Molecularly Imprinted Polymers for Gossypol via Sol–Gel, Bulk, and Surface Layer Imprinting—A Comparative Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
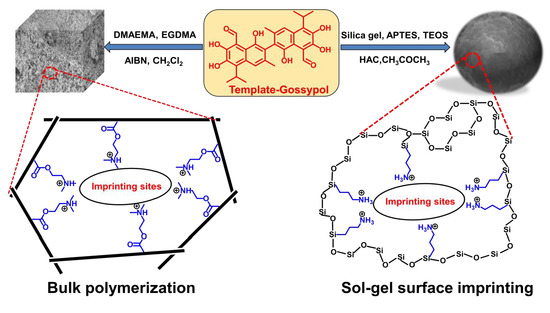
2.2. Preparation of MIPs and NIPs
2.2.1. Synthesis of MIP1 and NIP1 by Bulk Polymerization
2.2.2. Synthesis of MIP2 and NIP2 by Surface Layer Imprinting
2.2.3. Synthesis of MIP3 and NIP3 by Sol-Gel Process
2.3. Characterization
2.4. Binding Experiments
3. Results and Discussion
3.1. Preparation of the Gossypol-MIPs
3.2. Characterization of MIPs and NIPs
3.2.1. SEM Analysis
3.2.2. Surface Area Measurement via Nitrogen Adsorption-Desorption Analysis
3.3. Adsorption Kinetics of MIPs and NIPs
3.4. Adsorption Isotherms of MIPs
3.5. Adsorption Selectivity of the MIPs
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Withers, W.A.; Carruth, F.E. Gossypol—A toxic substance in cottonseed. A preliminary note. Science 1915, 41, 324. [Google Scholar] [CrossRef]
- Adams, R.; Geissman, T.A.; Edwards, J.D. Gossypol, a pigment of cottonseed. Chem. Rev. 1960, 60, 555–574. [Google Scholar] [CrossRef] [PubMed]
- Maugh, T.H. Male “pill” blocks sperm enzyme. Science 1981, 212, 314. [Google Scholar] [CrossRef] [PubMed]
- Randel, R.D.; Chase, C.C.; Wyse, S.J. Effects of gossypol and cottonseed products on reproduction of mammals. J. Anim. Sci. 1992, 70, 1628–1638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montamat, E.E.; Burgos, C.; De Burgos, N.M.G.; Rovai, L.E.; Blanco, A.; Segura, E.L. Inhibitory action of gossypol on enzymes and growth of Trypanosoma cruzi. Science 1982, 218, 288–289. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Xu, Z.; Zhao, S.; Sun, J.; Yang, X. Development of a microbial fermentation process for detoxification of gossypol in cottonseed meal. Anim. Feed. Sci. Technol. 2007, 135, 176–186. [Google Scholar] [CrossRef]
- Johnson, L.A.; Lusas, E.W. Comparison of alternative solvents for oils extraction. J. Am. Oil. Chem. Soc. 1983, 60, 229–242. [Google Scholar] [CrossRef]
- Barraza, M.L.; Coppock, C.E.; Brooks, K.N.; Wilks, D.L.; Saunders, R.G.; Latimer, G.W. Iron sulfate and feed pelleting to detoxify free gossypol in cottonseed diets for dairy cattle. J. Dairy Sci. 1991, 74, 3457–3467. [Google Scholar] [CrossRef]
- Zhi, K.; Wang, L.; Zhang, Y.; Zhang, X.; Zhang, L.; Liu, L.; Yao, J.; Xiang, W. Preparation and evaluation of molecularly imprinted polymer for selective recognition and adsorption of gossypol. J. Mol. Recognit. 2018, 31, e2627. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Romanova, E.V.; Sweedler, J.V. Distinguishing Endogenous d-Amino Acid-Containing Neuropeptides in Individual Neurons Using Tandem Mass Spectrometry. Anal. Chem. 2011, 83, 2794–2800. [Google Scholar] [CrossRef]
- Kuk, M.S.; Tetlow, R. Gossypol removal by adsorption from cottonseed miscella. J. Am. Oil. Chem. Soc. 2005, 82, 905–909. [Google Scholar] [CrossRef]
