In Silico Studies of Small Molecule Interactions with Enzymes Reveal Aspects of Catalytic Function
Abstract
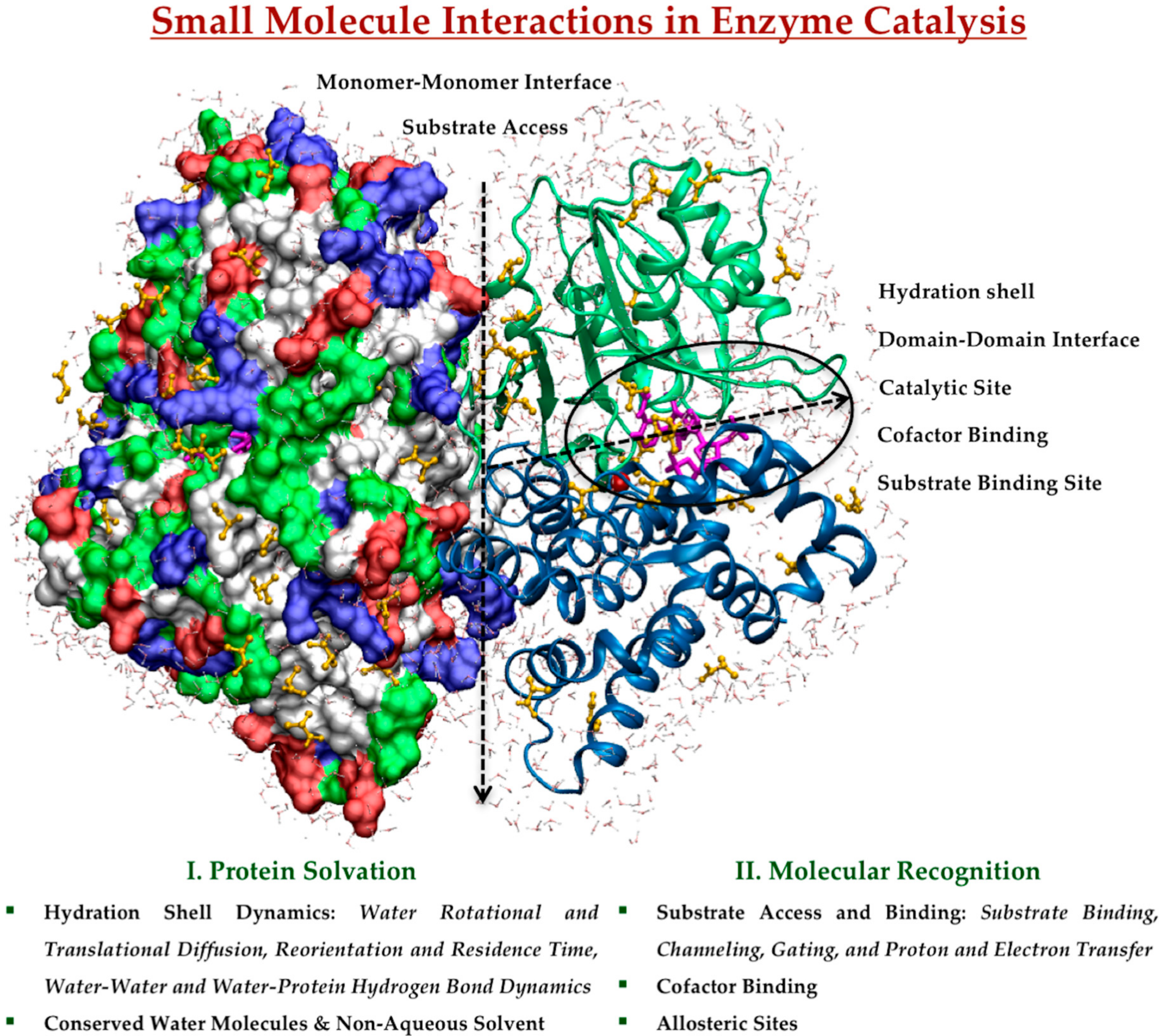
:1. Introduction
2. Effect of Small Molecule Interactions on Protein Structure, Function, and Dynamics
2.1. Protein Solvation in Enzyme Catalysis
2.1.1. Hydration Shell Dynamics
2.1.2. Conserved Water Molecules and Non-Aqueous Solvent
2.2. Molecular Recognition in Enzyme Catalysis
2.2.1. Substrate Access and Binding
2.2.2. Cofactor Binding
2.2.3. Allosteric Sites
3. Concluding Remarks
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Agarwal, P.K.; Doucet, N.; Chennubhotla, C.; Ramanathan, A.; Narayanan, C. Conformational Sub-states and Populations in Enzyme Catalysis. Methods Enzymol. 2016, 578, 273–297. [Google Scholar] [PubMed]
- Ramanathan, A.; Savol, A.; Burger, V.; Chennubhotla, C.S.; Agarwal, P.K. Protein conformational populations and functionally relevant substates. Acc. Chem. Res. 2014, 47, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Bruice, T.C. Computational Approaches: Reaction Trajectories, Structures, and Atomic Motions. Enzyme Reactions and Proficiency. Chem. Rev. 2006, 106, 3119–3139. [Google Scholar] [CrossRef] [PubMed]
- Fujisaki, H.; Moritsugu, K.; Matsunaga, Y.; Morishita, T.; Maragliano, L. Extended Phase-Space Methods for Enhanced Sampling in Molecular Simulations: A Review. Front. Bioeng. Biotechnol. 2015, 3, 125. [Google Scholar] [CrossRef] [PubMed]
- Iida, S.; Nakamura, H.; Higo, J. Enhanced conformational sampling to visualize a free-energy landscape of protein complex formation. Biochem. J. 2016, 473, 1651–1662. [Google Scholar] [CrossRef] [PubMed]
- Warshel, A.; Sharma, P.K.; Kato, M.; Xiang, Y.; Liu, H.; Olsson, M.H.M. Electrostatic Basis for Enzyme Catalysis. Chem. Rev. 2006, 106, 3210–3235. [Google Scholar] [CrossRef] [PubMed]
- Bahar, I.; Lezon, T.R.; Yang, L.-W.; Eyal, E. Global Dynamics of Proteins: Bridging Between Structure and Function. Annu. Rev. Biophys. 2010, 39, 23–42. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Li, H. Force spectroscopy studies on protein-ligand interactions: A single protein mechanics perspective. FEBS Lett. 2014, 588, 3613–3620. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, K.; Sako, Y. Recent advances in FRET for the study of protein interactions and dynamics. Curr. Opin. Struct. Biol. 2017, 46, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Grossman, M.; Born, B.; Heyden, M.; Tworowski, D.; Fields, G.B.; Sagi, I.; Havenith, M. Correlated structural kinetics and retarded solvent dynamics at the metalloprotease active site. Nat. Struct. Mol. Biol. 2011, 18, 1102–1108. [Google Scholar] [CrossRef] [PubMed]
- Kohen, A. Role of dynamics in enzyme catalysis: Substantial versus semantic controversies. Acc. Chem. Res. 2015, 48, 466–473. [Google Scholar] [CrossRef] [PubMed]
- Callender, R.; Dyer, R.B. The dynamical nature of enzymatic catalysis. Acc. Chem Res. 2015, 48, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Fogarty, A.C.; Laage, D. Water dynamics in protein hydration shells: The molecular origins of the dynamical perturbation. J. Phys. Chem. B 2014, 118, 7715–7729. [Google Scholar] [CrossRef] [PubMed]
- Ryde, U.; Söderhjelm, P. Ligand-Binding Affinity Estimates Supported by Quantum-Mechanical Methods. Chem. Rev. 2016, 116, 5520–5566. [Google Scholar] [CrossRef] [PubMed]
- Fischer, E. Einfluss der Configuration auf die Wirkung der Enzyme. Eur. J. Inorg. Chem. 1894, 27, 2985–2993. [Google Scholar] [CrossRef]
- Koshland, D.E. Application of a Theory of Enzyme Specificity to Protein Synthesis. Proc. Natl. Acad. Sci. USA 1958, 44, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Weikl, T.R.; Paul, F. Conformational selection in protein binding and function. Protein Sci. 2014, 23, 1508–1518. [Google Scholar] [CrossRef] [PubMed]
- Vogt, A.D.; Pozzi, N.; Chen, Z.; Di Cera, E. Essential role of conformational selection in ligand binding. Biophys. Chem. 2014, 186, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.D.; Schramm, V.L. Enzymatic transition states and dynamic motion in barrier crossing. Nat. Chem. Biol. 2009, 5, 551–558. [Google Scholar] [CrossRef] [PubMed]
- Schramm, V.L. Enzymatic transition states, transition-state analogs, dynamics, thermodynamics, and lifetimes. Annu. Rev. Biochem. 2011, 80, 703–732. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Li, Y.; Xia, Y.-L.; Ai, S.-M.; Liang, J.; Sang, P.; Ji, X.-L.; Liu, S.-Q. Insights into Protein–Ligand Interactions: Mechanisms, Models, and Methods. Int. J. Mol. Sci. 2016, 17, 144. [Google Scholar] [CrossRef] [PubMed]
- Konig, P.M.; Roth, R.; Dietrich, S. Lock and key model system. EPL 2008, 84, 68006. [Google Scholar] [CrossRef]
- Wlodarski, T.; Zagrovic, B. Conformational selection and induced fit mechanism underlie specificity in noncovalent interactions with ubiquitin. Proc. Natl. Acad. Sci. USA 2009, 106, 19346–19351. [Google Scholar] [CrossRef] [PubMed]
- Silva, D.-A.; Bowman, G.R.; Sosa-Peinado, A.; Huang, X. A Role for Both Conformational Selection and Induced Fit in Ligand Binding by the LAO Protein. PLoS Comput. Biol. 2011, 7, e1002054. [Google Scholar] [CrossRef] [PubMed]
- Prokop, Z.; Gora, A.; Brezovsky, J.; Chaloupkova, R.; Stepankova, V.; Damborsky, J. Engineering of protein tunnels: Keyhole-lock-key model for catalysis by the enzymes with buried active sites. In Protein Engineering Handbook; Lutz, S., Bornscheuer, U.T., Eds.; Wiley-VCH: Weinheim, Germany, 2012; Volume 3, pp. 421–464. [Google Scholar]
- Eisenmesser, E.Z.; Millet, O.; Labeikovsky, W.; Korzhnev, D.M.; Wolf-Watz, M.; Bosco, D.A.; Skalicky, J.J.; Kay, L.E.; Kern, D. Intrinsic dynamics of an enzyme underlies catalysis. Nature 2005, 438, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, C.; Bernard, D.N.; Doucet, N. Role of Conformational Motions in Enzyme Function: Selected Methodologies and Case Studies. Catalysts 2016, 6, 81. [Google Scholar] [CrossRef] [PubMed]
- Bhabha, G.; Biel, J.T.; Fraser, J.S. Keep on moving: Discovering and perturbing the conformational dynamics of enzymes. Acc. Chem. Res. 2015, 48, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Warshel, A.; Bora, R.P. Perspective: Defining and quantifying the role of dynamics in enzyme catalysis. J. Chem. Phys. 2016, 144, 180901. [Google Scholar] [CrossRef] [PubMed]
- Lonsdale, R.; Ranaghan, K.E.; Mulholland, A.J. Computational enzymology. Chem. Commun. 2010, 46, 2354–2372. [Google Scholar] [CrossRef] [PubMed]
