Links among Microbial Communities, Soil Properties and Functions: Are Fungi the Sole Players in Decomposition of Bio-Based and Biodegradable Plastic?
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Procedure: Soil, PBSA, and Experimental Conditions
2.2. Analyses of Microbial Communities in Soils
2.3. Soil Physicochemical Properties and Enzyme Analyses
2.4. Determination of Ergosterol
2.5. Statistical Analysis
3. Results
3.1. Bacteria and Fungi in Soils without PBSA and Soils of PBSA–Soil Systems: Who Is Who?
3.2. Effects of PBSA and N Addition on Soil Properties, Fungal Biomass
3.3. Microbial Communities Are Shaped by Soil Physicochemical Properties and Linked to Soil Functions
3.4. Enzymes Activity Patterns, Soil Properties, and Fungal Biomass: Are There Any Links?
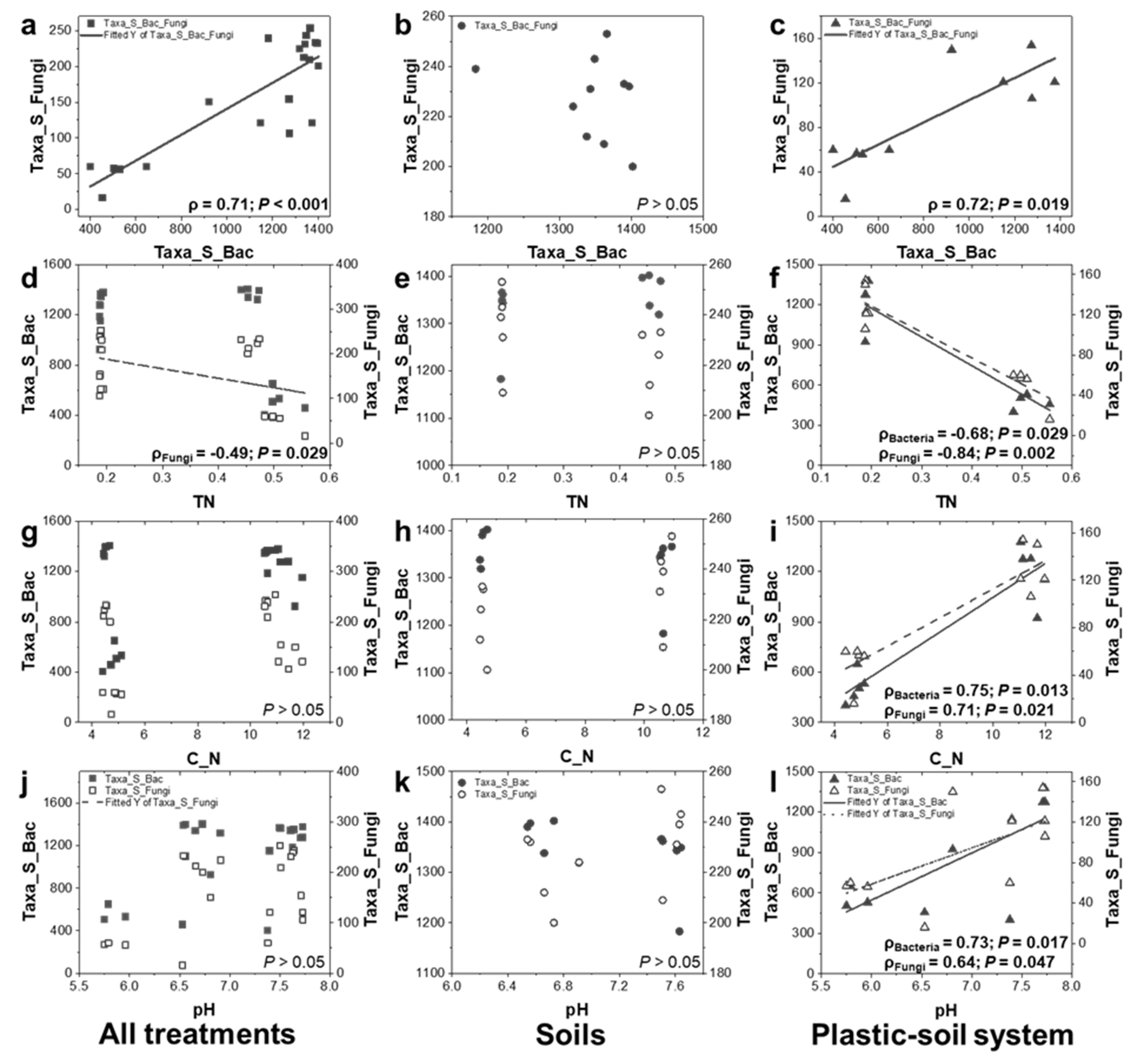
3.5. Relationships between Microbial Richness and Soil Properties: Significant Differences between Soils and PBSA–Soil Systems