- Kuk, M.S.; Hronsr, R.J.; Abraham, G. Adsorptive gossypol removal. J. Am. Oil. Chem. Soc. 1993, 70, 209–210. [Google Scholar] [CrossRef]
- Wuff, G.; Sarhan, A. The use of polymers with enzyme-analogous structures for the resolution of racemate. Angew. Chem. Int. Ed. 1972, 11, 341–345. [Google Scholar]
- Andersson, L.; Sellergren, B.; Mosbach, K. Imprinting of amino acid derivatives in macroporous polymers. Tetrahedron Lett. 1984, 25, 5211–5214. [Google Scholar] [CrossRef]
- Sellergren, B.; Shea, K.J. Influence of polymer morphology on the ability of imprinted network polymers to resolve enantiomers. J. Chromatogr. A 1993, 635, 31–49. [Google Scholar] [CrossRef]
- Vlatakis, G.; Andersson, L.I.; Müller, R.; Mosbach, K. Drug assay using antibody mimics made by molecular imprinting. Nature 1993, 361, 645–647. [Google Scholar] [CrossRef]
- Whitcombe, M.J.; Rodriguez, M.E.; Villar, P.; Vulfson, E.N. A new method for the introduction of recognition site functionality into polymers prepared by molecular imprinting: Synthesis and characterization of polymeric receptors for cholesterol. J. Am. Chem. Soc. 1995, 117, 7105–7111. [Google Scholar] [CrossRef]
- Mosbach, K. Preparation of Synthetic Enzymes and Synthetic Antibodies and Use of the Thus Prepared Enzymes and Antibodies. US Patent 5110833A, 5 May 1992. [Google Scholar]
- Zhu, J.L.; Chen, D.P.; Ai, Y.H.; Dang, X.P.; Huang, J.L.; Chen, H.X. A dummy molecularly imprinted monolith for selective solid-phase microextraction of vanillin and methyl vanillin prior to their determination by HPLC. Microchim. Acta 2017, 184, 1161–1167. [Google Scholar] [CrossRef]
- Deiminiat, B.; Rounaghi, G.H.; Arbab-Zavar, M.H. Development of a new electrochemical imprinted sensor based on poly-pyrrole, sol-gel and multiwall carbon nanotubes for determination of tramadol. Sens. Actuators B 2017, 238, 651–659. [Google Scholar] [CrossRef]
- Li, C.Y.; Ma, X.G.; Zhang, X.J.; Wang, R.; Chen, Y.; Li, Z.Y. Magnetic molecularly imprinted polymer nanoparticles-based solid-phase extraction coupled with gas chromatography-mass spectrometry for selective determination of trace di-(2-ethylhexyl) phthalate in water samples. Anal. Bioanal. Chem. 2016, 408, 7857–7864. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, X.; Lu, W.; Wu, X.; Li, J. Molecular imprinting: Perspectives and applications. Chem. Soc. Rev. 2016, 45, 2137. [Google Scholar] [CrossRef]
- Wulff, G.; Liu, J. Design of biomimetic catalysts by molecular imprinting in synthetic polymers: The role of transition state stabilization. Acc. Chem. Res. 2012, 45, 239. [Google Scholar] [CrossRef]
- Whitcombe, M.J.; Chianella, I.; Larcombe, L.; Piletsky, S.A.; Noble, J.; Porter, R.; Horgan, A. The rational development of molecularly imprinted polymer-based sensors for protein detection. Chem. Soc. Rev. 2011, 40, 1547–1571. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Xu, S.; Li, J. Recent advances in molecular imprinting technology: Current status, challenges and highlighted applications. Chem. Soc. Rev. 2011, 40, 2922–2942. [Google Scholar] [CrossRef]