- Gutteridge, A.; Thornton, J. Conformational change in substrate binding, catalysis and product release: An open and shut case? FEBS Lett. 2004, 567, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.K. Enzymes: An integrated view of structure, dynamics and function. Microb. Cell. Fact. 2006, 5, 2. [Google Scholar] [CrossRef] [PubMed]
- Henzler-Wildman, K.A.; Lei, M.; Thai, V.; Kerns, S.J.; Karplus, M.; Kern, D. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature 2007, 450, 913–916. [Google Scholar] [CrossRef] [PubMed]
- Maximova, T.; Moffatt, R.; Ma, B.; Nussinov, R.; Shehu, A. Principles and Overview of Sampling Methods for Modeling Macromolecular Structure and Dynamics. PLoS Comput. Biol. 2016, 12, e1004619. [Google Scholar] [CrossRef] [PubMed]
- Zewail, A.H. Diffraction, crystallography and microscopy beyond three dimensions: Structural dynamics in space and time. Philos. Trans. A Math. Phys. Eng. Sci. 2005, 363, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Egli, M. Diffraction Techniques in Structural Biology. Curr. Protoc. Nucleic Acid. Chem. 2016, 65, 7.13.11–17.13.41. [Google Scholar] [PubMed]
- Lisi, G.P.; Loria, J.P. Using NMR spectroscopy to elucidate the role of molecular motions in enzyme function. Prog. Nucl. Magn. Reson. Spectrosc. 2016, 92–93, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Lonsdale, R.; Harvey, J.N.; Mulholland, A.J. A practical guide to modelling enzyme-catalysed reactions. Chem. Soc. Rev. 2012, 41, 3025–3038. [Google Scholar] [CrossRef] [PubMed]
- Gherib, R.; Dokainish, H.M.; Gauld, J.W. Multi-Scale Computational Enzymology: Enhancing Our Understanding of Enzymatic Catalysis. Int. J. Mol. Sci. 2014, 15, 401–422. [Google Scholar] [CrossRef] [PubMed]
- Dror, R.O.; Dirks, R.M.; Grossman, J.P.; Xu, H.; Shaw, D.E. Biomolecular Simulation: A Computational Microscope for Molecular Biology. Annu. Rev. Biophys. 2012, 41, 429–452. [Google Scholar] [CrossRef] [PubMed]
- Vlachakis, D.; Bencurova, E.; Papangelopoulos, N.; Kossida, S. Current State-of-the-Art Molecular Dynamics Methods and Applications. Adv. Protein Chem. Struct. Biol. 2014, 94, 269–313. [Google Scholar] [PubMed]
- Frauenfelder, H.; Fenimore, P.W.; Young, R.D. Protein dynamics and function: Insights from the energy landscape and solvent slaving. IUBMB Life 2007, 59, 506–512. [Google Scholar] [CrossRef] [PubMed]
- Fenimore, P.W.; Frauenfelder, H.; McMahon, B.H.; Parak, F.G. Slaving: Solvent fluctuations dominate protein dynamics and functions. Proc. Natl. Acad. Sci. USA 2002, 99, 16047–16051. [Google Scholar] [CrossRef] [PubMed]
- Wood, K.; Plazanet, M.; Gabel, F.; Kessler, B.; Oesterhelt, D.; Tobias, D.J.; Zaccai, G.; Weik, M. Coupling of protein and hydration-water dynamics in biological membranes. Proc. Natl. Acad. Sci. USA 2007, 104, 18049–18054. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Li, W.; Tian, P. Reconciling Mediating and Slaving Roles of Water in Protein Conformational Dynamics. PLoS ONE 2013, 8, e60553. [Google Scholar] [CrossRef] [PubMed]
- Mobley, D.L.; Dill, K.A. Binding of Small-Molecule Ligands to Proteins: “What You See” Is Not Always “What You Get”. Structure 2009, 17, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Moritsugu, K.; Terada, T.; Kidera, A. Free-Energy Landscape of Protein–Ligand Interactions Coupled with Protein Structural Changes. J. Phys. Chem. B 2017, 121, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Syrén, P.-O.; Hammer, S.C.; Claasen, B.; Hauer, B. Entropy is Key to the Formation of Pentacyclic Terpenoids by Enzyme-Catalyzed Polycyclization. Angew. Chem. Int. Ed. 2014, 53, 4845–4849. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.M.; Kang, K.; Sastry, M.; Sherman, W.; Sankaran, B.; Zwart, P.H.; Whitesides, G.M. Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase. Angew. Chem. Int. Ed. 2017, 56, 3833–3837. [Google Scholar] [CrossRef] [PubMed]
- Fink, M.J.; Syrén, P.-O. Redesign of water networks for efficient biocatalysis. Curr. Opin. Chem. Biol. 2017, 37, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Hendil-Forssell, P.; Martinelle, M.; Syren, P.-O. Exploring water as building bricks in enzyme engineering. Chem. Commun. 2015, 51, 17221–17224. [Google Scholar] [CrossRef] [PubMed]
- Klepeis, J.L.; Lindorff-Larsen, K.; Dror, R.O.; Shaw, D.E. Long-timescale molecular dynamics simulations of protein structure and function. Curr. Opin. Struct. Biol. 2009, 19, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Zwier, M.C.; Chong, L.T. Reaching biological timescales with all-atom molecular dynamics simulations. Curr. Opin. Pharmacol. 2010, 10, 745–752. [Google Scholar] [CrossRef] [PubMed]
- Perilla, J.R.; Goh, B.C.; Cassidy, C.K.; Liu, B.; Bernardi, R.C.; Rudack, T.; Yu, H.; Wu, Z.; Schulten, K. Molecular dynamics simulations of large macromolecular complexes. Curr. Opin. Struct. Biol. 2015, 31, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Sulzenbacher, G.; Alvarez, K.; van den Heuvel, R.H.H.; Versluis, C.; Spinelli, S.; Campanacci, V.; Valencia, C.; Cambillau, C.; Eklund, H.; Tegoni, M. Crystal Structure of E. coli Alcohol Dehydrogenase YqhD: Evidence of a Covalently Modified NADP Coenzyme. J. Mol. Biol. 2004, 342, 489–502. [Google Scholar] [CrossRef] [PubMed]
- Ball, P. Water as an Active Constituent in Cell Biology. Chem. Rev. 2008, 108, 74–108. [Google Scholar] [CrossRef] [PubMed]
- Levy, Y.; Onuchic, J.N. Water mediation in protein folding and molecular recognition. Annu. Rev. Biophys. Biomol. Struct. 2006, 35, 389–415. [Google Scholar] [CrossRef] [PubMed]
- Schug, A.; Onuchic, J.N. From protein folding to protein function and biomolecular binding by energy landscape theory. Curr. Opin. Pharmacol. 2010, 10, 709–714. [Google Scholar] [CrossRef] [PubMed]
- Fogarty, A.C.; Duboue-Dijon, E.; Sterpone, F.; Hynes, J.T.; Laage, D. Biomolecular hydration dynamics: A jump model perspective. Chem. Soc. Rev. 2013, 42, 5672–5683. [Google Scholar] [CrossRef] [PubMed]
- Purkiss, A.; Skoulakis, S.; Goodfellow, J.M. The protein–solvent interface: A big splash. Philos. Trans. A Math. Phys. Eng. Sci. 2001, 359, 1515–1527. [Google Scholar] [CrossRef]
- Raschke, T.M. Water structure and interactions with protein surfaces. Curr. Opin. Struct. Biol. 2006, 16, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Hospital, A.; Candotti, M.; Gelpí, J.L.; Orozco, M. The Multiple Roles of Waters in Protein Solvation. J. Phys. Chem. B 2017. [Google Scholar] [CrossRef] [PubMed]
