google Scholar] [CrossRef]
- Jo, G.H.; Park, R.D.; Jung, W.J. Enzymatic production of chitin from crustacean shell waste. In Chitin, Chitosan, Oligosaccharides and Their Derivatives; Kim, S.K., Ed.; CRC Press: Boca Raton, FL, USA, 2011; pp. 37–45. [Google Scholar]
- Filion, D.; Lavertu, M.; Buschmann, M.D. Ionization and solubility of chitosan solutions related to thermosensitive chitosan/glycerol-phosphate systems. Biomacromolecules 2007, 8, 3224–3234. [Google Scholar] [CrossRef]
- Sorlier, P.; Denuziere, A.; Viton, C.; Domard, A. Relation between the degree of acetylation and the electrostatic properties of chitin and chitosan. Biomacromolecules 2001, 2, 765–772. [Google Scholar] [CrossRef]
- Strand, S.P.; Tommeraas, K.; Varum, K.M.; Ostgaard, K. Electrophoretic light scattering studies of chitosans with different degrees of N-acetylation. Biomacromolecules 2001, 2, 1310–1314. [Google Scholar] [CrossRef]
- Aam, B.B.; Heggset, E.B.; Norberg, A.L.; Sorlie, M.; Varum, K.M.; Eijsink, V.G. Production of chitooligosaccharides and their potential applications in medicine. Mar. Drugs 2010, 8, 1482–1517. [Google Scholar] [CrossRef]
- Romoren, K.; Pedersen, S.; Smistad, G.; Evensen, O.; Thu, B.J. The influence of formulation variables on in vitro transfection efficiency and physicochemical properties of chitosan-based polyplexes. Int. J. Pharm. 2003, 261, 115–127. [Google Scholar] [CrossRef]
- Strand, S.P.; Lelu, S.; Reitan, N.K.; de Lange Davies, C.; Artursson, P.; Vårum, K.M. Molecular design of chitosan gene delivery systems with an optimized balance between polyplex stability and polyplex unpacking. Biomaterials 2010, 31, 975–987. [Google Scholar] [CrossRef]
- Huang, M.; Fong, C.; Khor, E.; Lim, L. Transfection efficiency of chitosan vectors: Effect of polymer molecular weight and degree of deacetylation. J. Controll. Release 2005, 106, 391–406. [Google Scholar] [CrossRef]
- Lee, M.; Nah, J.W.; Kwon, Y.; Koh, J.J.; Ko, K.S.; Kim, S.W. Water-soluble and low molecular weight chitosan-based plasmid DNA delivery. Pharm. Res. 2001, 18, 427–431. [Google Scholar] [CrossRef]
- Duceppe, N.; Tabrizian, M. Factors influencing the transfection efficiency of ultra low molecular weight chitosan/hyaluronic acid nanoparticles. Biomaterials 2009, 30, 2625–2631. [Google Scholar] [CrossRef]
- Koping-Hoggard, M.; Varum, K.M.; Issa, M.; Danielsen, S.; Christensen, B.E.; Stokke, B.T.; Artursson, P. Improved chitosan-mediated gene delivery based on easily dissociated chitosan polyplexes of highly defined chitosan oligomers. Gene Ther. 2004, 11, 1441–1452. [Google Scholar] [CrossRef]
- Kiang, T.; Wen, J.; Lim, H.W.; Leong, K.W. The effect of the degree of chitosan deacetylation on the efficiency of gene transfection. Biomaterials 2004, 25, 5293–5301. [Google Scholar] [CrossRef]
- Ma, P.L.; Lavertu, M.; Winnik, F.M.; Buschmann, M.D. New insights into chitosan-DNA interactions using isothermal titration microcalorimetry. Biomacromolecules 2009, 10, 1490–1499. [Google Scholar] [CrossRef]
- Kim, S.; Rajapakse, N. Enzymatic production and biological activities of chitosan oligosaccharides (COS): A review. Carbohydr. Polym. 2005, 62, 357–368. [Google Scholar] [CrossRef]