- Yin, J.; Cui, Y.; Yang, G.; Wang, H. Molecularly imprinted nanotubes for enantioselective drug delivery and controlled release. Chem. Commun. 2010, 46, 7688–7690. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Jia, X.; Yang, Y.; Liu, X. Porous Carbon Microspheres: An Excellent Support To Prepare Surface Molecularly Imprinted Polymers for Selective Removal of Dibenzothiophene in Fuel Oil. Ind. Eng. Chem. Res. 2016, 55, 1710–1719. [Google Scholar] [CrossRef]
- Moczko, E.; Guerreiro, A.; Piletska, E.; Piletsky, S. PEG-Stabilized Core–Shell Surface-Imprinted Nanoparticles. Langmuir 2013, 29, 9891–9896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, X.; Xu, M.; Wang, Y.; Ran, D.; Yang, S.; Zhang, M. Polydopamine-based molecular imprinting on silica-modified magnetic nanoparticles for recognition and separation of bovine hemoglobin. Analyst 2013, 138, 651–658. [Google Scholar] [CrossRef]
- He, Y.; Huang, Y.; Jin, Y.; Liu, X.; Liu, G.; Zhao, R. Well-Defined Nanostructured Surface-Imprinted Polymers for Highly Selective Magnetic Separation of Fluoroquinolones in Human Urine. ACS Appl. Mater. Interfaces 2014, 6, 9634–9642. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Mu, X.; Zhang, J.; Tang, Y. Specific recognition of bovine serum albumin using superparamagnetic molecularly imprinted nanomaterials prepared by two-stage core–shell sol–gel polymerization. J. Mater. Chem. B 2014, 2, 783–792. [Google Scholar] [CrossRef]
- Duan, F.; Chen, C.; Chen, L.; Sun, Y.; Wang, Y.; Yang, Y.; Liu, X.; Qin, Y. Preparation and Evaluation of Water-Compatible Surface Molecularly Imprinted Polymers for Selective Adsorption of Bisphenol A from Aqueous Solution. Ind. Eng. Chem. Res. 2014, 53, 14291–14300. [Google Scholar] [CrossRef]
- Qin, Y.P.; Li, D.Y.; He, X.W.; Li, W.Y.; Zhang, Y.K. Preparation of High-Efficiency Cytochrome c-Imprinted Polymer on the Surface of Magnetic Carbon Nanotubes by Epitope Approach via Metal Chelation and Six-Membered Ring. ACS Appl. Mater. Interfaces 2016, 8, 10155–10163. [Google Scholar] [CrossRef] [PubMed]
- Arfaoui, F.; Khlifi, A.; Bargaoui, M.; Khalfaoui, M.; Kalfat, R. Thin Melamine Imprinted Sol Gel Coating on Silica Beads: Experimental and Statistical Physics Study. Chem. Afr. 2018, 1, 175–185. [Google Scholar] [CrossRef]
- Chrzanowska, A.M.; Poliwoda, A.; Wieczorek, P.P. Surface molecularly imprinted silica for selective solid-phase extraction of biochanin A, daidzein and genistein from urine samples. J. Chromatogr. A 2015, 1392, 1–9. [Google Scholar] [CrossRef]