- Prakash, P.; Sayyed-Ahmad, A.; Gorfe, A.A. The Role of Conserved Waters in Conformational Transitions of Q61H K-ras. PLoS Comput. Biol. 2012, 8, e1002394. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.K.; Zewail, A.H. Dynamics of Water in Biological Recognition. Chem. Rev. 2004, 104, 2099–2124. [Google Scholar] [CrossRef] [PubMed]
- Schoenborn, B.P.; Garcia, A.; Knott, R. Hydration in protein crystallography. Prog. Biophys. Mol. Biol. 1995, 64, 105–119. [Google Scholar] [CrossRef]
- Nakasako, M. Large-scale networks of hydration water molecules around proteins investigated by cryogenic X-ray crystallography. Cell. Mol. Biol. 2001, 47, 767–790. [Google Scholar] [PubMed]
- Svergun, D.I.; Richard, S.; Koch, M.H.; Sayers, Z.; Kuprin, S.; Zaccai, G. Protein hydration in solution: Experimental observation by X-ray and neutron scattering. Proc. Natl. Acad. Sci. USA 1998, 95, 2267–2272. [Google Scholar] [CrossRef] [PubMed]
- Perticaroli, S.; Ehlers, G.; Stanley, C.B.; Mamontov, E.; O'Neill, H.; Zhang, Q.; Cheng, X.; Myles, D.A.; Katsaras, J.; Nickels, J.D. Description of Hydration Water in Protein (Green Fluorescent Protein) Solution. J. Am. Chem. Soc. 2017, 139, 1098–1105. [Google Scholar] [CrossRef] [PubMed]
- Russo, D.; Ollivier, J.; Teixeira, J. Water hydrogen bond analysis on hydrophilic and hydrophobic biomolecule sites. Phys. Chem. Chem. Phys. 2008, 10, 4968–4974. [Google Scholar] [CrossRef] [PubMed]
- Zanotti, J.-M.; Bellissent-Funel, M.-C.; Parello, J. Hydration-Coupled Dynamics in Proteins Studied by Neutron Scattering and NMR: The Case of the Typical EF-Hand Calcium-Binding Parvalbumin. Biophys. J. 1999, 76, 2390–2411. [Google Scholar] [CrossRef]
- Martini, S.; Bonechi, C.; Foletti, A.; Rossi, C. Water-Protein Interactions: The Secret of Protein Dynamics. Sci. World J. 2013, 2013, 138916. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Hochstrasser, R.M. Applications of 2D IR spectroscopy to peptides, proteins, and hydrogen-bond dynamics. J. Phys. Chem. B 2009, 113, 8231–8251. [Google Scholar] [CrossRef] [PubMed]
- Koch, M.H.; Vachette, P.; Svergun, D.I. Small-angle scattering: A view on the properties, structures and structural changes of biological macromolecules in solution. Q. Rev. Biophys. 2003, 36, 147–227. [Google Scholar] [CrossRef] [PubMed]
- Petoukhov, M.V.; Svergun, D.I. Applications of small-angle X-ray scattering to biomacromolecular solutions. Int. J. Biochem. Cell Biol. 2013, 45, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Palamini, M.; Canciani, A.; Forneris, F. Identifying and Visualizing Macromolecular Flexibility in Structural Biology. Front. Mol. Biosci. 2016, 3, 47. [Google Scholar] [CrossRef] [PubMed]
- Laage, D.; Elsaesser, T.; Hynes, J.T. Water Dynamics in the Hydration Shells of Biomolecules. Chem. Rev. 2017. [Google Scholar] [CrossRef] [PubMed]
- McCammon, J.A.; Gelin, B.R.; Karplus, M. Dynamics of folded proteins. Nature 1977, 267, 585–590. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, J. Dynamics of hydration water in proteins. Gen. Physiol. Biophys. 2009, 28, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Knight, J.D.R.; Hamelberg, D.; McCammon, J.A.; Kothary, R. The role of conserved water molecules in the catalytic domain of protein kinases. Proteins 2009, 76, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Almazán, C.; Arreola, R.; Rodríguez-Larrea, D.; Aguirre-López, B.; de Gómez-Puyou, M.T.; Pérez-Montfort, R.; Costas, M.; Gómez-Puyou, A.; Torres-Larios, A. Structural Basis of Human Triosephosphate Isomerase Deficiency: Mutation E104d Is Related to Alterations of a Conserved Water Network At The Dimer Interface. J. Biol. Chem. 2008, 283, 23254–23263. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, B.D.; Choi, J.; López, C.; Wesener, D.A.; Hubbell, W.; Cavagnero, S.; Han, S. Site-Specific Hydration Dynamics in the Nonpolar Core of a Molten Globule by Dynamic Nuclear Polarization of Water. J. Am. Chem. Soc. 2011, 133, 5987–5995. [Google Scholar] [CrossRef] [PubMed]
- Halle, B. Protein hydration dynamics in solution: A critical survey. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004, 359, 1207. [Google Scholar] [CrossRef] [PubMed]
- Engen, J.R. Analysis of Protein Conformation and Dynamics by Hydrogen/Deuterium Exchange MS. Anal. Chem. 2009, 81, 7870–7875. [Google Scholar] [CrossRef] [PubMed]
- Hamuro, Y.; Coales, S.J.; Southern, M.R.; Nemeth-Cawley, J.F.; Stranz, D.D.; Griffin, P.R. Rapid Analysis of Protein Structure and Dynamics by Hydrogen/Deuterium Exchange Mass Spectrometry. J. Biomol. Tech. 2003, 14, 171–182. [Google Scholar] [PubMed]
- Wang, L.; Chance, M.R. Protein Footprinting Comes of Age: Mass Spectrometry for Biophysical Structure Assessment. Mol. Cell. Proteom. 2017, 16, 706–716. [Google Scholar] [CrossRef] [PubMed]
- Benesch, J.L.P.; Ruotolo, B.T. Mass Spectrometry: An Approach Come-of-Age for Structural and Dynamical Biology. Curr. Opin. Struct. Biol. 2011, 21, 641–649. [Google Scholar] [CrossRef] [PubMed]
- Olshina, M.A.; Sharon, M. Mass Spectrometry: A Technique of Many Faces. Q. Rev. Biophys. 2016, 49, e18. [Google Scholar] [CrossRef] [PubMed]
- Faini, M.; Stengel, F.; Aebersold, R. The Evolving Contribution of Mass Spectrometry to Integrative Structural Biology. J. Am. Soc. Mass Spectrom. 2016, 27, 966–974. [Google Scholar] [CrossRef] [PubMed]
- Konermann, L.; Pan, J.; Liu, Y.-H. Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem. Soc. Rev. 2011, 40, 1224–1234. [Google Scholar] [CrossRef] [PubMed]
- Kiselar, J.G.; Chance, M.R. Future Directions of Structural Mass Spectrometry using Hydroxyl Radical Footprinting. J. Mass Spectrom. 2010, 45, 1373–1382. [Google Scholar] [CrossRef] [PubMed]
- King, J.T.; Kubarych, K.J. Site-specific coupling of hydration water and protein flexibility studied in solution with ultrafast 2D-IR spectroscopy. J. Am. Chem Soc. 2012, 134, 18705–18712. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Bagchi, B. Free Energy Barriers for Escape of Water Molecules from Protein Hydration Layer. J. Phys. Chem. B 2012, 116, 2958–2968. [Google Scholar] [CrossRef] [PubMed]
- Makarov, V.A.; Andrews, B.K.; Smith, P.E.; Pettitt, B.M. Residence Times of Water Molecules in the Hydration Sites of Myoglobin. Biophys. J. 2000, 79, 2966–2974. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, L.; Kao, Y.-T.; Qiu, W.; Yang, Y.; Okobiah, O.; Zhong, D. Mapping hydration dynamics around a protein surface. Proc. Natl. Acad. Sci. USA 2007, 104, 18461–18466. [Google Scholar] [CrossRef] [PubMed]
