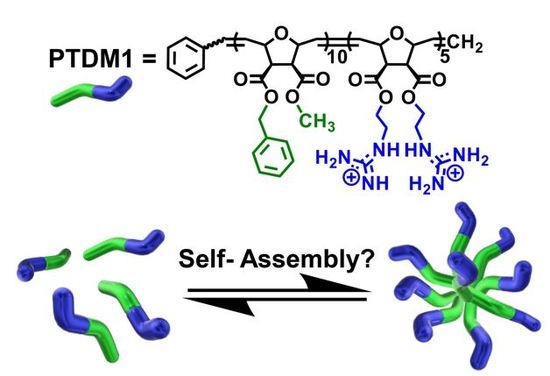
Protein Transduction Domain Mimic (PTDM) Self-Assembly?
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Instrumentation and Software
2.3. PTDM Selection and Stock Solution Preparation
2.4. Pendant Drop Interfacial Tensiometry (IFT)
2.5. Transmission Electron Microscopy (TEM)
2.6. Dynamic Light Scattering (DLS)
2.7. DLS Titration Experiment
2.8. Transmittance (%T) Assays and UV-Visible Spectra
3. Results and Discussion
3.1. Interfacial Tension (IFT)
3.2. Transmission Electron Microscopy (TEM)
3.3. Dynamic Light Scattering (DLS) and Transmittance Assays (%T)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vives, E.; Brodin, P.; Lebleu, B. A Truncated HIV-1 Tat Protein Basic Domain Rapidly Translocates through the Plasma Membrane and Accumulates in the Cell Nucleus. J. Biol. Chem. 1997, 272, 16010–16017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frankel, A.D.; Pabo, C.O. Cellular Uptake of the Tat Protein from Human Immunodeficiency Virus. Cell 1988, 55, 1189–1193. [Google Scholar] [CrossRef]
- Walrant, A.; Cardon, S.; Burlina, F.; Sagan, S. Membrane Crossing and Membranotropic Activity of Cell-Penetrating Peptides: Dangerous Liaisons? Acc. Chem. Res. 2017, 50, 2968–2975. [Google Scholar] [CrossRef] [PubMed]
- De Ronde, B.M.; Tew, G.N. Development of Protein Mimics for Intracellular Delivery. Biopolymers 2015, 104, 265–280. [Google Scholar] [CrossRef] [PubMed]
- Deshayes, S.; Morris, M.; Heitz, F.; Divita, G. Delivery of Proteins and Nucleic Acids Using a Non-Covalent Peptide-Based Strategy. Adv. Drug Deliv. Rev. 2008, 60, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Heitz, F.; Morris, M.C.; Divita, G. Twenty Years of Cell-Penetrating Peptides: from Molecular Mechanisms to Therapeutics. Br. J. Pharmacol. 2009, 157, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Derossi, D.; Joliot, A.H.; Chassaing, G.; Prochiantz, A. The Third Helix of the Antennapedia Homeodomain Translocates Through Biological Membranes. J. Biol. Chem. 1994, 269, 10444–10450. [Google Scholar] [PubMed]
- Derossi, D.; Chassaing, G.; Prochiantz, A. Trojan Peptides: The Penetratin System for Intracellular Delivery. Trends Cell Biol. 1998, 8, 84–87. [Google Scholar] [CrossRef]
- Pooga, M.; Hallbrink, M.; Zorko, M.; Langel, U. Cell Penetration by Transportan. FASEB J. 1998, 12, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wang, Y.; Zhang, X.; Zhang, W.; Guo, S.; Jin, F. Recent Progress of Cell-Penetrating Peptides as New Carriers for Intracellular Cargo Delivery. J. Control. Release 2014, 174, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Sgolastra, F.; DeRonde, B.M.; Sarapas, J.M.; Som, A.; Tew, G.N. Designing Mimics of Membrane Active Proteins. Acc. Chem. Res. 2013, 46, 2977–2987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wender, P.A.; Mitchell, D.J.; Pattabiraman, K.; Pelkey, E.T.; Steinman, L.; Rothbard, J.B. The Design, Synthesis, and Evaluation of Molecules that Enable or Enhance Cellular Uptake: Peptoid Molecular Transporters. Proc. Natl. Acad. Sci. USA 2000, 97, 13003–13008. [Google Scholar] [CrossRef] [PubMed]
- Wender, P.A.; Cooley, C.B.; Geihe, E.I. Beyond cell penetrating peptides: Designed molecular transporters. Drug Discov. Today Technol. 2012, 9, e49–e55. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, D.J.; Kim, D.T.; Steinman, L.; Fathman, C.G.; Rothbard, J.B. Polyarginine Enters Cells More Efficiently than other Polycationic Homopolymers. J. Pept. Res. 2000, 56, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Futaki, S.; Suzuki, T.; Ohashi, W.; Yagami, T.; Tanaka, S.; Ueda, K.; Sugiura, Y. Arginine-Rich peptides An abundant Source of Membrane-Permeable Peptides Having Potential as Carriers for Intracellular Protein Delivery. J. Biol. Chem. 2001, 276, 5836–5840. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.; Peltier, R.; Kuroki, A.; Town, J.; Perrier, S. Investigating the Cell-Uptake of Guanidinium-Rich RAFT Polymers: Impact of Comonomer and Monomer Distribution. Biomacromolecules 2018, 19, 3190–3200. [Google Scholar] [CrossRef] [PubMed]