- Meng, M.; Wang, Z.; Ma, L.; Zhang, M.; Wang, J.; Dai, X.; Yan, Y. Selective Adsorption of Methylparaben by Submicrosized Molecularly Imprinted Polymer: Batch and Dynamic Flow Mode Studies. Ind. Eng. Chem. Res. 2012, 51, 14915–14924. [Google Scholar] [CrossRef]
- Ren, Y.; Ma, W.; Ma, J.; Wen, Q.; Wang, J.; Zhao, F. Synthesis and properties of bisphenol A molecular imprinted particle for selective recognition of BPA from water. J. Colloid Interface Sci. 2012, 367, 355–361. [Google Scholar] [CrossRef]
- Wang, S.; Xu, Z.; Fang, G.; Duan, Z.; Zhang, Y.; Chen, S. Synthesis and characterization of a molecularly imprinted silica gel sorbent for the on-line determination of trace Sudan I in Chilli powder through high-performance liquid chromatography. J. Agric. Food Chem. 2007, 55, 3869–3876. [Google Scholar] [CrossRef] [PubMed]
- Zhu, R.; Zhao, W.; Zhai, M.; Wei, F.; Cai, Z.; Sheng, N.; Hu, Q. Molecularly imprinted layer-coated silica nanoparticles for selective solid-phase extraction of bisphenol A from chemical cleansing and cosmetics samples. Anal. Chim. Acta 2010, 658, 209–216. [Google Scholar] [CrossRef]
- Zhao, C.; Wu, D. Rapid detection assay for the molecular imprinting of gossypol using a two-layer PMAA/SiO2 bulk structure with a piezoelectric imprinting sensor. Sens. Actuators B 2013, 181, 104–113. [Google Scholar] [CrossRef]
- Zhi, K.; Wang, L.; Zhang, Y.; Jiang, Y.; Zhang, L.; Yasin, A. Influence of Size and Shape of Silica Supports on the Sol–Gel Surface Molecularly Imprinted Polymers for Selective Adsorption of Gossypol. Materials 2018, 11, 777. [Google Scholar] [CrossRef]
- Arabi, M.; Ghaedi, M.; Ostovan, A. Development of a Lower Toxic Approach Based on Green Synthesis of Water-Compatible Molecularly Imprinted Nanoparticles for the Extraction of Hydrochlorothiazide from Human Urine. ACS Sustain. Chem. Eng. 2017, 5, 3775–3785. [Google Scholar] [CrossRef]
- Luo, J.; Gao, Y.; Tan, K.; Wei, W.; Liu, X. Preparation of a Magnetic Molecularly Imprinted Graphene Composite Highly Adsorbent for 4-Nitrophenol in Aqueous Medium. ACS Sustain. Chem. Eng. 2016, 4, 3316–3326. [Google Scholar] [CrossRef]
- Lofgreen, J.E.; Ozin, G.A. Controlling morphology and porosity to improve performance of molecularly imprinted sol-gel silica. Chem. Soc. Rev. 2014, 43, 911–933. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, J.; Wang, X.; Shen, D.; Chen, L. Quantum Dots Based Mesoporous Structured Imprinting Microspheres for the Sensitive Fluorescent Detection of Phycocyanin. ACS Appl. Mater. Interfaces 2015, 7, 9118–9127. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Yang, Y.; Xu, L.; Zhang, J. Bisphenol A sensing based on surface molecularly imprinted, ordered mesoporous silica. Electrochim. Acta 2011, 56, 2105–2109. [Google Scholar] [CrossRef]
- Jiang, X.; Tian, W.; Zhao, C.; Zhang, H.; Liu, M. A novel sol-gel-material prepared by a surface imprinting technique for the selective solid-phase extraction of bisphenol A. Talanta 2007, 72, 119–125. [Google Scholar] [CrossRef]