- Sciortino, F.; Fabbian, L.; Chen, S.-H.; Tartaglia, P. Supercooled water and the kinetic glass transition. II. Collective dynamics. Phys. Rev. E 1997, 56, 5397–5404. [Google Scholar] [CrossRef]
- Halle, B.; Davidovic, M. Biomolecular hydration: From water dynamics to hydrodynamics. Proc. Natl. Acad. Sci. USA 2003, 100, 12135–12140. [Google Scholar] [CrossRef] [PubMed]
- Duboue-Dijon, E.; Laage, D. Comparative study of hydration shell dynamics around a hyperactive antifreeze protein and around ubiquitin. J. Chem. Phys. 2014, 141, 22D529. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Hu, B.; Lill, M.A. Analysis of factors influencing hydration site prediction based on molecular dynamics simulations. J. Chem. Inf. Model. 2014, 54, 2987–2995. [Google Scholar] [CrossRef] [PubMed]
- De Beer, S.B.; Vermeulen, N.P.; Oostenbrink, C. The role of water molecules in computational drug design. Curr. Top. Med. Chem. 2010, 10, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Maricarmen, H.-R.; Martha, C.R.-H.; Jessica, E.M.-W.; Marlet, M.-A.; José Correa, B. Current Tools and Methods in Molecular Dynamics (MD) Simulations for Drug Design. Curr. Top. Med. Chem. 2016, 23, 3909–3924. [Google Scholar]
- Spyrakis, F.; Ahmed, M.H.; Bayden, A.S.; Cozzini, P.; Mozzarelli, A.; Kellogg, G.E. The Roles of Water in the Protein Matrix: A Largely Untapped Resource for Drug Discovery. J. Med. Chem. 2017. [Google Scholar] [CrossRef] [PubMed]
- Abel, R.; Young, T.; Farid, R.; Berne, B.J.; Friesner, R.A. Role of the Active-Site Solvent in the Thermodynamics of Factor Xa Ligand Binding. J. Am. Chem. Soc. 2008, 130, 2817–2831. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Lill, M.A. WATsite: Hydration site prediction program with PyMOL interface. J. Comp. Chem. 2014, 35, 1255–1260. [Google Scholar] [CrossRef] [PubMed]
- Barron, L.D.; Hecht, L.; Wilson, G. The Lubricant of Life: A Proposal That Solvent Water Promotes Extremely Fast Conformational Fluctuations in Mobile Heteropolypeptide Structure. Biochemistry 1997, 36, 13143–13147. [Google Scholar] [CrossRef] [PubMed]
- Laage, D.; Hynes, J.T. A Molecular Jump Mechanism of Water Reorientation. Science 2006, 311, 832–835. [Google Scholar] [CrossRef] [PubMed]
- Kılıç, M.; Ensing, B. First and Second One-Electron Reduction of Lumiflavin in Water—A First Principles Molecular Dynamics Study. J. Chem. Theory Comput. 2013, 9, 3889–3899. [Google Scholar] [CrossRef] [PubMed]
- Marcus, R.A. H and Other Transfers in Enzymes and in Solution: Theory and Computations, a Unified View. 2. Applications to Experiment and Computations. J. Phys. Chem. B 2007, 111, 6643–6654. [Google Scholar] [CrossRef] [PubMed]
- Hammes-Schiffer, S. Hydrogen Tunneling and Protein Motion in Enzyme Reactions. Acc. Chem. Res. 2006, 39, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Hammes-Schiffer, S. Proton-Coupled Electron Transfer: Theoretical Formulation and Applications. In Hydrogen-Transfer Reactions; Hynes, J.T., Klinman, J.P., Limbach, H.-H., Schowen, R.L., Eds.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2007; pp. 479–502. [Google Scholar]
- Hammes-Schiffer, S. Theory of proton-coupled electron transfer in energy conversion processes. Acc. Chem. Res. 2009, 42, 1881–1889. [Google Scholar] [CrossRef] [PubMed]
- Born, B.; Kim, S.J.; Ebbinghaus, S.; Gruebele, M.; Havenith, M. The terahertz dance of water with the proteins: The effect of protein flexibility on the dynamical hydration shell of ubiquitin. Faraday Discuss. 2009, 141, 161–173. [Google Scholar] [CrossRef]
- Qin, Y.; Wang, L.; Zhong, D. Dynamics and mechanism of ultrafast water–protein interactions. Proc. Natl. Acad. Sci. USA 2016, 113, 8424–8429. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.; Gu, W.; Geyer, T.; Helms, V. Adhesive water networks facilitate binding of protein interfaces. Nat. Commun. 2011, 2, 261. [Google Scholar] [CrossRef] [PubMed]
- Tirion, M.M. Large Amplitude Elastic Motions in Proteins from a Single-Parameter, Atomic Analysis. Phys. Rev. Lett. 1996, 77, 1905–1908. [Google Scholar] [CrossRef] [PubMed]
- Schirò, G.; Fichou, Y.; Gallat, F.-X.; Wood, K.; Gabel, F.; Moulin, M.; Härtlein, M.; Heyden, M.; Colletier, J.-P.; Orecchini, A.; et al. Translational diffusion of hydration water correlates with functional motions in folded and intrinsically disordered proteins. Nat. Commun. 2015, 6, 6490. [Google Scholar] [CrossRef] [PubMed]
- Mattea, C.; Qvist, J.; Halle, B. Dynamics at the Protein-Water Interface from (17)O Spin Relaxation in Deeply Supercooled Solutions. Biophys. J. 2008, 95, 2951–2963. [Google Scholar] [CrossRef] [PubMed]
- Garcia, A.E.; Hummer, G. Water penetration and escape in proteins. Proteins 2000, 38, 261–272. [Google Scholar] [CrossRef]
- Hunt, N.T.; Kattner, L.; Shanks, R.P.; Wynne, K. The dynamics of water-protein interaction studied by ultrafast optical Kerr-effect spectroscopy. J. Am. Chem. Soc. 2007, 129, 3168–3172. [Google Scholar] [CrossRef] [PubMed]
- Mazur, K.; Heisler, I.A.; Meech, S.R. Ultrafast dynamics and hydrogen-bond structure in aqueous solutions of model peptides. J. Phys. Chem. B 2010, 114, 10684–10691. [Google Scholar] [CrossRef] [PubMed]
- Ebbinghaus, S.; Kim, S.J.; Heyden, M.; Yu, X.; Heugen, U.; Gruebele, M.; Leitner, D.M.; Havenith, M. An extended dynamical hydration shell around proteins. Proc. Natl. Acad. Sci. USA 2007, 104, 20749–20752. [Google Scholar] [CrossRef] [PubMed]
- Dielmann-Gessner, J.; Grossman, M.; Conti Nibali, V.; Born, B.; Solomonov, I.; Fields, G.B.; Havenith, M.; Sagi, I. Enzymatic turnover of macromolecules generates long-lasting protein–water-coupled motions beyond reaction steady state. Proc. Natl. Acad. Sci. USA 2014, 111, 17857–17862. [Google Scholar] [CrossRef] [PubMed]
- Stals, P.J.M.; Cheng, C.-Y.; van Beek, L.; Wauters, A.C.; Palmans, A.R.A.; Han, S.; Meijer, E.W. Surface water retardation around single-chain polymeric nanoparticles: Critical for catalytic function? Chem. Sci. 2016, 7, 2011–2015. [Google Scholar] [CrossRef]
- Khodadadi, S.; Roh, J.H.; Kisliuk, A.; Mamontov, E.; Tyagi, M.; Woodson, S.A.; Briber, R.M.; Sokolov, A.P. Dynamics of Biological Macromolecules: Not a Simple Slaving by Hydration Water. Biophys. J. 2010, 98, 1321–1326. [Google Scholar] [CrossRef] [PubMed]
- Rahaman, O.; Kalimeri, M.; Melchionna, S.; Hénin, J.; Sterpone, F. On the Role of Internal Water on Protein Thermal Stability: The Case of Homologous G-domains. J. Phys. Chem. B 2015, 119, 8939–8949. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, D.; Taly, A.; Sterpone, F. Stay Wet, Stay Stable? How Internal Water Helps Stability of Thermophilic Proteins. J. Phys. Chem. B 2015, 119, 12760–12770. [Google Scholar] [CrossRef] [PubMed]