3.6. Relationships between Fungal Biomass and Soil Functions: Consistent for C and N Cycles
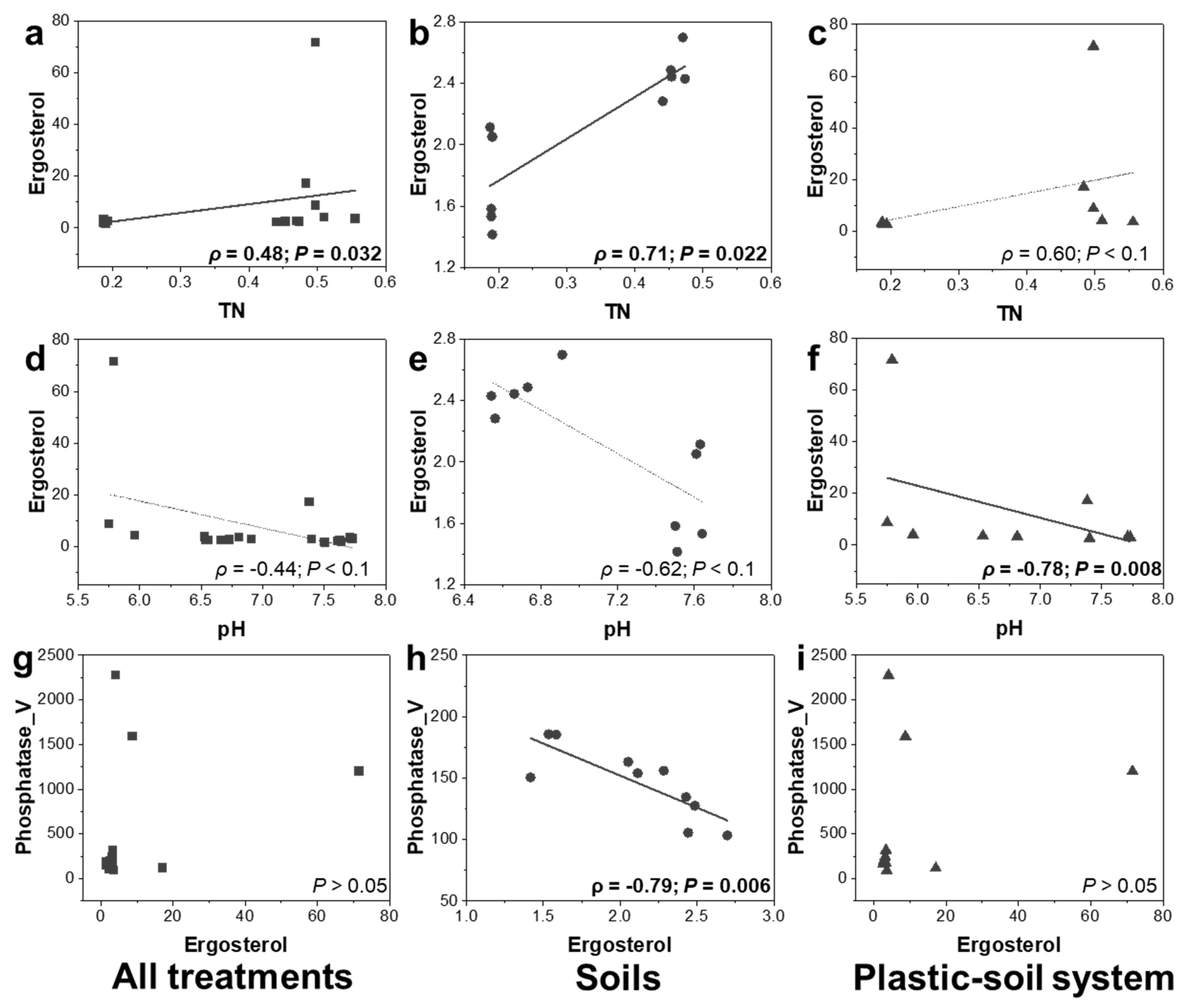
3.7. Relationships between Ergosterol Content and Soil Properties
4. Discussion
4.1. Presence of PBSA Alters Link between Bacterial and Fungal Richness and Its Relationships with Soil Properties
4.2. Soil Nutrient Cycling Is Still Functioning despite the High Load of PBSA but How about Soil Health?
4.3. Functional Redundancy, Competitions, and Degradation Efficiency
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Van den Oever, M.; Molenveld, K.; Zee, M.; Bos, H. Bio-Based and Biodegradable Plastics—Facts and Figures. Focus on Food Packaging in the Netherlands; Wageningen Food & Biobased Research: Wageningen, The Netherlands, 2017. [Google Scholar] [CrossRef] [Green Version]
- Di Bartolo, A.; Infurna, G.; Dintcheva, N.T. A Review of Bioplastics and Their Adoption in the Circular Economy. Polymers 2021, 13, 1229. [Google Scholar] [CrossRef]
- Haider, T.P.; Völker, C.; Kramm, J.; Landfester, K.; Wurm, F.R. Plastics of the Future? The Impact of Biodegradable Polymers on the Environment and on Society. Angew. Chem. Int. Ed. 2019, 58, 50–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, Y.; Wu, C.; Xue, Q.; Hui, X. Effects of plastic contamination on water evaporation and desiccation cracking in soil. Sci. Total Environ. 2019, 654, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Liwarska-Bizukojc, E. Effect of (bio)plastics on soil environment: A review. Sci. Total Environ. 2021, 795, 148889. [Google Scholar] [CrossRef] [PubMed]
- Purahong, W.; Wahdan, S.F.M.; Heinz, D.; Jariyavidyanont, K.; Sungkapreecha, C.; Tanunchai, B.; Sansupa, C.; Sadubsarn, D.; Alaneed, R.; Heintz-Buschart, A.; et al. Back to the Future: Decomposability of a Biobased and Biodegradable Plastic in Field Soil Environments and Its Microbiome under Ambient and Future Climates. Environ. Sci. Technol. 2021, 55, 12337–12351. [Google Scholar] [CrossRef]
- Scheid, S.-M.; Juncheed, K.; Tanunchai, B.; Wahdan, S.F.M.; Buscot, F.; Noll, M.; Purahong, W. Interactions Between High Load of a Bio-based and Biodegradable Plastic and Nitrogen Fertilizer Affect Plant Biomass and Health: A Case Study with Fusarium solani and Mung Bean (Vigna radiata L.). J. Polym. Environ. 2022. [Google Scholar] [CrossRef]
- Brizga, J.; Hubacek, K.; Feng, K. The Unintended Side Effects of Bioplastics: Carbon, Land, and Water Footprints. One Earth 2020, 3, 45–53. [Google Scholar] [CrossRef]
- Cheroennet, N.; Pongpinyopap, S.; Leejarkpai, T.; Suwanmanee, U. A trade-off between carbon and water impacts in bio-based box production chains in Thailand: A case study of PS, PLAS, PLAS/starch, and PBS. J. Clean. Prod. 2017, 167, 987–1001. [Google Scholar] [CrossRef]