- Lavertu, M.; Methot, S.; Buschmann, M. Composition Method for Efficient Delivery of Nucleic Acids to Cells using Chitosan. Patents. WO2007059605-A1; EP1948810-A1; US2009075383-A1; CA26283131-A1, 31 May 2007. [Google Scholar]
- Strand, S.P.; Danielsen, S.; Christensen, B.E.; Varum, K.M. Influence of chitosan structure on the formation and stability of DNA-chitosan polyelectrolyte complexes. Biomacromolecules 2005, 6, 3357–3366. [Google Scholar] [CrossRef]
- Klausner, E.A.; Zhang, Z.; Chapman, R.L.; Multack, R.F.; Volin, M.V. Ultrapure chitosan oligomers as carriers for corneal gene transfer. Biomaterials 2010, 31, 1814–1820. [Google Scholar] [CrossRef]
- Puras, G.; Zarate, J.; Aceves, M.; Murua, A.; Díaz, A.R.; Avilés-Triguero, M.; Fernández, E.; Pedraz, J.L. Low Molecular Weight Oligochitosans for Non-Viral Retinal Gene Therapy. Eur. J. Pharm. Biopharm. 2012, 2012. [Google Scholar] [CrossRef]
- Agirre, M.; Zarate, J.; Puras, G.; Ojeda, E.; Pedraz, J.L. Improving transfection efficiency of ultrapure oligochitosan/DNA polyplexes by medium acidification. Drug Deliv. 2014, in press. [Google Scholar]
- Anchordoquy, T.J.; Koe, G.S. Physical stability of nonviral plasmid-based therapeutics. J. Pharm. Sci. 2000, 89, 289–296. [Google Scholar] [CrossRef]
- Pfeifer, C.; Hasenpusch, G.; Uezguen, S.; Aneja, M.K.; Reinhardt, D.; Kirch, J.; Schneider, M.; Claus, S.; Friess, W.; Rudolph, C. Dry powder aerosols of polyethylenimine (PEI)-based gene vectors mediate efficient gene delivery to the lung. J. Controll. Release 2011, 154, 69–76. [Google Scholar] [CrossRef]
- Puras, G.; Zarate, J.; Díaz-Tahoces, A.; Avilés-Trigueros, M.; Fernández, E.; Pedraz, J.L. Oligochitosan polyplexes as carriers for retinal gene delivery. Eur. J. Pharm. Sci. 2013, 48, 323–331. [Google Scholar] [CrossRef]
- Klausner, E.A.; Zhang, Z.; Wong, S.P.; Chapman, R.L.; Volin, M.V.; Harbottle, R.P. Corneal gene delivery: Chitosan oligomer as a carrier of CpG rich, CpG free or S/MAR plasmid DNA. J. Gene Med. 2012, 14, 100–108. [Google Scholar] [CrossRef]
- Rungsardthong, U.; Ehtezazi, T.; Bailey, L.; Armes, S.P.; Garnett, M.C.; Stolnik, S. Effect of polymer ionization on the interaction with DNA in nonviral gene delivery systems. Biomacromolecules 2003, 4, 683–690. [Google Scholar] [CrossRef]
- Köping-Höggård, M.; Mel'nikova, Y.S.; Vårum, K.M.; Lindman, B.; Artursson, P. Relationship between the physical shape and the efficiency of oligomeric chitosan as a gene delivery system in Vitro and in Vivo. J. Gene Med. 2003, 5, 130–141. [Google Scholar]
- Lou, Y.; Peng, Y.; Chen, B.; Wang, L.; Leong, K.W. Poly(ethylene imine)-g-chitosan using EX-810 as a spacer for nonviral gene delivery vectors. J. Biomed. Mater. Res. A 2009, 88A, 1058–1068. [Google Scholar] [CrossRef]
- Erbacher, P.; Zou, S.; Bettinger, T.; Steffan, A.M.; Remy, J.S. Chitosan-based vector/DNA complexes for gene delivery: Biophysical characteristics and transfection efficiency. Pharm. Res. 1998, 15, 1332–1339. [Google Scholar]