- Treat, N.J.; Smith, D.; Teng, C.; Flores, J.D.; Abel, B.A.; York, A.W.; Huang, F.; McCormick, C.L. Guanidine-Containing Methacrylamide (Co)polymers via aRAFT: Toward a Cell-Penetrating Peptide Mimic. ACS Macro Lett. 2012, 1, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Kolonko, E.M.; Pontrello, J.K.; Mangold, S.L.; Kiessling, L.L. General Synthetic Route to Cell-Permeable Block Copolymers via ROMP. J. Am. Chem. Soc. 2009, 131, 7327–7333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geihe, E.I.; Cooley, C.B.; Simon, J.R.; Kiesewetter, M.K.; Edward, J.A.; Hickerson, R.P.; Kaspar, R.L.; Hedrick, J.L.; Waymouth, R.M.; Wender, P.A. Designed Guanidinium-Rich Amphipathic Oligocarbonate Molecular Transporters Complex, Deliver and Release siRNA in Cells. Proc. Natl. Acad. Sci. USA 2012, 109, 13171–13176. [Google Scholar] [CrossRef] [PubMed]
- Backlund, C.M.; Sgolastra, F.; Otter, R.; Minter, L.M.; Takeuchi, T.; Futaki, S.; Tew, G.N. Increased Hydrophobic Block Length of PTDMs Promotes Protein Internalization. Polym. Chem. 2016, 7, 7514–7521. [Google Scholar] [CrossRef] [PubMed]
- Posey, N.D.; Caffrey, L.M.; Minter, L.M.; Tew, G.N. Protein Mimic Hydrophobicity Affects Intracellular Delivery but not Cargo Binding. ChemistrySelect 2016, 1, 6146–6150. [Google Scholar] [CrossRef]
- deRonde, B.M.; Torres, J.A.; Minter, L.M.; Tew, G.N. Development of Guanidinium-Rich Protein Mimics for Efficient siRNA Delivery into Human T Cells. Biomacromolecules 2015, 16, 3172–3179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tezgel, A.Ö.; Jacobs, P.; Backlund, C.M.; Telfer, J.C.; Tew, G.N. Synthetic Protein Mimics for Functional Protein Delivery. Biomacromolecules 2017, 18, 819–825. [Google Scholar] [CrossRef] [PubMed]
- Tezgel, A.Ö.; Gonzalez-Perez, G.; Telfer, J.C.; Osborne, B.A.; Minter, L.M.; Tew, G.N. Novel Protein Transduction Domain Mimics as Nonviral Delivery Vectors for siRNA Targeting NOTCH1 in Primary Human T cells. Mol. Ther. 2012, 21, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Tezgel, A.Ö.; Telfer, J.C.; Tew, G.N. De novo designed protein transduction domain mimics from simple synthetic polymers. Biomacromolecules 2011, 12, 3078–3083. [Google Scholar] [CrossRef] [PubMed]
- Sarapas, J.M.; Backlund, C.M.; deRonde, B.M.; Minter, L.M.; Tew, G.N. ROMP- and RAFT-Based Guanidinium-Containing Polymers as Scaffolds for Protein Mimic Synthesis. Chem. Eur. J. 2017, 23, 6858–6863. [Google Scholar] [CrossRef] [PubMed]
- Sgolastra, F.; Minter, L.M.; Osborne, B.A.; Tew, G.N. Importance of Sequence Specific Hydrophobicity in Synthetic Protein Transduction Domain Mimics. Biomacromolecules 2014, 15, 812–820. [Google Scholar] [CrossRef] [PubMed]
- Sgolastra, F.; Backlund, C.M.; Ozay, E.I.; DeRonde, B.M.; Minter, L.M.; Tew, G.N. Sequence Segregation Improves Non-Covalent Protein Delivery. J. Control. Release 2017, 254, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Backlund, C.M.; Takeuchi, T.; Futaki, S.; Tew, G.N. Relating Structure and Internalization for ROMP-Based Protein Mimics. Biochim. Biophys. Acta 2016, 1858, 1443–1450. [Google Scholar] [CrossRef] [PubMed]