- Tanunchai, B.; Juncheed, K.; Wahdan, S.F.M.; Guliyev, V.; Udovenko, M.; Lehnert, A.-S.; Alves, E.G.; Glaser, B.; Noll, M.; Buscot, F.; et al. Analysis of microbial populations in plastic–soil systems after exposure to high poly(butylene succinate-co-adipate) load using high-resolution molecular technique. Environ. Sci. Eur. 2021, 33, 105. [Google Scholar] [CrossRef]
- Qi, Y.; Yang, X.; Pelaez, A.M.; Huerta Lwanga, E.; Beriot, N.; Gertsen, H.; Garbeva, P.; Geissen, V. Macro- and micro- plastics in soil-plant system: Effects of plastic mulch film residues on wheat (Triticum aestivum) growth. Sci. Total Environ. 2018, 645, 1048–1056. [Google Scholar] [CrossRef]
- Powlson, D.S.; Hirsch, P.; Brookes, P. The role of soil microorganisms in soil organic matter conservation in the tropics. Nutr. Cycl. Agroecosyst. 2001, 61, 41–51. [Google Scholar] [CrossRef]
- Abe, M.; Kobayashi, K.; Honma, N.; Nakasaki, K. Microbial degradation of poly(butylene succinate) by Fusarium solani in soil environments. Polym. Degrad. Stab. 2010, 95, 138–143. [Google Scholar] [CrossRef]
- Yamamoto-Tamura, K.; Hoshino, Y.T.; Tsuboi, S.; Huang, C.; Kishimoto-Mo, A.W.; Sameshima-Yamashita, Y.; Kitamoto, H. Fungal community dynamics during degradation of poly(butylene succinate-co-adipate) film in two cultivated soils in Japan. Biosci. Biotechnol. Biochem. 2020, 84, 1077–1087. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Brandón, M.; Probst, M.; Siles, J.A.; Peintner, U.; Bardelli, T.; Egli, M.; Insam, H.; Ascher-Jenull, J. Fungal communities and their association with nitrogen-fixing bacteria affect early decomposition of Norway spruce deadwood. Sci. Rep. 2020, 10, 8025. [Google Scholar] [CrossRef]
- Zumstein, M.T.; Schintlmeister, A.; Nelson, T.F.; Baumgartner, R.; Woebken, D.; Wagner, M.; Kohler, H.-P.E.; McNeill, K.; Sander, M. Biodegradation of synthetic polymers in soils: Tracking carbon into CO2 and microbial biomass. Sci. Adv. 2018, 4, eaas9024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Gui, H.; Banfield, C.C.; Wen, Y.; Zang, H.; Dippold, M.A.; Charlton, A.; Jones, D.L. The microplastisphere: Biodegradable microplastics addition alters soil microbial community structure and function. Soil Biol. Biochem. 2021, 156, 108211. [Google Scholar] [CrossRef]
- Adhikari, D.; Masaki, M.; Kubota, K.; Kai, T.; Kaneko, N.; Araki, K.; Kubo, M. Degradation of Bioplastics in Soil and Their Degradation Effects on Environmental Microorganisms. J. Agric. Chem. Environ. 2016, 05, 23–34. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Myrold, D.D.; Shi, L.; Kuzyakov, Y.; Dai, H.; Thu Hoang, D.T.; Dippold, M.A.; Meng, X.; Song, X.; Li, Z.; et al. Resistance of microbial community and its functional sensitivity in the rhizosphere hotspots to drought. Soil Biol. Biochem. 2021, 161, 108360. [Google Scholar] [CrossRef]
- Riah, W.; Trinsoutrot Gattin, I.; Martin-Laurent, F.; Laroche-Ajzenberg, E.; Norini, M.-P.; Latour, X.; Laval, K. Soil microbial community structure and function relationships: A heat stress experiment. Appl. Soil Ecol. 2015, 86, 121–130. [Google Scholar] [CrossRef]