- Barillari, C.; Taylor, J.; Viner, R.; Essex, J.W. Classification of water molecules in protein binding sites. J. Am. Chem. Soc. 2007, 129, 2577–2587. [Google Scholar] [CrossRef] [PubMed]
- Ross, G.A.; Morris, G.M.; Biggin, P.C. Rapid and Accurate Prediction and Scoring of Water Molecules in Protein Binding Sites. PLoS ONE 2012, 7, e32036. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Sosa, A.T.; Mancera, R.L.; Dean, P.M. WaterScore: A novel method for distinguishing between bound and displaceable water molecules in the crystal structure of the binding site of protein-ligand complexes. J. Mol. Model. 2003, 9, 172–182. [Google Scholar] [CrossRef] [PubMed]
- Patel, H.; Gruning, B.A.; Gunther, S.; Merfort, I. PyWATER: A PyMOL plug-in to find conserved water molecules in proteins by clustering. Bioinformatics 2014, 30, 2978–2980. [Google Scholar] [CrossRef] [PubMed]
- Ogata, K.; Wodak, S.J. Conserved water molecules in MHC class-I molecules and their putative structural and functional roles. Protein Eng. 2002, 15, 697–705. [Google Scholar] [CrossRef] [PubMed]
- Wallnoefer, H.G.; Handschuh, S.; Liedl, K.R.; Fox, T. Stabilizing of a Globular Protein by a Highly Complex Water Network: A Molecular Dynamics Simulation Study on Factor Xa. J. Phys. Chem. B 2010, 114, 7405–7412. [Google Scholar] [CrossRef] [PubMed]
- Teze, D.; Hendrickx, J.; Dion, M.; Tellier, C.; Woods, V.L.; Tran, V.; Sanejouand, Y.-H. Conserved Water Molecules in Family 1 Glycosidases: A DXMS and Molecular Dynamics Study. Biochemistry 2013, 52, 5900–5910. [Google Scholar] [CrossRef] [PubMed]
- Chovancova, E.; Pavelka, A.; Benes, P.; Strnad, O.; Brezovsky, J.; Kozlikova, B.; Gora, A.; Sustr, V.; Klvana, M.; Medek, P.; et al. CAVER 3.0: A Tool for the Analysis of Transport Pathways in Dynamic Protein Structures. PLoS Comput. Biol. 2012, 8, e1002708. [Google Scholar] [CrossRef] [PubMed]
- Roccatano, D. Computer simulations study of biomolecules in non-aqueous or cosolvent/water mixture solutions. Curr. Protein Pept. Sci. 2008, 9, 407–426. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Meng, X.; Zhou, H.; Liu, Y.; Secundo, F.; Liu, Y. Enzyme Stability and Activity in Non-Aqueous Reaction Systems: A Mini Review. Catalysts 2016, 6, 32. [Google Scholar] [CrossRef]
- Micaelo, N.M.; Soares, C.M. Modeling hydration mechanisms of enzymes in nonpolar and polar organic solvents. FEBS J. 2007, 274, 2424–2436. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Dordick, J.S.; Garde, S. Hydration of enzyme in nonaqueous media is consistent with solvent dependence of its activity. Biophys. J. 2004, 87, 812–821. [Google Scholar] [CrossRef] [PubMed]
- Dahanayake, J.N.; Gautam, D.N.; Verma, R.; Mitchell-Koch, K.R. To keep or not to keep? the question of crystallographic waters for enzyme simulations in organic solvent. Mol. Simul. 2016, 42, 1001–1013. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Yang, W.; Meng, Y.Y.; Xiao, X.; Guo, Y.; Pu, X.; Li, M. Effects of organic solvent and crystal water on gamma-chymotrypsin in acetonitrile media: Observations from molecular dynamics simulation and DFT calculation. J. Phys. Chem. B 2012, 116, 3292–3304. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Yuan, Y.; Zhu, Y.; Guo, Y.; Li, M.; Wang, Z.; Pu, X.; Jiang, L. Effects of organic solvents and substrate binding on trypsin in acetonitrile and hexane media. J. Mol. Model. 2013, 19, 3749–3766. [Google Scholar] [CrossRef] [PubMed]
- Allen, K.N.; Bellamacina, C.R.; Ding, X.; Jeffery, C.J.; Mattos, C.; Petsko, G.A.; Ringe, D. An Experimental Approach to Mapping the Binding Surfaces of Crystalline Proteins. J. Phys. Chem. A 1996, 100, 2605–2611. [Google Scholar] [CrossRef]
- Mattos, C.; Ringe, D. Locating and characterizing binding sites on proteins. Nat. Biotechnol. 1996, 14, 595–599. [Google Scholar] [CrossRef] [PubMed]
- Bakan, A.; Nevins, N.; Lakdawala, A.S.; Bahar, I. Druggability Assessment of Allosteric Proteins by Dynamics Simulations in the Presence of Probe Molecules. J. Chem. Theory Comput. 2012, 8, 2435–2447. [Google Scholar] [CrossRef] [PubMed]
- Mattos, C.; Bellamacina, C.R.; Peisach, E.; Pereira, A.; Vitkup, D.; Petsko, G.A.; Ringe, D. Multiple solvent crystal structures: Probing binding sites, plasticity and hydration. J. Mol. Biol. 2006, 357, 1471–1482. [Google Scholar] [CrossRef] [PubMed]
- Ghanakota, P.; Carlson, H.A. Driving Structure-Based Drug Discovery through Cosolvent Molecular Dynamics. J. Med. Chem. 2016, 59, 10383–10399. [Google Scholar] [CrossRef] [PubMed]
- Ghanakota, P.; Carlson, H.A. Moving Beyond Active-Site Detection: MixMD Applied to Allosteric Systems. J. Phys. Chem. B 2016, 120, 8685–8695. [Google Scholar] [CrossRef] [PubMed]
- Mancera, R.L. Molecular modeling of hydration in drug design. Curr. Opin. Drug Discov. Dev. 2007, 10, 275–280. [Google Scholar] [PubMed]
- Callender, R.; Dyer, R.B. Advances in Time-Resolved Approaches To Characterize the Dynamical Nature of Enzymatic Catalysis. Chem. Rev. 2006, 106, 3031–3042. [Google Scholar] [CrossRef] [PubMed]
- Radkiewicz, J.L.; Brooks, C.L. Protein Dynamics in Enzymatic Catalysis: Exploration of Dihydrofolate Reductase. J. Am. Chem. Soc. 2000, 122, 225–231. [Google Scholar] [CrossRef]
- McGowan, L.C.; Hamelberg, D. Conformational plasticity of an enzyme during catalysis: Intricate coupling between cyclophilin A dynamics and substrate turnover. Biophys. J. 2013, 104, 216–226. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.K. Role of Protein Dynamics in Reaction Rate Enhancement by Enzymes. J. Am. Chem. Soc. 2005, 127, 15248–15256. [Google Scholar] [CrossRef] [PubMed]
- Bahnson, B.J.; Colby, T.D.; Chin, J.K.; Goldstein, B.M.; Klinman, J.P. A link between protein structure and enzyme catalyzed hydrogen tunneling. Proc. Natl. Acad. Sci. USA 1997, 94, 12797–12802. [Google Scholar] [CrossRef] [PubMed]
- Van der Kamp, M.W.; Mulholland, A.J. Combined Quantum Mechanics/Molecular Mechanics (QM/MM) Methods in Computational Enzymology. Biochemistry 2013, 52, 2708–2728. [Google Scholar] [CrossRef] [PubMed]
- Kamerlin, S.C.L.; Haranczyk, M.; Warshel, A. Progresses in Ab Initio QM/MM Free Energy Simulations of Electrostatic Energies in Proteins: Accelerated QM/MM Studies of pK(a), Redox Reactions and Solvation Free Energies. J. Phys. Chem. B 2009, 113, 1253–1272. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, S.; Ruiz-Pernía, J.; Martí, S.; Moliner, V.; Tuñón, I.; Bertrán, J.; Andrés, J. Hybrid Schemes Based on Quantum Mechanics/Molecular Mechanics Simulations: Goals to Success, Problems, and Perspectives. In Advances in Protein Chemistry and Structural Biology; Christo, C., Ed.; Academic Press: Cambridge, MA, USA, 2011; Volume 85, pp. 81–142. [Google Scholar]