- MacLaughlin, F.C.; Mumper, R.J.; Wang, J.; Tagliaferri, J.M.; Gill, I.; Hinchcliffe, M.; Rolland, A.P. Chitosan and depolymerized chitosan oligomers as condensing carriers for in vivo plasmid delivery. J. Controll. Release 1998, 56, 259–272. [Google Scholar]
- Schroeder, A.; Heller, D.; Winslow, M.; Dahlman, J.; Pratt, G.; Langer, R.; Jacks, T.; Anderson, D. Treating metastatic cancer with nanotechnology. Nat. Rev. Cancer 2011, 12, 39–50. [Google Scholar] [CrossRef]
- Nimesh, S.; Thibault, M.; Lavertu, M.; Thibault, M. Enhanced gene delivery mediated by low molecular weight chitosan/DNA complexes: Effect of pH and serum. 2010, 46, 182–196. [Google Scholar]
- Danielsen, S.; Varum, K.M.; Stokke, B.T. Structural analysis of chitosan mediated DNA condensation by AFM: Influence of chitosan molecular parameters. Biomacromolecules 2004, 5, 928–936. [Google Scholar] [CrossRef]
- Corsi, K.; Chellat, F.; Yahia, L.; Fernandes, J.C. Mesenchymal stem cells, MG63 and HEK293 transfection using chitosan-DNA nanoparticles. Biomaterials 2003, 24, 1255–1264. [Google Scholar] [CrossRef]
- Lu, B.; Wang, C.; Wu, D.; Li, C.; Zhang, X.; Zhuo, R. Chitosan based oligoamine polymers: Synthesis, characterization, and gene delivery. J. Control. Release 2009, 137, 54–62. [Google Scholar] [CrossRef]
- Richard, I.; Thibault, M.; De Crescenzo, G.; Buschmann, M.; Lavertu, M. Ionization behavior of chitosan and chitosan-DNA polyplexes indicate that chitosan has a similar capability to induce a proton-sponge effect as PEI. Biomacromolecules 2013, 14, 1732–1740. [Google Scholar] [CrossRef]
- Strand, S.P.; Issa, M.M.; Christensen, B.E.; Varum, K.M.; Artursson, P. Tailoring of chitosans for gene delivery: novel self-branched glycosylated chitosan oligomers with improved functional properties. Biomacromolecules 2008, 9, 3268–3276. [Google Scholar] [CrossRef]
- Ruponen, M.; Honkakoski, P.; Tammi, M.; Urtti, A. Cell-surface glycosaminoglycans inhibit cation-mediated gene transfer. J. Gene Med. 2004, 6, 405–414. [Google Scholar]
- Danielsen, S.; Strand, S.; de Lange Davies, C.; Stokke, B.T. Glycosaminoglycan destabilization of DNA–chitosan polyplexes for gene delivery depends on chitosan chain length and GAG properties. Biochim. Biophys. Acta 2005, 1721, 44–54. [Google Scholar] [CrossRef]
- Hashimoto, M.; Morimoto, M.; Saimoto, H.; Shigemasa, Y.; Sato, T. Lactosylated chitosan for DNA delivery into hepatocytes: The effect of lactosylation on the physicochemical properties and intracellular trafficking of pDNA/chitosan complexes. Bioconjug.Chem. 2006, 17, 309–316. [Google Scholar] [CrossRef]
- Jiang, X.; Dai, H.; Leong, K.W.; Goh, S.; Mao, H.; Yang, Y. Chitosan-g-PEG/DNA complexes deliver gene to the rat liver via intrabiliary and intraportal infusions. J. Gene Med. 2006, 8, 477–487. [Google Scholar] [CrossRef]
- Thanou, M.; Florea, B.I.; Geldof, M.; Junginger, H.E.; Borchard, G. Quaternized chitosan oligomers as novel gene delivery vectors in epithelial cell lines. Biomaterials 2002, 23, 153–159. [Google Scholar] [CrossRef]
- Issa, M.M.; Köping-Höggård, M.; Tømmeraas, K.; Vårum, K.M.; Christensen, B.E.; Strand, S.P.; Artursson, P. Targeted gene delivery with trisaccharide-substituted chitosan oligomers in vitro and after lung administration in vivo. J. Controll. Release 2006, 115, 103–112. [Google Scholar] [CrossRef]