- Kieft, B.; Li, Z.; Bryson, S.; Crump, B.C.; Hettich, R.; Pan, C.; Mayali, X.; Mueller, R.S. Microbial Community Structure–Function Relationships in Yaquina Bay Estuary Reveal Spatially Distinct Carbon and Nitrogen Cycling Capacities. Front. Microbiol. 2018, 9, 1282. [Google Scholar] [CrossRef] [Green Version]
- Purahong, W.; Schloter, M.; Pecyna, M.J.; Kapturska, D.; Däumlich, V.; Mital, S.; Buscot, F.; Hofrichter, M.; Gutknecht, J.L.M.; Krüger, D. Uncoupling of microbial community structure and function in decomposing litter across beech forest ecosystems in Central Europe. Sci. Rep. 2014, 4, 7014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rizzarelli, P.; Impallomeni, G.; Montaudo, G. Evidence for Selective Hydrolysis of Aliphatic Copolyesters Induced by Lipase Catalysis. Biomacromolecules 2003, 5, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Siracusa, V. Microbial Degradation of Synthetic Biopolymers Waste. Polymers 2019, 11, 1066. [Google Scholar] [CrossRef] [Green Version]
- Dorodnikov, M.; Blagodatskaya, E.; Blagodatsky, S.; Marhan, S.; Fangmeier, A.; Kuzyakov, Y. Stimulation of microbial extracellular enzyme activities by elevated CO2 depends on soil aggregate size. Glob. Chang. Biol. 2009, 15, 1603–1614. [Google Scholar] [CrossRef]
- Ananbeh, H.; Stojanović, M.; Pompeiano, A.; Voběrková, S.; Trasar-Cepeda, C. Use of soil enzyme activities to assess the recovery of soil functions in abandoned coppice forest systems. Sci. Total Environ. 2019, 694, 133692. [Google Scholar] [CrossRef] [PubMed]
- Elemuo, N.; Ikiensikimama, S.; Ubani, C.; Egwim, E.; Osuoha, J. Production and Characterization of Lipase Enzyme Expressed by Crude Oil Contaminated Soil Isolates. Univers. J. Microbiol. Res. 2019, 7, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Bharathi, D.; Rajalakshmi, G.; Komathi, S. Optimization and production of lipase enzyme from bacterial strains isolated from petrol spilled soil. J. King Saud Univ.-Sci. 2019, 31, 898–901. [Google Scholar] [CrossRef]
- Khan, I.; Nagarjuna, R.; Dutta, J.R.; Ganesan, R. Enzyme-Embedded Degradation of Poly(ε-caprolactone) using Lipase-Derived from Probiotic Lactobacillus plantarum. ACS Omega 2019, 4, 2844–2852. [Google Scholar] [CrossRef]
- Kumar, A.; Dorodnikov, M.; Splettstößer, T.; Kuzyakov, Y.; Pausch, J. Effects of maize roots on aggregate stability and enzyme activities in soil. Geoderma 2017, 306, 50–57. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Lambers, H.; Bolland, M.; Veneklaas, E. Distribution of Carboxylates and Acid Phosphatase and Depletion of Different Phosphorus Fractions in the Rhizosphere of a Cereal and Three Grain Legumes. Plant Soil 2006, 281, 109–120. [Google Scholar] [CrossRef]