- Ozay, E.I.; Gonzalez-Perez, G.; Torres, J.A.; Vijayaraghavan, J.; Lawlor, R.; Sherman, H.L.; Garrigan, D.T.; Burnside, A.S.; Osborne, B.A.; Tew, G.N.; et al. Intracellular Delivery of Anti-pPKCθ (Thr538) via Protein Transduction Domain Mimics for Immunomodulation. Mol. Ther. 2016, 24, 2118–2130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Posey, N.D.; Hango, C.R.; Minter, L.M.; Tew, G.N. The Role of Cargo Binding Strength in Polymer-Mediated Intracellular Protein Delivery. Bioconj. Chem. 2018, 29, 2679–2690. [Google Scholar] [CrossRef] [PubMed]
- Mai, Y.; Eisenberg, A. Self-Assembly of Block Copolymers. Chem. Soc. Rev. 2012, 41, 5969–5985. [Google Scholar] [CrossRef] [PubMed]
- Skinner, M.; Selhorst, R.; Emrick, T. Synthesis of Water-Soluble Zwitterionic Polysiloxanes. J. Polym. Sci. Part A Polym. Chem. 2016, 54, 127–134. [Google Scholar] [CrossRef]
- Zhao, W.; Fonsny, P.; Fitzgerald, P.; Warr, G.G.; Perrier, S. Unexpected Behavior of Polydimethylsiloxane/Poly(2-(dimethylamino)ethyl acrylate) (Charged) Amphiphilic Block Copolymers in Aqueous Solution. Polym. Chem. 2013, 4, 2140–2150. [Google Scholar] [CrossRef]
- Jan, J.Z.; Huang, B.H.; Lin, J.-J. Facile Preparation of Amphiphilic Oxyethylene-Oxypropylene Block Copolymers by Selective Triazine Coupling. Polymer 2003, 44, 1003–1011. [Google Scholar] [CrossRef]
- Prokop, R.M.; Hair, M.L.; Neumann, A.W. Interfacial Tension of a Polystyrene-Poly(ethylene oxide) Diblock Copolymer at the Water-Toluene Interface. Macromolecules 1996, 29, 5902–5906. [Google Scholar] [CrossRef]
- Pollard, T.D. A Guide to Simple and Informative Binding Assays. Mol. Biol. Cell 2010, 21, 4061–4067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berry, J.D.; Neeson, M.J.; Dagastine, R.R.; Chan, D.Y.C.; Tabor, R.F. Measurement of surface and interfacial tension using pendant drop tensiometry. J. Colloid Interface Sci. 2015, 454, 226–237. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.N. Surfactant Interactions with Biomembranes and Proteins. Chem. Soc. Rev. 1992, 21, 127–136. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Harris, L.J.; Larson, S.B.; Hasel, K.W.; McPherson, A. Refined Structure of an Intact IgG2a Monoclonal Antibody. Biochemistry 1997, 36, 1581–1597. [Google Scholar] [CrossRef] [PubMed]
- Saien, J.; Akbari, S. Interfacial Tension of Toluene + Water + Sodium Dodecyl Sulfate from (20 to 50) °C and pH between 4 and 9. J. Chem. Eng. Data 2006, 51, 1832–1835. [Google Scholar] [CrossRef]
- Hubert, H.F.; Thies, C. Adsorption of Toluene-Soluble Polymers at the Toluene-Water Interface. J. Polym. Sci. Part A-2 Polym. Phys. 1970, 8, 71–80. [Google Scholar] [CrossRef]
- Lucas, E.F.; Oliveira, C.M.; Gomes, A.S. Surface Properties of Graft Copolymers Surfactants: Behavior at the Water/Toluene Interface. J. Appl. Polym. Sci. 1992, 46, 733–737. [Google Scholar] [CrossRef]
- Azzam, T.; Eisenberg, A. Fully Collapsed (Kippah) Vesicles: Preparation and Characterization. Langmuir 2010, 26, 10513–10523. [Google Scholar] [CrossRef] [PubMed]
- Franken, L.E.; Boekema, E.J.; Stuart, M.C.A. Transmission Electron Microscopy as a Tool for the Characterization of Soft Materials: Application and Interpretation. Adv. Sci. 2017, 4, 1600476. [Google Scholar] [CrossRef] [PubMed]





© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
D. Posey, N.; N. Tew, G. Protein Transduction Domain Mimic (PTDM) Self-Assembly? Polymers 2018, 10, 1039. https://doi.org/10.3390/polym10091039
D. Posey N, N. Tew G. Protein Transduction Domain Mimic (PTDM) Self-Assembly? Polymers. 2018; 10(9):1039. https://doi.org/10.3390/polym10091039
Chicago/Turabian StyleD. Posey, Nicholas, and Gregory N. Tew. 2018. "Protein Transduction Domain Mimic (PTDM) Self-Assembly?" Polymers 10, no. 9: 1039. https://doi.org/10.3390/polym10091039