- Lu, X.; Fang, D.; Ito, S.; Okamoto, Y.; Ovchinnikov, V.; Cui, Q. QM/MM free energy simulations: Recent progress and challenges. Mol. Simul. 2016, 42, 1056–1078. [Google Scholar] [CrossRef] [PubMed]
- Trzesniak, D.; Kunz, A.-P.E.; van Gunsteren, W.F. A Comparison of Methods to Compute the Potential of Mean Force. ChemPhysChem 2007, 8, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Chakravorty, D.K.; Merz, K.M. Role of Substrate Dynamics in Protein Prenylation Reactions. Acc. Chem. Res. 2015, 48, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Strickland, C.L.; Windsor, W.T.; Syto, R.; Wang, L.; Bond, R.; Wu, Z.; Schwartz, J.; Le, H.V.; Beese, L.S.; Weber, P.C. Crystal Structure of Farnesyl Protein Transferase Complexed with a CaaX Peptide and Farnesyl Diphosphate Analogue. Biochemistry 1998, 37, 16601–16611. [Google Scholar] [CrossRef] [PubMed]
- Kingsley, L.J.; Lill, M.A. Substrate Tunnels in Enzymes: Structure-Function Relationships and Computational Methodology. Proteins 2015, 83, 599–611. [Google Scholar] [CrossRef] [PubMed]
- Stank, A.; Kokh, D.B.; Fuller, J.C.; Wade, R.C. Protein Binding Pocket Dynamics. Acc. Chem. Res. 2016, 49, 809–815. [Google Scholar] [CrossRef] [PubMed]
- Gora, A.; Brezovsky, J.; Damborsky, J. Gates of Enzymes. Chem. Rev. 2013, 113, 5871–5923. [Google Scholar] [CrossRef] [PubMed]
- Weeks, A.; Lund, L.; Raushel, F.M. Tunneling of intermediates in enzyme-catalyzed reactions. Curr. Opin. Chem. Biol. 2006, 10, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Rydzewski, J.; Nowak, W. Ligand diffusion in proteins via enhanced sampling in molecular dynamics. Phys. Life Rev. 2017. [Google Scholar] [CrossRef] [PubMed]
- Poshyvailo, L.; von Lieres, E.; Kondrat, S. Does metabolite channeling accelerate enzyme-catalyzed cascade reactions? PLoS ONE 2017, 12, e0172673. [Google Scholar] [CrossRef] [PubMed]
- Drummond, M.L.; Wilson, A.K.; Cundari, T.R. Nature of Protein–CO2 Interactions as Elucidated via Molecular Dynamics. J. Phys. Chem. B 2012, 116, 11578–11593. [Google Scholar] [CrossRef] [PubMed]
- Case, C.L.; Concar, E.M.; Boswell, K.L.; Mukhopadhyay, B. Roles of Asp75, Asp78, and Glu83 of GTP-dependent phosphoenolpyruvate carboxykinase from Mycobacterium smegmatis. J. Biol. Chem. 2006, 281, 39262–39272. [Google Scholar] [CrossRef] [PubMed]
- Sepulveda, C.; Poch, A.; Espinoza, R.; Cardemil, E. Electrostatic interactions play a significant role in the affinity of Saccharomyces cerevisiae phosphoenolpyruvate carboxykinase for Mn2+. Biochimie 2010, 92, 814–819. [Google Scholar] [CrossRef] [PubMed]
- Yevenes, A.; Espinoza, R.; Rivas-Pardo, J.A.; Villarreal, J.M.; Gonzalez-Nilo, F.D.; Cardemil, E. Site-directed mutagenesis study of the microenvironment characteristics of Lys213 of Saccharomyces cerevisiae phosphoenolpyruvate carboxykinase. Biochimie 2006, 88, 663–672. [Google Scholar] [CrossRef] [PubMed]
- Yevenes, A.; Gonzalez-Nilo, F.D.; Cardemil, E. Relevance of phenylalanine 216 in the affinity of Saccharomyces cerevisiae phosphoenolpyruvate carboxykinase for Mn(II). Protein J. 2007, 26, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Laurie, A.T.R.; Jackson, R.M. Q-SiteFinder: An energy-based method for the prediction of protein–ligand binding sites. Bioinformatics 2005, 21, 1908–1916. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.-h.; Bruschi, M.; De Gioia, L.; Blumberger, J. Uncovering a Dynamically Formed Substrate Access Tunnel in Carbon Monoxide Dehydrogenase/Acetyl-CoA Synthase. J. Am. Chem. Soc. 2013, 135, 9493–9502. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Lund, L.; Shao, Q.; Gao, Y.-Q.; Raushel, F.M. A Combined Theoretical and Experimental Study of the Ammonia Tunnel in Carbamoyl Phosphate Synthetase. J. Am. Chem. Soc. 2009, 131, 10211–10219. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Lund, L.; Yang, L.; Raushel, F.M.; Gao, Y.-Q. Mechanism for the Transport of Ammonia within Carbamoyl Phosphate Synthetase Determined by Molecular Dynamics Simulations. Biochemistry 2008, 47, 2935–2944. [Google Scholar] [CrossRef] [PubMed]
- Lund, L.; Fan, Y.; Shao, Q.; Gao, Y.Q.; Raushel, F.M. Carbamate Transport in Carbamoyl Phosphate Synthetase: A Theoretical and Experimental Investigation. J. Am. Chem. Soc. 2010, 132, 3870–3878. [Google Scholar] [CrossRef] [PubMed]
- Miles, E.W.; Rhee, S.; Davies, D.R. The Molecular Basis of Substrate Channeling. J. Biol. Chem. 1999, 274, 12193–12196. [Google Scholar] [CrossRef] [PubMed]
- Nadeau, O.W.; Falick, A.M.; Woodworth, R.C. Structural evidence for an anion-directing track in the hen ovotransferrin N-lobe: Implications for transferrin synergistic anion binding. Biochemistry 1996, 35, 14294–14303. [Google Scholar] [CrossRef] [PubMed]
- Moxley, M.A.; Sanyal, N.; Krishnan, N.; Tanner, J.J.; Becker, D.F. Evidence for hysteretic substrate channeling in the proline dehydrogenase and ∆1-pyrroline-5-carboxylate dehydrogenase coupled reaction of proline utilization A (PutA). J. Biol. Chem. 2014, 289, 3639–3651. [Google Scholar] [CrossRef] [PubMed]
- Smith, N.E.; Vrielink, A.; Attwood, P.V.; Corry, B. Binding and Channeling of Alternative Substrates in the Enzyme DmpFG: A Molecular Dynamics Study. Biophys. J. 2014, 106, 1681–1690. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.I.; Lan, D.; Durrani, R.; Huan, W.; Zhao, Z.; Wang, Y. The Lid Domain in Lipases: Structural and Functional Determinant of Enzymatic Properties. Front. Bioeng. Biotechnol. 2017, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Anobom, C.D.; Pinheiro, A.S.; De-Andrade, R.A.; Aguieiras, E.C.; Andrade, G.C.; Moura, M.V.; Almeida, R.V.; Freire, D.M. From structure to catalysis: Recent developments in the biotechnological applications of lipases. Biomed. Res. Int. 2014, 2014, 684506. [Google Scholar] [CrossRef] [PubMed]
- Rehm, S.; Trodler, P.; Pleiss, J. Solvent-induced lid opening in lipases: A molecular dynamics study. Protein Sci. 2010, 19, 2122–2130. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.Z.A.; Salleh, A.B.; Rahman, R.N.Z.R.A.; Rahman, M.B.A.; Basri, M.; Leow, T.C. Unlocking the mystery behind the activation phenomenon of T1 lipase: A molecular dynamics simulations approach. Protein Sci. 2012, 21, 1210–1221. [Google Scholar] [CrossRef] [PubMed]