- Brzezinska, M.S.; Jankiewicz, U.; Walczak, M. Biodegradation of chitinous substances and chitinase production by the soil actinomycete Streptomyces rimosus. Int. Biodeterior. Biodegrad. 2013, 84, 104–110. [Google Scholar] [CrossRef]
- Ch, S.; Grover, M.; Kundu, S.; Desai, S. Soil Enzymes; Taylor & Francis Group: Abingdon, UK, 2017; pp. 2100–2107. [Google Scholar] [CrossRef]
- Ferraz-Almeida, R.; Naves, E.; Mota, R. Soil quality: Enzymatic activity of soil β-glucosidase. Glob. J. Agric. Res. Rev. 2015, 3, 146–150. [Google Scholar]
- Kaiser, C.; Koranda, M.; Kitzler, B.; Fuchslueger, L.; Schnecker, J.; Schweiger, P.; Rasche, F.; Zechmeister-Boltenstern, S.; Sessitsch, A.; Richter, A. Belowground carbon allocation by trees drives seasonal patterns of extracellular enzyme activities by altering microbial community composition in a beech forest soil. New Phytol. 2010, 187, 843–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGuire, K.L.; Treseder, K.K. Microbial communities and their relevance for ecosystem models: Decomposition as a case study. Soil Biol. Biochem. 2010, 42, 529–535. [Google Scholar] [CrossRef] [Green Version]
- Schädler, M.; Buscot, F.; Klotz, S.; Reitz, T.; Durka, W.; Bumberger, J.; Merbach, I.; Michalski, S.G.; Kirsch, K.; Remmler, P.; et al. Investigating the consequences of climate change under different land-use regimes: A novel experimental infrastructure. Ecosphere 2019, 10, e02635. [Google Scholar] [CrossRef] [Green Version]
- Puchalski, M.; Szparaga, G.; Biela, T.; Gutowska, A.; Sztajnowski, S.; Krucińska, I. Molecular and Supramolecular Changes in Polybutylene Succinate (PBS) and Polybutylene Succinate Adipate (PBSA) Copolymer during Degradation in Various Environmental Conditions. Polymers 2018, 10, 251. [Google Scholar] [CrossRef] [Green Version]
- Tanunchai, B.; Kalkhof, S.; Guliyev, V.; Wahdan, S.F.M.; Krstic, D.; Schädler, M.; Geissler, A.; Glaser, B.; Buscot, F.; Blagodatskaya, E. Nitrogen fixing bacteria facilitate microbial biodegradation of a bio-based and biodegradable plastic in soils under ambient and future climatic conditions. Environ. Sci. Processes Impacts 2022, 24, 233–241. [Google Scholar] [CrossRef]
- Caporaso, J.; Lauber, C.; Walters, W.; Berg-Lyons, D.; Lozupone, C.; Turnbaugh, P.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4516–4522. [Google Scholar] [CrossRef] [Green Version]
- Ihrmark, K.; Bödeker, I.T.M.; Cruz-Martinez, K.; Friberg, H.; Kubartova, A.; Schenck, J.; Strid, Y.; Stenlid, J.; Brandström-Durling, M.; Clemmensen, K.E.; et al. New primers to amplify the fungal ITS2 region—Evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 2012, 82, 666–677. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. 38—Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- ISO 10694:1996-08; Soil Quality—Determination of Organic and Total Carbon after dry Combustion (Elementary Analysis) (ISO 10694:1995). Beuth-Verlag: Berlin, Germany, 1996.