- Dai, J.; Zhang, Y.; Pan, M.; Kong, L.; Wang, S. Development and Application of Quartz Crystal Microbalance Sensor Based on Novel Molecularly Imprinted Sol–Gel Polymer for Rapid Detection of Histamine in Foods. J. Agric. Food Chem. 2014, 62, 5269–5274. [Google Scholar] [CrossRef]
- Xu, S.; Lu, H.; Li, J.; Song, X.; Wang, A.; Chen, L.; Han, S. Dummy Molecularly Imprinted Polymers-Capped CdTe Quantum Dots for the Fluorescent Sensing of 2,4,6-Trinitrotoluene. ACS Appl. Mater. Interfaces 2013, 5, 8146–8154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reyes, J.; Wyrick, S.D.; Borriero, L.; Benos, D.J. Membrane actions of male contraceptive gossypol tautomers. BBA Biomembr. 1986, 863, 101–109. [Google Scholar] [CrossRef]
- Ho, Y.S.; McKay, G. Pseudo-second order model for sorption processes. Process Biochem. 1999, 34, 451–465. [Google Scholar] [CrossRef]
- Ho, Y.S. Review of second-order models for adsorption systems. J. Hazard. Mater. 2006, 136, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Mu, X.; Hao, Y.; Zhang, L.; Zhang, J.; Tang, Y. Combination of surface imprinting and immobilized template techniques for preparation of core–shell molecularly imprinted polymers based on directly amino-modified Fe3O4 nanoparticles for specific recognition of bovine hemoglobin. J. Mater. Chem. B 2014, 2, 1733–1741. [Google Scholar] [CrossRef]
- Langmuir, I. The constitution and fundamental properties of solids and liquids. Part I. Solids. J. Am. Chem. Soc. 1916, 38, 2221–2295. [Google Scholar] [CrossRef]
- Freundlich, H. Over the adsorption in solution. J. Phys. Chem. B 1906, 57, 1100–1107. [Google Scholar]







| Polymer | Template (mmol) | FM (mmol) | Cross-linker (mmol) | Initiator/Catalyst | Support (g) | Solvent (mL) |
|---|---|---|---|---|---|---|
| MIP1 a | Gossypol (0.083) | DMAEMA (1.0) | EGDMA (5.0) | AIBN (0.27 mmol) | CH2Cl2 (4.0) | |
| NIP1 a | DMAEMA (1.0) | EGDMA (5.0) | AIBN (0.27 mmol) | CH2Cl2 (4.0) | ||
| MIP2 b | Gossypol (0.083) | APTES (1.25) | TEOS (2.6) | 1.0 mol L−1 HAc (0.15mL) | activated silica (0.058) | Acetone (3.0) |
| NIP2 b | APTES (1.25) | TEOS (2.6) | 1.0 mol L−1 HAc (0.15mL) | activated silica (0.058) | Acetone (3.0) | |
| MIP3 b | Gossypol (0.083) | APTES (1.25) | TEOS (2.6) | 1.0 mol L−1 HAc (0.15mL) | Acetone (3.0) | |
| NIP3 b | APTES (1.25) | TEOS (2.6) | 1.0 mol L−1 HAc (0.15mL) | Acetone (3.0) |
| Samples | BET Surface Area (m2 g−1) | Average Pore Diameter (nm) |
|---|---|---|
| NIP1 | 91.1 | 6.57 |
| MIP1 | 13.9 | 40.4 |
| Silica gel (spherical 500 nm) | 14.29 | 23.07 |
| NIP2 | 31.53 | 22.29 |
| MIP2 | 268.2 | 5.542 |
| Samples | te (min) | QMIP-e (mg·g−1) | QNIP-e (mg·g−1) | IF |
|---|---|---|---|---|
| MIP1 | 720 | 139.00 | 122.00 | 1.14 |
| MIP2 | 40 | 77.05 | 30.50 | 2.53 |
| MIP3 | 140 | 50.95 | 41.95 | 1.22 |
| Sample | Qe, exp (mg g−1) | Pseudo-First-Order | Pseudo -Second-Order | ||||