- Zisis, T.; Freddolino, P.L.; Turunen, P.; van Teeseling, M.C.F.; Rowan, A.E.; Blank, K.G. Interfacial activation of Candida antarctica lipase B: Combined evidence from experiment and simulation. Biochemistry 2015, 54, 5969–5979. [Google Scholar] [CrossRef] [PubMed]
- Stauch, B.; Fisher, S.J.; Cianci, M. Open and closed states of Candida antarctica lipase B: Protonation and the mechanism of interfacial activation. J. Lipid Res. 2015, 56, 2348–2358. [Google Scholar] [CrossRef] [PubMed]
- Shirts, M.R.; Chodera, J.D. Statistically optimal analysis of samples from multiple equilibrium states. J. Chem. Phys. 2008, 129, 124105. [Google Scholar] [CrossRef] [PubMed]
- Tyukhtenko, S.; Karageorgos, I.; Rajarshi, G.; Zvonok, N.; Pavlopoulos, S.; Janero, D.R.; Makriyannis, A. Specific Inter-residue Interactions as Determinants of Human Monoacylglycerol Lipase Catalytic Competency: A Role for Global Conformational Changes. J. Biol. Chem. 2016, 291, 2556–2565. [Google Scholar] [CrossRef] [PubMed]
- Roccatano, D.; Wong, T.S.; Schwaneberg, U.; Zacharias, M. Structural and dynamic properties of cytochrome P450 BM-3 in pure water and in a dimethylsulfoxide/water mixture. Biopolymers 2005, 78, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Roccatano, D.; Wong, T.S.; Schwaneberg, U.; Zacharias, M. Toward understanding the inactivation mechanism of monooxygenase P450 BM-3 by organic cosolvents: A molecular dynamics simulation study. Biopolymers 2006, 83, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Kuper, J.; Wong, T.S.; Roccatano, D.; Wilmanns, M.; Schwaneberg, U. Understanding a Mechanism of Organic Cosolvent Inactivation in Heme Monooxygenase P450 BM-3. J. Am. Chem. Soc. 2007, 129, 5786–5787. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Taraphder, S. Role of protein motions on proton transfer pathways in human carbonic anhydrase II. Biochim. Biophys. Acta Proteins Proteom. 2010, 1804, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Domsic, J.F.; Williams, W.; Fisher, S.Z.; Tu, C.; Agbandje-McKenna, M.; Silverman, D.N.; McKenna, R. Structural and Kinetic Study of the Extended Active Site for Proton Transfer in Human Carbonic Anhydrase II. Biochemistry 2010, 49, 6394–6399. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Taraphder, S. Determination of the Reaction Coordinate for a Key Conformational Fluctuation in Human Carbonic Anhydrase II. J. Phys. Chem. B 2015, 119, 11403–11415. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Taraphder, S. Transition Path Sampling Study of the Conformational Fluctuation of His-64 in Human Carbonic Anhydrase II. J. Phys. Chem. B 2009, 113, 12555–12564. [Google Scholar] [CrossRef] [PubMed]
- Maupin, C.M.; McKenna, R.; Silverman, D.N.; Voth, G.A. Elucidation of the Proton Transport Mechanism in Human Carbonic Anhydrase II. J. Am. Chem. Soc. 2009, 131, 7598–7608. [Google Scholar] [CrossRef] [PubMed]
- Taraphder, S.; Maupin, C.M.; Swanson, J.M.; Voth, G.A. Coupling Protein Dynamics with Proton Transport in Human Carbonic Anhydrase II. J. Phys. Chem. B 2016, 120, 8389–8404. [Google Scholar] [CrossRef] [PubMed]
- Balabin, I.A.; Hu, X.; Beratan, D.N. Exploring biological electron transfer pathway dynamics with the Pathways Plugin for VMD. J. Comp. Chem. 2012, 33, 906–910. [Google Scholar] [CrossRef] [PubMed]
- Beratan, D.N.; Betts, J.N.; Onuchic, J.N. Protein electron transfer rates set by the bridging secondary and tertiary structure. Science 1991, 252, 1285–1288. [Google Scholar] [CrossRef] [PubMed]
- Nazor, J.; Dannenmann, S.; Adjei, R.O.; Fordjour, Y.B.; Ghampson, I.T.; Blanusa, M.; Roccatano, D.; Schwaneberg, U. Laboratory evolution of P450 BM3 for mediated electron transfer yielding an activity-improved and reductase-independent variant. Protein Eng. Des. Sel. 2008, 21, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Verma, R.; Schwaneberg, U.; Roccatano, D. Insight into the redox partner interaction mechanism in cytochrome P450BM-3 using molecular dynamics simulations. Biopolymers 2014, 101, 197–209. [Google Scholar] [CrossRef] [PubMed]
- Sevrioukova, I.; Shaffer, C.; Ballou, D.P.; Peterson, J.A. Equilibrium and Transient State Spectrophotometric Studies of the Mechanism of Reduction of the Flavoprotein Domain of P450BM-3. Biochemistry 1996, 35, 7058–7068. [Google Scholar] [CrossRef] [PubMed]
- Beratan, D.N.; Skourtis, S.S.; Balabin, I.A.; Balaeff, A.; Keinan, S.; Venkatramani, R.; Xiao, D. Steering Electrons on Moving Pathways. Acc. Chem. Res. 2009, 42, 1669–1678. [Google Scholar] [CrossRef] [PubMed]
- Narth, C.; Gillet, N.; Cailliez, F.; Lévy, B.; de la Lande, A. Electron Transfer, Decoherence, and Protein Dynamics: Insights from Atomistic Simulations. Acc. Chem. Res. 2015, 48, 1090–1097. [Google Scholar] [CrossRef] [PubMed]
- Verma, R.; Schwaneberg, U.; Roccatano, D. Conformational Dynamics of the FMN-Binding Reductase Domain of Monooxygenase P450BM-3. J. Chem. Theory Comput. 2013, 9, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Cummins, P.L.; Ramnarayan, K.; Singh, U.C.; Gready, J.E. Molecular dynamics/free energy perturbation study on the relative affinities of the binding of reduced and oxidized NADP to dihydrofolate reductase. J. Am. Chem. Soc. 1991, 113, 8247–8256. [Google Scholar] [CrossRef]
- Schmidtke, P.; Bidon-Chanal, A.; Luque, F.J.; Barril, X. MDpocket: Open-source cavity detection and characterization on molecular dynamics trajectories. Bioinformatics 2011, 27, 3276–3285. [Google Scholar] [CrossRef] [PubMed]
- Sibille, N.; Blackledge, M.; Brutscher, B.; Coves, J.; Bersch, B. Solution structure of the sulfite reductase flavodoxin-like domain from Escherichia coli. Biochemistry 2005, 44, 9086–9095. [Google Scholar] [CrossRef] [PubMed]
- Longbotham, J.E.; Hardman, S.J.O.; Görlich, S.; Scrutton, N.S.; Hay, S. Untangling Heavy Protein and Cofactor Isotope Effects on Enzyme-Catalyzed Hydride Transfer. J. Am. Chem. Soc. 2016, 138, 13693–13699. [Google Scholar] [CrossRef] [PubMed]
- Blikstad, C.; Dahlstrom, K.M.; Salminen, T.A.; Widersten, M. Substrate scope and selectivity in offspring to an enzyme subjected to directed evolution. FEBS J. 2014, 281, 2387–2398. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Bruice, T.C. Dynamic Structures of Horse Liver Alcohol Dehydrogenase (HLADH): Results of Molecular Dynamics Simulations of HLADH-NAD+-PhCH2OH, HLADH-NAD+-PhCH2O-, and HLADH-NADH-PhCHO. J. Am. Chem. Soc. 2001, 123, 11952–11959. [Google Scholar] [CrossRef] [PubMed]