- Razavi, B.S.; Blagodatskaya, E.; Kuzyakov, Y. Nonlinear temperature sensitivity of enzyme kinetics explains canceling effect—A case study on loamy haplic Luvisol. Front. Microbiol. 2015, 6, 1126. [Google Scholar] [CrossRef] [Green Version]
- Amend, A.S.; Seifert, K.A.; Bruns, T.D. Quantifying microbial communities with 454 pyrosequencing: Does read abundance count? Mol. Ecol. 2010, 19, 5555–5565. [Google Scholar] [CrossRef]
- Hammer, O.; Harper, D.; Ryan, P. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Venkatesh, S.; Mahboob, S.; Govindarajan, M.; Al-Ghanim, K.A.; Ahmed, Z.; Al-Mulhm, N.; Gayathri, R.; Vijayalakshmi, S. Microbial degradation of plastics: Sustainable approach to tackling environmental threats facing big cities of the future. J. King Saud Univ.-Sci. 2021, 33, 101362. [Google Scholar] [CrossRef]
- Kamiya, M.; Asakawa, S.; Kimura, M. Molecular analysis of fungal communities of biodegradable plastics in two Japanese soils. Soil Sci. Plant Nutr. 2007, 53, 568–574. [Google Scholar] [CrossRef]
- Emadian, S.M.; Onay, T.T.; Demirel, B. Biodegradation of bioplastics in natural environments. Waste Manag. 2017, 59, 526–536. [Google Scholar] [CrossRef] [PubMed]
- Lavelle, P.; Bignell, D.; Lepage, M.; Wolters, V.; Roger, P.; Ineson, P.; Heal, O.; Dhillion, S.P. Soil function in a changing world: The role of invertebrate ecosystem engineers. Eur. J. Soil Sci. 1997, 33, 159–193. [Google Scholar]
- Vogel, H.-J.; Balseiro-Romero, M.; Kravchenko, A.; Otten, W.; Pot, V.; Schlüter, S.; Weller, U.; Baveye, P.C. A holistic perspective on soil architecture is needed as a key to soil functions. Eur. J. Soil Sci. 2022, 73, e13152. [Google Scholar] [CrossRef]
- Piotrowska-Długosz, A.; Charzyński, P. The impact of the soil sealing degree on microbial biomass, enzymatic activity, and physicochemical properties in the Ekranic Technosols of Toruń (Poland). J. Soils Sediments 2015, 15, 47–59. [Google Scholar] [CrossRef] [Green Version]
- Ananyeva, N.D.; Demkina, T.S.; Jones, W.J.; Cabrera, M.L.; Steen, W.C. Microbial biomass in soils of Russia under long-term management practices. Biol. Fertil. Soils 1999, 29, 291–299. [Google Scholar] [CrossRef]
- Wahdan, S.F.M.; Heintz-Buschart, A.; Sansupa, C.; Tanunchai, B.; Wu, Y.-T.; Schädler, M.; Noll, M.; Purahong, W.; Buscot, F. Targeting the Active Rhizosphere Microbiome of Trifolium pratense in Grassland Evidences a Stronger-Than-Expected Belowground Biodiversity-Ecosystem Functioning Link. Front. Microbiol. 2021, 12, 629169. [Google Scholar] [CrossRef]
- Põlme, S.; Abarenkov, K.; Nilsson, R.H.; Lindahl, B.; Clemmensen, K.; Kauserud, H.; Nguyen, N.; Kjøller, R.; Bates, S.; Baldrian, P.; et al. FungalTraits: A user-friendly traits database of fungi and fungus-like stramenopiles. Fungal Divers. 2020, 105, 1–16. [Google Scholar] [CrossRef]
- Maia, M.M.; Heasley, A.; Camargo de Morais, M.M.; Melo, E.H.; Morais, M.A., Jr.; Ledingham, W.M.; Lima Filho, J.L. Effect of culture conditions on lipase production by Fusarium solani in batch fermentation. Bioresour. Technol. 2001, 76, 23–27. [Google Scholar] [CrossRef]