- Thibault, M.; Nimesh, S.; Lavertu, M.; Buschmann, M. Intracellular trafficking and decondensation kinetics of chitosan-pDNA polyplexes. Mol. Ther. 2010, 18, 1787–1795. [Google Scholar] [CrossRef]
- Yue, Z.; Wei, W.; Lv, P.; Yue, H.; Wang, L.; Su, Z.; Ma, G. Surface charge affects cellular uptake and intracellular trafficking of Chitosan-Based Nanoparticles. Biomacromolecules 2011, 12, 2440–2446. [Google Scholar] [CrossRef]
- Khalil, I.A.; Kogure, K.; Akita, H.; Harashima, H. Uptake pathways and subsequent intracellular trafficking in nonviral gene delivery. Pharmacol. Rev. 2006, 58, 32–45. [Google Scholar] [CrossRef]
- Xiang, S.; Tong, H.; Shi, Q.; Fernandes, J.C.; Jin, T.; Dai, K.; Zhang, X. Uptake mechanisms of non-viral gene delivery. J. Controll. Release 2012, 158, 371–378. [Google Scholar] [CrossRef]
- Garaiova, Z.; Strand, S.P.; Reitan, N.K.; Lélu, S.; Størset, S.Ø; Berg, K.; Malmo, J.; Folasire, O.; Bjørkøy, A.; de Lange Davies, C. Cellular uptake of DNA–chitosan nanoparticles: The role of clathrin- and caveolae-mediated pathways. Int. J. Biol. Macromol. 2012, 51, 1043–1051. [Google Scholar] [CrossRef]
- Conner, S.D.; Schmid, S.L. Regulated portals of entry into the cell. Nature 2003, 422, 37–44. [Google Scholar] [CrossRef]
- Douglas, K.L.; Piccirillo, C.A.; Tabrizian, M. Cell line-dependent internalization pathways and intracellular trafficking determine transfection efficiency of nanoparticle vectors. Eur. J. Pharm. Biopharm. 2008, 68, 676–687. [Google Scholar] [CrossRef]
- Fang, N.; Chan, V.; Mao, H.; Leong, K. Interactions of phospholipid bilayer with chitosan: Effect of molecular weight and pH. Biomacromolecules 2001, 2, 1161–1168. [Google Scholar] [CrossRef]
- Hsu, C.; Uludağ, H. Nucleic-acid based gene therapeutics: Delivery challenges and modular design of nonviral gene carriers and expression cassettes to overcome intracellular barriers for sustained targeted expression. J Drug Target 2012, 20, 301–328. [Google Scholar] [CrossRef]
- Douglas, K.L. Toward development of artificial viruses for gene therapy: A comparative evaluation of viral and non-viral transfection. Biotechnol. Prog. 2008, 24, 871–883. [Google Scholar]
- Chiu, Y.; Ho, Y.; Chen, Y.; Peng, S.; Ke, C.; Chen, K.; Mi, F.; Sung, H. The characteristics, cellular uptake and intracellular trafficking of nanoparticles made of hydrophobically-modified chitosan. J. Controll. Release 2010, 146, 152–159. [Google Scholar] [CrossRef]
- Contreras-Ruiz, L.; de la Fuente, M.; Párraga, J.; López-García, A.; Fernández, I.; Seijo, B.; Sánchez, A.; Calonge, M.; Diebold, Y. Intracellular trafficking of hyaluronic acid-chitosan oligomer-based nanoparticles in cultured human ocular surface cells. Mol. Vis. 2011, 17, 279–290. [Google Scholar]
- Thibault, M.; Astolfi, M.; Tran-Khanh, N.; Lavertu, M.; Darras, V.; Merzouki, A.; Buschmann, M.D. Excess polycation mediates efficient chitosan-based gene transfer by promoting lysosomal release of the polyplexes. Biomaterials 2011, 32, 4639–4646. [Google Scholar] [CrossRef]