|---|---|---|---|---|---|---|---|
| Qe, cal (mg g−1) | k1 (min−1) | R2 | Qe (mg g−1) | k2 (g mg−1 min−1) | R2 | ||
| MIP1 | 139.00 | 126.82 | 0.01869 | 0.9006 | 138.56 | 1.84×10−4 | 0.9633 |
| MIP2 | 77.05 | 74.94 | 0.5490 | 0.9936 | 76.54 | 0.02523 | 0.9976 |
| MIP3 | 50.95 | 39.36 | 0.3924 | 0.9750 | 49.30 | 0.01772 | 0.9915 |
| Sample | Qe,exp (mg g−1) | Langmuir | Freundlich | ||||
|---|---|---|---|---|---|---|---|
| Qm (mg g−1) | KL (L mg−1) | R2 | KF (mg g−1) | n | R2 | ||
| MIP1 | 564 | 606.9 | 0.00869 | 0.9527 | 27.54 | 2.751 | 0.9926 |
| NIP1 | 506.5 | 635.3 | 0.00384 | 0.9979 | 49.78 | 2.155 | 0.9838 |
| MIP2 | 120 | 110.6 | 0.02401 | 0.9287 | 27.54 | 4.842 | 0.9984 |
| NIP2 | 42 | 47.37 | 0.00725 | 0.9891 | 6.915 | 3.853 | 0.9357 |
| MIP3 | 61.8 | 69.02 | 0.01661 | 0.9959 | 13.53 | 4.000 | 0.9923 |
| NIP3 | 50.8 | 57.50 | 0.01543 | 0.9943 | 11.15 | 4.013 | 0.9958 |
| Samples | C0 (mg L−1) | Regression Equation Y = A + BX | KD (mg L−1) KD = −1/B | Qm (mg g−1) Qm = −A/B | R2 |
|---|---|---|---|---|---|
| MIP1 | 200–500 | Qe/Ce = 19.6 − 0.0776Q | 12.9 | 252 | 0.9761 |
| 600–2000 | Qe/Ce = 4.04 − 0.00617Q | 162.1 | 654 | 0.9222 | |
| NIP1 | 200–2000 | Qe/Ce = 2.59 − 0.0042Q | 238.1 | 616.7 | 0.9675 |
| MIP2 | 120–260 | Qe/Ce = 5.71 − 0.0688Q | 14.5 | 83.0 | 0.9223 |
| 300–1600 | Qe/Ce = 0.95 − 0.00730Q | 137.0 | 130.1 | 0.9099 | |
| NIP2 | 120–1600 | Qe/Ce = 0.33 − 0.0068Q | 147.1 | 48.5 | 0.9493 |
| MIP3 | 140–600 | Qe/Ce = 1.12 − 0.0161Q | 62.1 | 69.6 | 0.9477 |
| NIP3 | 140–600 | Qe/Ce = 0.89 − 0.0155Q | 64.5 | 57.4 | 0.9355 |
| Q (mg g−1) | Adsorbates (12 h) | Adsorbates (1 h) | ||||
|---|---|---|---|---|---|---|
| Gossypol | Ellagic Acid | Quercetin | Gossypol | Ellagic Acid | Quercetin | |
| MIP1 | 144.5 | 75 | 21.9 | 82.5 | 42.8 | 12.5 |
| NIP1 | 121.1 | 64.5 | 16.7 | 60.5 | 32.2 | 8.3 |
| MIP2 | 77.3 | 40.1 | 11.7 | 77.0 | 40.1 | 11.7 |
| NIP2 | 30.5 | 33.2 | 12.1 | 30.1 | 32.7 | 12.1 |
| MIP3 | 50.9 | 26.5 | 7.7 | 47.2 | 24.6 | 7.1 |
| NIP3 | 41.8 | 17.3 | 5.3 | 39.3 | 16.3 | 4.9 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Zhi, K.; Zhang, Y.; Liu, Y.; Zhang, L.; Yasin, A.; Lin, Q. Molecularly Imprinted Polymers for Gossypol via Sol–Gel, Bulk, and Surface Layer Imprinting—A Comparative Study. Polymers 2019, 11, 602. https://doi.org/10.3390/polym11040602
Wang L, Zhi K, Zhang Y, Liu Y, Zhang L, Yasin A, Lin Q. Molecularly Imprinted Polymers for Gossypol via Sol–Gel, Bulk, and Surface Layer Imprinting—A Comparative Study. Polymers. 2019; 11(4):602. https://doi.org/10.3390/polym11040602
Chicago/Turabian StyleWang, Lulu, Keke Zhi, Yagang Zhang, Yanxia Liu, Letao Zhang, Akram Yasin, and Qifeng Lin. 2019. "Molecularly Imprinted Polymers for Gossypol via Sol–Gel, Bulk, and Surface Layer Imprinting—A Comparative Study" Polymers 11, no. 4: 602. https://doi.org/10.3390/polym11040602