- Bhatti, H.N.; Batool, S.; Afzal, N. Production and Characterization of a Novel β-Glucosidase from Fusarium solani. Int. J. Agric. Biol. 2013, 15, 140–144. [Google Scholar]
- Nuero, O.M. Production of chitinase by Fusarium species. Curr. Microbiol. 1995, 30, 287–289. [Google Scholar] [CrossRef] [PubMed]
- Louca, S.; Polz, M.; Mazel, F.; Nelson Albright, M.; Huber, J.; O’Connor, M.; Ackermann, M.; Hahn, A.; Srivastava, D.; Crowe, S.; et al. Function and functional redundancy in microbial systems. Nat. Ecol. Evol. 2018, 2, 936–943. [Google Scholar] [CrossRef]
- Wertz, S.; Degrange, V.; Prosser, J.I.; Poly, F.; Commeaux, C.; Freitag, T.; Guillaumaud, N.; Roux, X.L. Maintenance of soil functioning following erosion of microbial diversity. Environ. Microbiol. 2006, 8, 2162–2169. [Google Scholar] [CrossRef] [PubMed]
- Xun, W.; Liu, Y.; Li, W.; Ren, Y.; Xiong, W.; Xu, Z.; Zhang, N.; Miao, Y.; Shen, Q.; Zhang, R. Specialized metabolic functions of keystone taxa sustain soil microbiome stability. Microbiome 2021, 9, 35. [Google Scholar] [CrossRef] [PubMed]
- Fukami, T.; Dickie, I.A.; Paula Wilkie, J.; Paulus, B.C.; Park, D.; Roberts, A.; Buchanan, P.K.; Allen, R.B. Assembly history dictates ecosystem functioning: Evidence from wood decomposer communities. Ecol. Lett. 2010, 13, 675–684. [Google Scholar] [CrossRef]
- Hoppe, B.; Purahong, W.; Wubet, T.; Kahl, T.; Bauhus, J.; Arnstadt, T.; Hofrichter, M.; Buscot, F.; Krüger, D. Linking molecular deadwood-inhabiting fungal diversity and community dynamics to ecosystem functions and processes in Central European forests. Fungal Divers. 2016, 77, 367–379. [Google Scholar] [CrossRef]
- Wagg, C.; Hautier, Y.; Pellkofer, S.; Banerjee, S.; Schmid, B.; van der Heijden, M.G. Diversity and asynchrony in soil microbial communities stabilizes ecosystem functioning. eLife 2021, 10, e62813. [Google Scholar] [CrossRef]




| Bacteria | Fungi | |||
|---|---|---|---|---|
| R2 | p | R2 | p | |
| Treatment | 0.60 | 0.001 | 0.54 | 0.001 |
| Total organic carbon (TOC) | 0.68 | 0.001 | 0.88 | 0.001 |
| Total nitrogen (TN) | 0.45 | 0.001 | 0.43 | 0.004 |
| C: N ratio | 0.30 | 0.034 | 0.32 | 0.015 |
| pH | 0.58 | 0.003 | 0.34 | 0.016 |
| PBSA amendment | 0.33 | 0.033 | 0.41 | 0.003 |
| N amendment | 0.33 | 0.001 | 0.31 | 0.013 |
| Bacteria | Fungi | |||
|---|---|---|---|---|
| R2 | p | R2 | p | |
| Vmax, Chitinase | 0.71 | 0.003 | 0.91 | 0.001 |
| Vmax, Lipase | 0.42 | 0.008 | 0.08 | 0.522 |
| Vmax, Phosphatase | 0.90 | 0.001 | 0.47 | 0.020 |
| Vmax, β–Glucosidase | 0.35 | 0.024 | 0.15 | 0.249 |
| Km, Chitinase | 0.04 | 0.759 | 0.05 | 0.640 |
| Km, Lipase | 0.14 | 0.215 | 0.18 | 0.181 |
| Km, Phosphatase | 0.61 | 0.003 | 0.48 | 0.008 |
| Km, β–Glucosidase | 0.23 | 0.121 | 0.12 | 0.348 |
| Microbial Taxa | β-Glucosidase (p) | β-Glucosidase (S) | Chitinase (p) | Chitinase (S) | Lipase (p) | Lipase (S) | Phosphatase (p) | Phosphatase (S) |
|---|---|---|---|---|---|---|---|---|