- Männistö, M.; Rönkkö, S.; Mättö, M.; Honkakoski, P.; Hyttinen, M.; Pelkonen, J.; Urtti, A. The role of cell cycle on polyplex-mediated gene transfer into a retinal pigment epithelial cell line. J.Gene Med. 2005, 7, 466–476. [Google Scholar] [CrossRef]
- Ishii, T.; Okahata, Y.; Sato, T. Mechanism of cell transfection with plasmid/chitosan complexes. Biochim. Biophys. Acta 2001, 1514, 51–64. [Google Scholar] [CrossRef]
- Sato, T.; Ishii, T.; Okahata, Y. In vitro gene delivery mediated by chitosan. Effect of pH, serum, and molecular mass of chitosan on the transfection efficiency. Biomaterials 2001, 22, 2075–2080. [Google Scholar] [CrossRef]
- Lavertu, M.; Méthot, S.; Tran-Khanh, N.; Buschmann, M.D. High efficiency gene transfer using chitosan/DNA nanoparticles with specific combinations of molecular weight and degree of deacetylation. Biomaterials 2006, 27, 4815–4824. [Google Scholar] [CrossRef]
- Kean, T.; Roth, S.; Thanou, M. Trimethylated chitosans as non-viral gene delivery vectors: cytotoxicity and transfection efficiency. J. Controll. Release 2005, 103, 643–653. [Google Scholar] [CrossRef]
- Seong, H.; Whang, H.S.; Ko, S. Synthesis of a quaternary ammonium derivative of chito-oligosaccharide as antimicrobial agent for cellulosic fibers. J. Appl. Polym. Sci. 2000, 76, 2009–2015. [Google Scholar] [CrossRef]
- Hermanson, G. Bioconjugate Techniques; Academic Press: Waltham, MA, USA, 2008. [Google Scholar]
- Kim, T.H.; Ihm, J.E.; Choi, Y.J.; Nah, J.W.; Cho, C.S. Efficient gene delivery by urocanic acid-modified chitosan. J. Controll. Release 2003, 93, 389–402. [Google Scholar] [CrossRef]
- Chang, K.L.; Higuchi, Y.; Kawakami, S.; Yamashita, F.; Hashida, M. Efficient gene transfection by histidine-modified chitosan through enhancement of endosomal escape. Bioconjug. Chem. 2010, 21, 1087–1095. [Google Scholar] [CrossRef]
- Pires, L.R.; Oliveira, H.; Barrias, C.C.; Sampaio, P.; Pereira, A.J.; Maiato, H.; Simoes, S.; Pego, A.P. Imidazole-grafted chitosan-mediated gene delivery: In vitro study on transfection, intracellular trafficking and degradation. Nanomedicine 2011, 6, 1499–1512. [Google Scholar] [CrossRef]
- Morris, V.B.; Sharma, C.P. Folate mediated in vitro targeting of depolymerised trimethylated chitosan having arginine functionality. J. Colloid Interface Sci. 2010, 348, 360–368. [Google Scholar] [CrossRef]
- Tømmeraas, K.; Strand, S.P.; Tian, W.; Kenne, L.; Vårum, K.M. Preparation and characterisation of fluorescent chitosans using 9-anthraldehyde as fluorophore. Carbohydr. Res. 2001, 336, 291–296. [Google Scholar] [CrossRef]
- Tiera, M.J.; Qiu, X.P.; Bechaouch, S.; Shi, Q.; Fernandes, J.C.; Winnik, F.M. Synthesis and characterization of phosphorylcholine-substituted chitosans soluble in physiological pH conditions. Biomacromolecules 2006, 7, 3151–3156. [Google Scholar] [CrossRef]
- Case, A.H.; Dalla Picola, I.P.; Zaniquelli, M.E.; Fernandes, J.C.; Taboga, S.R.; Winnik, F.M.; Tiera, M.J. Physicochemical characterization of nanoparticles formed between DNA and phosphorylcholine substituted chitosans. J. Colloid Interface Sci. 2009, 336, 125–133. [Google Scholar] [CrossRef]