- Oyen, D.; Fenwick, R.B.; Stanfield, R.L.; Dyson, H.J.; Wright, P.E. Cofactor-Mediated Conformational Dynamics Promote Product Release From Escherichia coli Dihydrofolate Reductase via an Allosteric Pathway. J. Am. Chem. Soc. 2015, 137, 9459–9468. [Google Scholar] [CrossRef] [PubMed]
- Boehr, D.D.; McElheny, D.; Dyson, H.J.; Wright, P.E. The Dynamic Energy Landscape of Dihydrofolate Reductase Catalysis. Science 2006, 313, 1638. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Li, H. Dynamics of Protein Folding and Cofactor Binding Monitored by Single-Molecule Force Spectroscopy. Biophys. J. 2011, 101, 2009–2017. [Google Scholar] [CrossRef] [PubMed]
- Stigler, J.; Rief, M. Calcium-dependent folding of single calmodulin molecules. Proc. Natl. Acad. Sci USA 2012, 109, 17814–17819. [Google Scholar] [CrossRef] [PubMed]
- Shukla, D.; Peck, A.; Pande, V.S. Conformational heterogeneity of the calmodulin binding interface. Nat. Commun. 2016, 7, 10910. [Google Scholar] [CrossRef] [PubMed]
- Catherine, L.H.; Muralidhara, B.K.; Pernilla, W.-S. How Do Cofactors Modulate Protein Folding? Protein Pept. Lett. 2005, 12, 165–170. [Google Scholar]
- Wilson, C.J.; Apiyo, D.; Wittung-Stafshede, P. Role of cofactors in metalloprotein folding. Q. Rev. Biophys. 2005, 37, 285–314. [Google Scholar] [CrossRef] [PubMed]
- Chodera, J.D.; Noé, F. Markov state models of biomolecular conformational dynamics. Curr. Opin. Struct. Biol. 2014, 25, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Negri, A.; Rodriguez-Larrea, D.; Marco, E.; Jimenez-Ruiz, A.; Sanchez-Ruiz, J.M.; Gago, F. Protein-protein interactions at an enzyme-substrate interface: Characterization of transient reaction intermediates throughout a full catalytic cycle of Escherichia coli thioredoxin reductase. Proteins 2010, 78, 36–51. [Google Scholar] [CrossRef] [PubMed]
- Goodey, N.M.; Benkovic, S.J. Allosteric regulation and catalysis emerge via a common route. Nat. Chem. Biol. 2008, 4, 474–482. [Google Scholar] [CrossRef] [PubMed]
- Lisi, G.P.; Loria, J.P. Solution NMR Spectroscopy for the Study of Enzyme Allostery. Chem. Rev. 2016, 116, 6323–6369. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Zhou, H.-X. Protein Allostery and Conformational Dynamics. Chem. Rev. 2016, 116, 6503–6515. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, W.; Takada, S. Energy landscape views for interplays among folding, binding, and allostery of calmodulin domains. Proc. Natl. Acad. Sci. USA 2014, 111, 10550–10555. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.E.; Weber, D.K.; Downton, M.T.; Wagner, J.; Perugini, M.A. Dynamic Modelling Reveals ‘Hotspots’ on the Pathway to Enzyme-Substrate Complex Formation. PLoS Comput. Biol. 2016, 12, e1004811. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Pang, X.; Zhou, H.-X. Two Pathways Mediate Interdomain Allosteric Regulation in Pin1. Structure 2015, 23, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Doshi, U.; Holliday, M.J.; Eisenmesser, E.Z.; Hamelberg, D. Dynamical network of residue-residue contacts reveals coupled allosteric effects in recognition, catalysis, and mutation. Proc. Natl. Acad. Sci. USA 2016, 113, 4735–4740. [Google Scholar] [CrossRef] [PubMed]
- Maciag, J.J.; Mackenzie, S.H.; Tucker, M.B.; Schipper, J.L.; Swartz, P.; Clark, A.C. Tunable allosteric library of caspase-3 identifies coupling between conserved water molecules and conformational selection. Proc. Natl. Acad. Sci. USA 2016, 113, E6080–E6088. [Google Scholar] [CrossRef] [PubMed]
- Bahar, I.; Lezon, T.R.; Bakan, A.; Shrivastava, I.H. Normal Mode Analysis of Biomolecular Structures: Functional Mechanisms of Membrane Proteins. Chem. Rev. 2010, 110, 1463–1497. [Google Scholar] [CrossRef] [PubMed]
- David, C.C.; Jacobs, D.J. Principal Component Analysis: A Method for Determining the Essential Dynamics of Proteins. Methods Mol. Biol. 2014, 1084, 193–226. [Google Scholar] [PubMed]
- Shukla, D.; Hernández, C.X.; Weber, J.K.; Pande, V.S. Markov State Models Provide Insights into Dynamic Modulation of Protein Function. Acc. Chem. Res. 2015, 48, 414–422. [Google Scholar] [CrossRef] [PubMed]
- E, W.; Vanden-Eijnden, E. Transition-Path Theory and Path-Finding Algorithms for the Study of Rare Events. Annu. Rev. Phys. Chem. 2010, 61, 391–420. [Google Scholar] [CrossRef] [PubMed]
- Noé, F.; Fischer, S. Transition networks for modeling the kinetics of conformational change in macromolecules. Curr. Opin. Struct. Biol. 2008, 18, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, R.C.; Melo, M.C.R.; Schulten, K. Enhanced Sampling Techniques in Molecular Dynamics Simulations of Biological Systems. Biochim. Biophys. Acta 2015, 1850, 872–877. [Google Scholar] [CrossRef] [PubMed]
- Schlick, T. Molecular dynamics-based approaches for enhanced sampling of long-time, large-scale conformational changes in biomolecules. F1000 Biol. Rep. 2009, 1, 51. [Google Scholar] [CrossRef] [PubMed]
- Brandsdal, B.O.; Österberg, F.; Almlöf, M.; Feierberg, I.; Luzhkov, V.B.; Åqvist, J. Free Energy Calculations and Ligand Binding. Adv. Protein Chem. 2003, 66, 123–158. [Google Scholar] [PubMed]
- Aqvist, J.; Medina, C.; Samuelsson, J.E. A new method for predicting binding affinity in computer-aided drug design. Protein Eng. 1994, 7, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Johan, A.; John, M. The Linear Interaction Energy Method for Predicting Ligand Binding Free Energies. Comb. Chem. High Throughput Screen. 2001, 4, 613–626. [Google Scholar]
- Gutiérrez-de-Terán, H.; Åqvist, J. Linear Interaction Energy: Method and Applications in Drug Design. In Computational Drug Discovery and Design; Baron, R., Ed.; Springer: New York, NY, USA, 2012; pp. 305–323. [Google Scholar]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.; Wang, J.; Li, Y.; Wang, W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Model. 2011, 51, 69–82. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verma, R.; Mitchell-Koch, K. In Silico Studies of Small Molecule Interactions with Enzymes Reveal Aspects of Catalytic Function. Catalysts 2017, 7, 212. https://doi.org/10.3390/catal7070212
Verma R, Mitchell-Koch K. In Silico Studies of Small Molecule Interactions with Enzymes Reveal Aspects of Catalytic Function. Catalysts. 2017; 7(7):212. https://doi.org/10.3390/catal7070212
Chicago/Turabian StyleVerma, Rajni, and Katie Mitchell-Koch. 2017. "In Silico Studies of Small Molecule Interactions with Enzymes Reveal Aspects of Catalytic Function" Catalysts 7, no. 7: 212. https://doi.org/10.3390/catal7070212