| Bacteria | ||||||||
| Achromobacter | 0.37 | 0.87 | 0.81 | 0.89 | 0.07 | 0.20 | −0.10 | −0.74 |
| Sphingomonas | 0.02 | 0.73 | −0.04 | 0.77 | −0.04 | 0.16 | −0.08 | −0.43 |
| RB41 | −0.58 | −0.77 | −0.68 | −0.95 | −0.36 | −0.33 | −0.22 | 0.59 |
| Luteimonas | 0.82 | 0.67 | 0.82 | 0.78 | 0.62 | 0.55 | 0.44 | −0.72 |
| Streptomyces | 0.18 | 0.65 | 0.68 | 0.53 | −0.12 | 0.38 | −0.27 | −0.58 |
| Rhodanobacter | 0.80 | 0.50 | 0.59 | 0.37 | 0.81 | 0.22 | 0.81 | −0.23 |
| Lysobacter | −0.39 | 0.79 | −0.45 | 0.93 | −0.37 | 0.52 | −0.33 | −0.68 |
| Gaiella | −0.47 | 0.14 | −0.68 | 0.33 | −0.13 | −0.26 | 0.03 | −0.21 |
| Chitinophaga | 0.75 | 0.05 | 0.54 | −0.08 | 0.77 | 0.07 | 0.77 | −0.38 |
| Gemmatimonas | 0.26 | 0.71 | −0.08 | 0.75 | 0.47 | 0.48 | 0.70 | −0.84 |
| Bradyrhizobium | −0.62 | 0.59 | −0.83 | 0.50 | −0.49 | −0.03 | −0.38 | −0.39 |
| Fungi | ||||||||
| Fusarium | 0.68 | −0.76 | 0.92 | −0.64 | 0.28 | −0.25 | 0.08 | 0.62 |
| Chaetomium | −0.31 | 0.79 | −0.76 | 0.85 | −0.02 | 0.38 | 0.12 | −0.56 |
| Exophiala | 0.42 | −0.13 | −0.08 | 0.05 | 0.61 | 0.60 | 0.55 | −0.10 |
| Tetracladium | −0.58 | 0.60 | −0.84 | 0.64 | −0.21 | 0.35 | 0.01 | −0.53 |
| Gibellulopsis | −0.47 | 0.55 | −0.65 | 0.78 | −0.24 | 0.26 | −0.26 | −0.21 |
| Schizothecium | −0.19 | 0.55 | −0.66 | 0.43 | 0.10 | 0.13 | 0.19 | −0.77 |
| Ilyonectria | −0.35 | −0.85 | −0.65 | −0.68 | −0.25 | −0.21 | −0.18 | 0.68 |
| Mortierella | −0.43 | 0.19 | −0.76 | 0.03 | −0.19 | −0.48 | −0.18 | 0.13 |
| Clonostachys | 0.82 | −0.68 | 0.94 | −0.55 | 0.44 | 0.09 | 0.25 | 0.36 |
| Preussia | −0.34 | −0.83 | −0.80 | −0.68 | −0.07 | −0.42 | 0.01 | 0.50 |
| Rhizophlyctis | −0.17 | −0.16 | −0.64 | −0.50 | 0.12 | −0.55 | 0.18 | 0.25 |
| Stachybotrys | −0.44 | 0.59 | −0.68 | 0.65 | −0.20 | 0.32 | −0.24 | −0.52 |
| Fusicolla | 0.74 | 0.88 | 0.20 | 0.89 | 0.84 | 0.39 | 0.85 | −0.58 |
| Acremonium | −0.29 | 0.81 | −0.71 | 0.64 | −0.04 | 0.19 | 0.17 | −0.50 |
| Sistotrema | −0.17 | −0.41 | −0.17 | −0.73 | −0.06 | −0.79 | −0.06 | 0.63 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guliyev, V.; Tanunchai, B.; Noll, M.; Buscot, F.; Purahong, W.; Blagodatskaya, E. Links among Microbial Communities, Soil Properties and Functions: Are Fungi the Sole Players in Decomposition of Bio-Based and Biodegradable Plastic? Polymers 2022, 14, 2801. https://doi.org/10.3390/polym14142801
Guliyev V, Tanunchai B, Noll M, Buscot F, Purahong W, Blagodatskaya E. Links among Microbial Communities, Soil Properties and Functions: Are Fungi the Sole Players in Decomposition of Bio-Based and Biodegradable Plastic? Polymers. 2022; 14(14):2801. https://doi.org/10.3390/polym14142801
Chicago/Turabian StyleGuliyev, Vusal, Benjawan Tanunchai, Matthias Noll, François Buscot, Witoon Purahong, and Evgenia Blagodatskaya. 2022. "Links among Microbial Communities, Soil Properties and Functions: Are Fungi the Sole Players in Decomposition of Bio-Based and Biodegradable Plastic?" Polymers 14, no. 14: 2801. https://doi.org/10.3390/polym14142801