- Tommeraas, K.; Koping-Hoggard, M.; Varum, K.M.; Christensen, B.E.; Artursson, P.; Smidsrod, O. Preparation and characterisation of chitosans with oligosaccharide branches. Carbohydr. Res. 2002, 337, 2455–2462. [Google Scholar] [CrossRef]
- Park, I.K.; Park, Y.H.; Shin, B.A.; Choi, E.S.; Kim, Y.R.; Akaike, T.; Cho, C.S. Galactosylated chitosan-graft-dextran as hepatocyte-targeting DNA carrier. J. Controll. Release 2000, 69, 97–108. [Google Scholar] [CrossRef]
- Ercelen, S.; Zhang, X.; Duportail, G.; Grandfils, C.; Desbrières, J.; Karaeva, S.; Tikhonov, V.; Mély, Y.; Babak, V. Physicochemical properties of low molecular weight alkylated chitosans: A new class of potential nonviral vectors for gene delivery. Colloids Surf. B Biointerfaces 2006, 51, 140–148. [Google Scholar] [CrossRef]
- Clement, J.; Kiefer, K.; Kimpfler, A.; Garidel, P.; Peschka-Süss, R. Large-scale production of lipoplexes with long shelf-life. Eur. J. Pharm. Biopharm. 2005, 59, 35–43. [Google Scholar] [CrossRef]
- Kasper, J.C.; Schaffert, D.; Ogris, M.; Wagner, E.; Friess, W. Development of a Lyophilized Plasmid/LPEI Polyplex Formulation with Long-Term stability—A Step Closer from Promising Technology to Application. J. Controlled Release 2011, 151, 246–255. [Google Scholar] [CrossRef]
- Kasper, J.C.; Troiber, C.; Küchler, S.; Wagner, E.; Friess, W. Formulation development of lyophilized, long-term stable siRNA/oligoaminoamide polyplexes. Eur. J. Pharm. Biopharm. 2013, 85, 294–305. [Google Scholar] [CrossRef]
- Abdelwahed, W.; Degobert, G.; Stainmesse, S.; Fessi, H. Freeze-drying of nanoparticles: formulation, process and storage considerations. Adv. Drug Deliv. Rev. 2006, 58, 1688–1713. [Google Scholar] [CrossRef]
- Anchordoquy, T.J.; Armstrong, T.K.; Molina, M.d.C. Low molecular weight dextrans stabilize nonviral vectors during lyophilization at low osmolalities: Concentrating suspensions by rehydration to reduced volumes. J. Pharm. Sci. 2005, 94, 1226–1236. [Google Scholar] [CrossRef]
- Vauthier, C.; Cabane, B.; Labarre, D. How to concentrate nanoparticles and avoid aggregation? Eur. J. Pharm. Biopharm. 2008, 69, 466–475. [Google Scholar] [CrossRef]
- Csaba, N.; Garcia-Fuentes, M.; Alonso, M.J. The performance of nanocarriers for transmucosal drug delivery. Expert Opin. Drug Deliv. 2006, 3, 463–478. [Google Scholar] [CrossRef]
- De la Fuente, M.; Ravina, M.; Paolicelli, P.; Sanchez, A.; Seijo, B.; Alonso, M.J. Chitosan-based nanostructures: A delivery platform for ocular therapeutics. Adv. Drug Deliv. Rev. 2010, 62, 100–117. [Google Scholar] [CrossRef]
- De la Fuente, M.; Seijo, B.; Alonso, M. Bioadhesive hyaluronan-chitosan nanoparticles can transport genes across the ocular mucosa and transfect ocular tissue. Gene Ther. 2008, 15, 668–676. [Google Scholar] [CrossRef]
- Mohammadi, Z.; Dorkoosh, F.A.; Hosseinkhani, S.; Gilani, K.; Amini, T.; Najafabadi, A.R.; Tehrani, M.R. In vivo transfection study of chitosan-DNA-FAP-B nanoparticles as a new non viral vector for gene delivery to the lung. Int. J. Pharm. 2011, 421, 183–188. [Google Scholar] [CrossRef]
