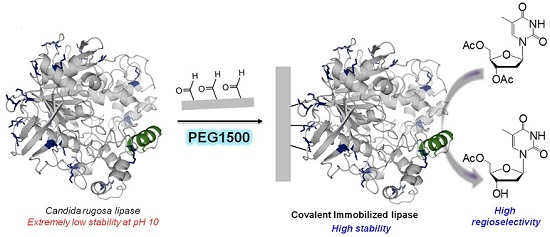
Covalent Immobilization of Candida rugosa Lipase at Alkaline pH and Their Application in the Regioselective Deprotection of Per-O-acetylated Thymidine
Abstract
:1. Introduction
2. Results and Discussions
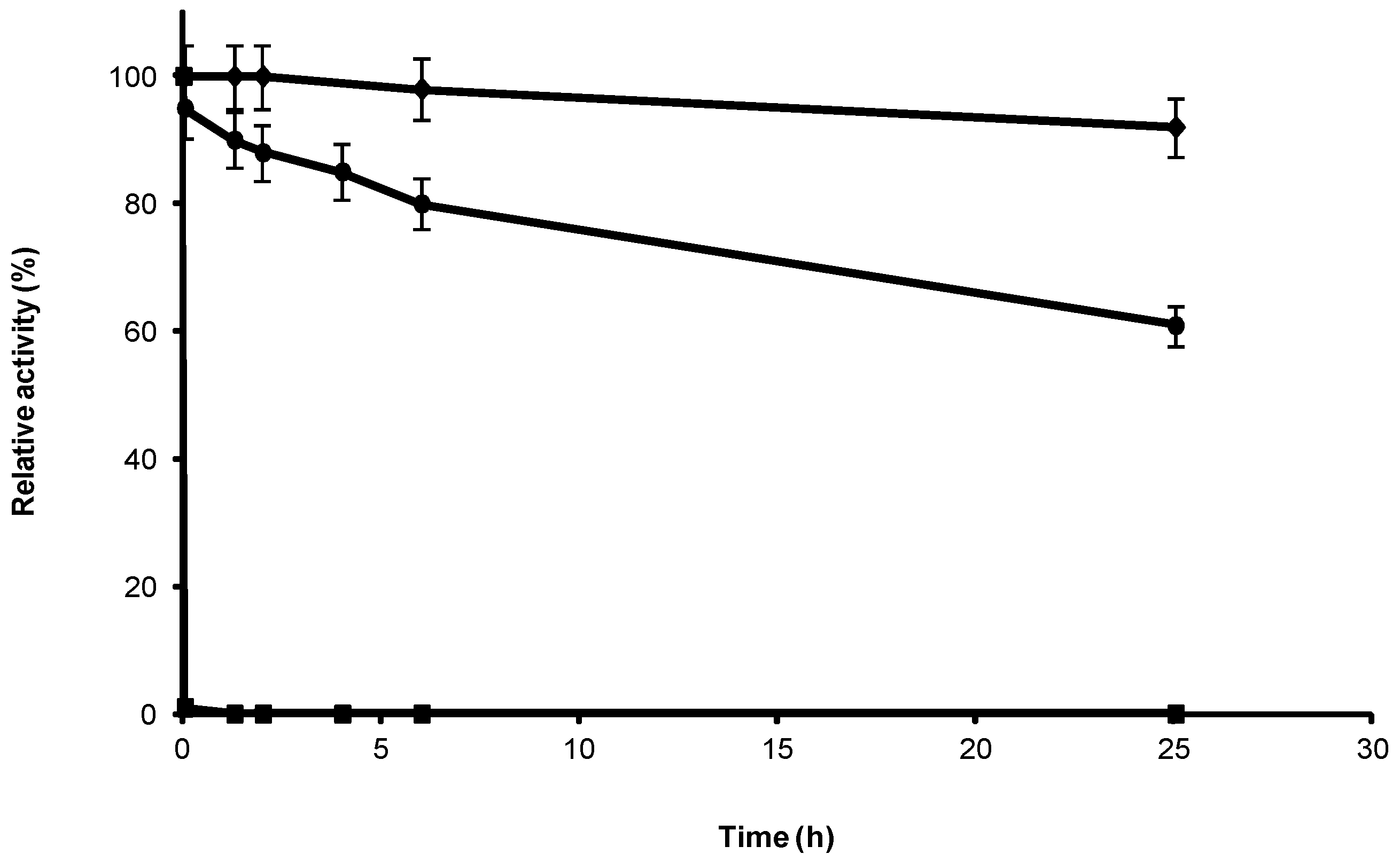