- Yang, X.; Yuan, X.; Cai, D.; Wang, S.; Zong, L. Low molecular weight chitosan in DNA vaccine delivery via mucosa. Int. J. Pharm. 2009, 375, 123–132. [Google Scholar] [CrossRef]
- Yao, W.; Peng, Y.; Du, M.; Luo, J.; Zong, L. Preventative vaccine-loaded mannosylated chitosan nanoparticles intended for nasal mucosal delivery enhance immune responses and potent tumor immunity. Mol. Pharm. 2013, 10, 2904–2914. [Google Scholar] [CrossRef]
- Jean, M.; Smaoui, F.; Lavertu, M.; Methot, S.; Bouhdoud, L.; Buschmann, M.D.; Merzouki, A. Chitosan-plasmid nanoparticle formulations for IM and SC delivery of recombinant FGF-2 and PDGF-BB or generation of antibodies. Gene Ther. 2009, 16, 1097–1110. [Google Scholar]
- Jean, M.; Alameh, M.; De Jesus, D.; Thibault, M.; Lavertu, M.; Darras, V.; Nelea, M.; Buschmann, M.D.; Merzouki, A. Chitosan-based therapeutic nanoparticles for combination gene therapy and gene silencing of in vitro cell lines relevant to type 2 diabetes. Eur. J. Pharm.Sci. 2012, 45, 138–149. [Google Scholar] [CrossRef]
- Yang, S.; Chang, S.; Tsai, K.; Chen, W.; Lin, F.; Shieh, M. Effect of chitosan-alginate nanoparticles and ultrasound on the efficiency of gene transfection of human cancer cells. J. Gene Med. 2010, 12, 168–179. [Google Scholar]
- Zhang, X.; Ercelen, S.; Duportail, G.; Schaub, E.; Tikhonov, V.; Slita, A.; Zarubaev, V.; Babak, V.; Mély, Y. Hydrophobically modified low molecular weight chitosans as efficient and nontoxic gene delivery vectors. J. Gene Med. 2008, 10, 527–539. [Google Scholar] [CrossRef]
- Tripathi, S.K.; Goyal, R.; Kashyap, M.P.; Pant, A.B.; Haq, W.; Kumar, P.; Gupta, K.C. Depolymerized chitosans functionalized with bPEI as carriers of nucleic acids and tuftsin-tethered conjugate for macrophage targeting. Biomaterials 2012, 33, 4204–4219. [Google Scholar] [CrossRef]
- United States Pharmacopeial Convention. Chitosan. In United States Pharmacopeial and The National Formulary (USP–NF); United States Pharmacopeial Convention: Rockville, MD, USA, 2011; Volume 29, pp. S5361–S5365. [Google Scholar]
- American Society for Testing and Materials. Standard Guide for Characterization and Testing of Chitosan Salts as Starting Materials Intended for Use in Biomedical and Tissue-Engineered Medical Product Applications; ASTM Standard F2103; American Society for Testing and Materials (ASTM): West Conshohocken, PA, USA, 2011. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Agirre, M.; Zarate, J.; Ojeda, E.; Puras, G.; Desbrieres, J.; Pedraz, J.L. Low Molecular Weight Chitosan (LMWC)-based Polyplexes for pDNA Delivery: From Bench to Bedside. Polymers 2014, 6, 1727-1755. https://doi.org/10.3390/polym6061727
Agirre M, Zarate J, Ojeda E, Puras G, Desbrieres J, Pedraz JL. Low Molecular Weight Chitosan (LMWC)-based Polyplexes for pDNA Delivery: From Bench to Bedside. Polymers. 2014; 6(6):1727-1755. https://doi.org/10.3390/polym6061727
Chicago/Turabian StyleAgirre, Mireia, Jon Zarate, Edilberto Ojeda, Gustavo Puras, Jacques Desbrieres, and Jose Luis Pedraz. 2014. "Low Molecular Weight Chitosan (LMWC)-based Polyplexes for pDNA Delivery: From Bench to Bedside" Polymers 6, no. 6: 1727-1755. https://doi.org/10.3390/polym6061727