2.1. Stabilization of CRL at Alkaline pH
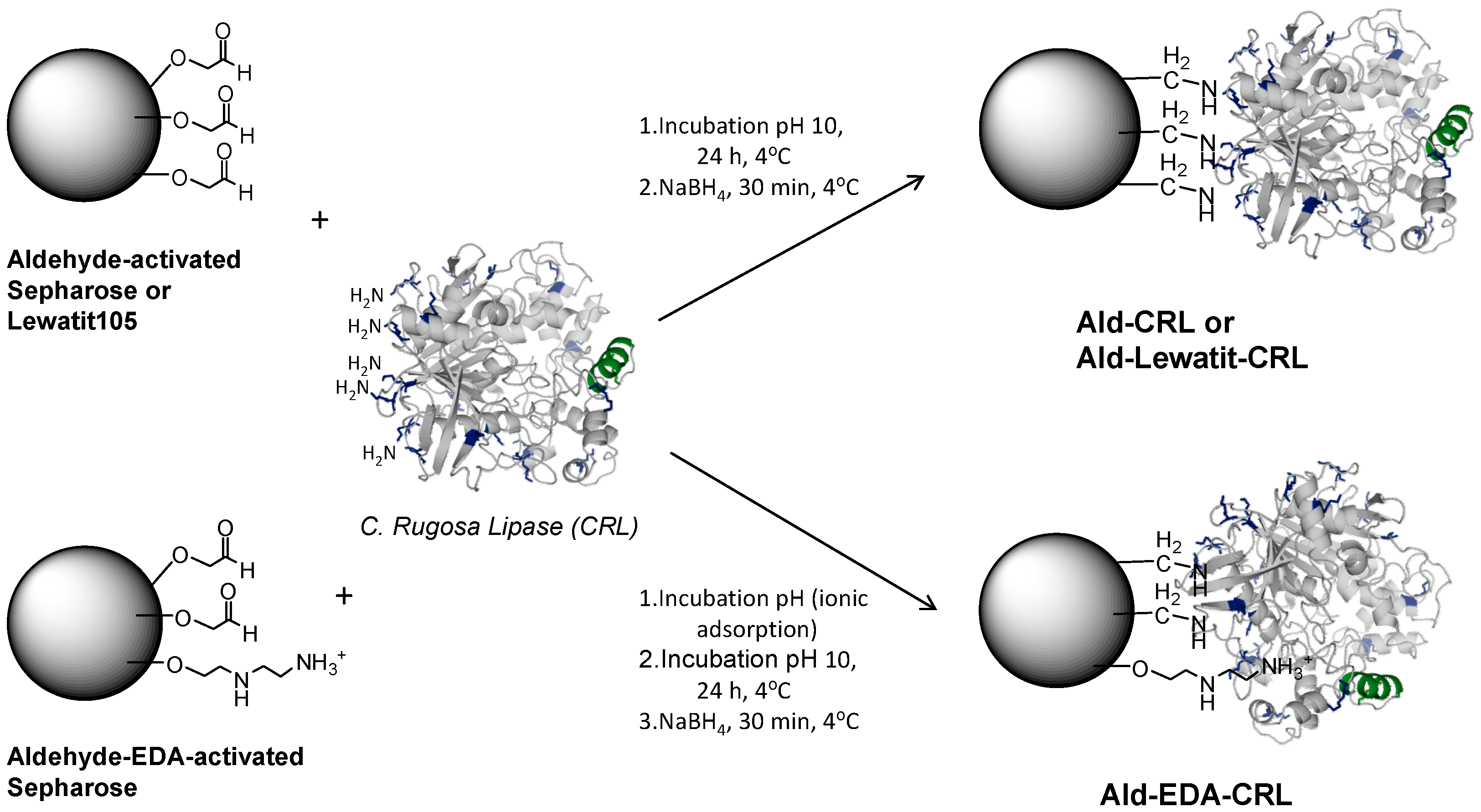
2.2. Multipoint Covalent Immobilization of CRL
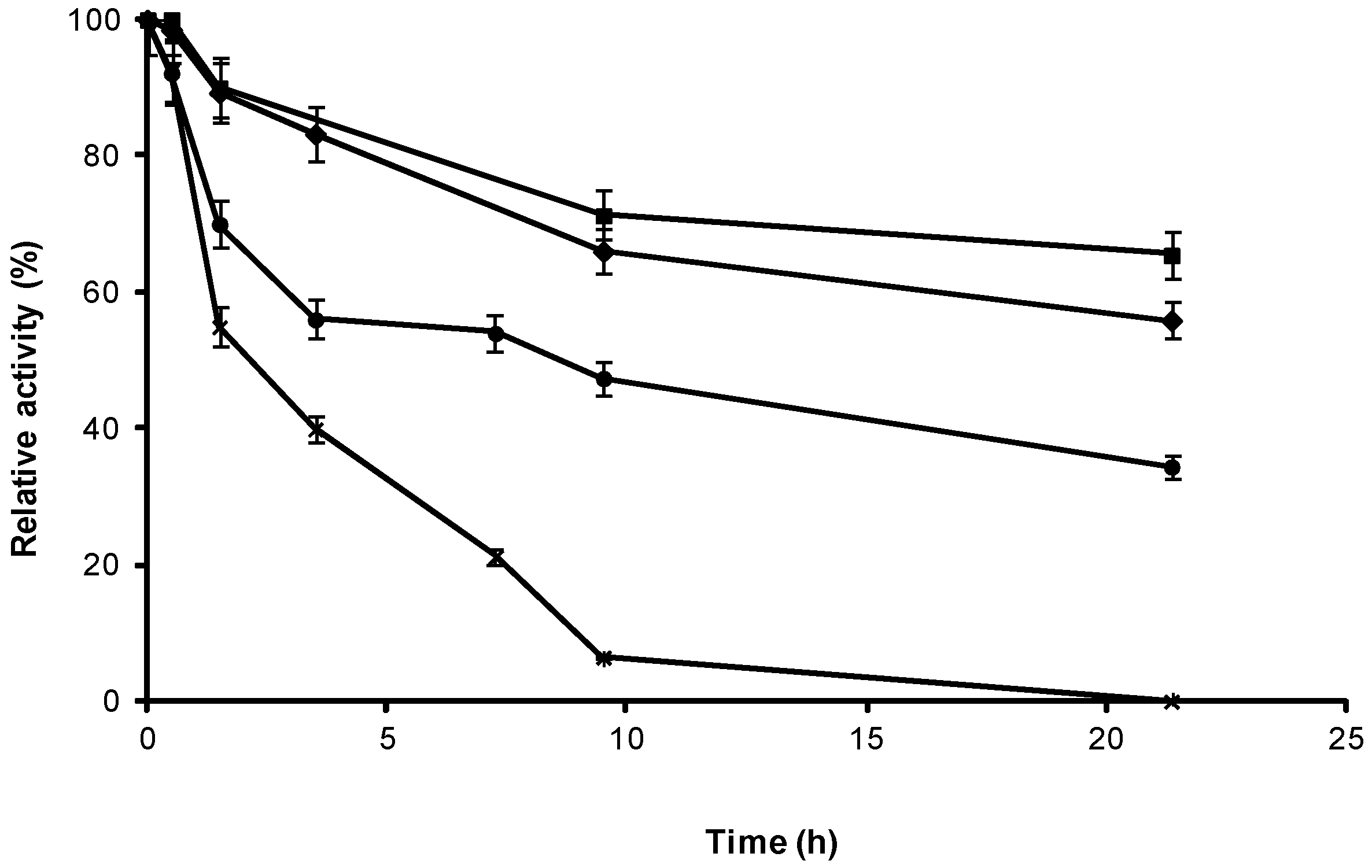
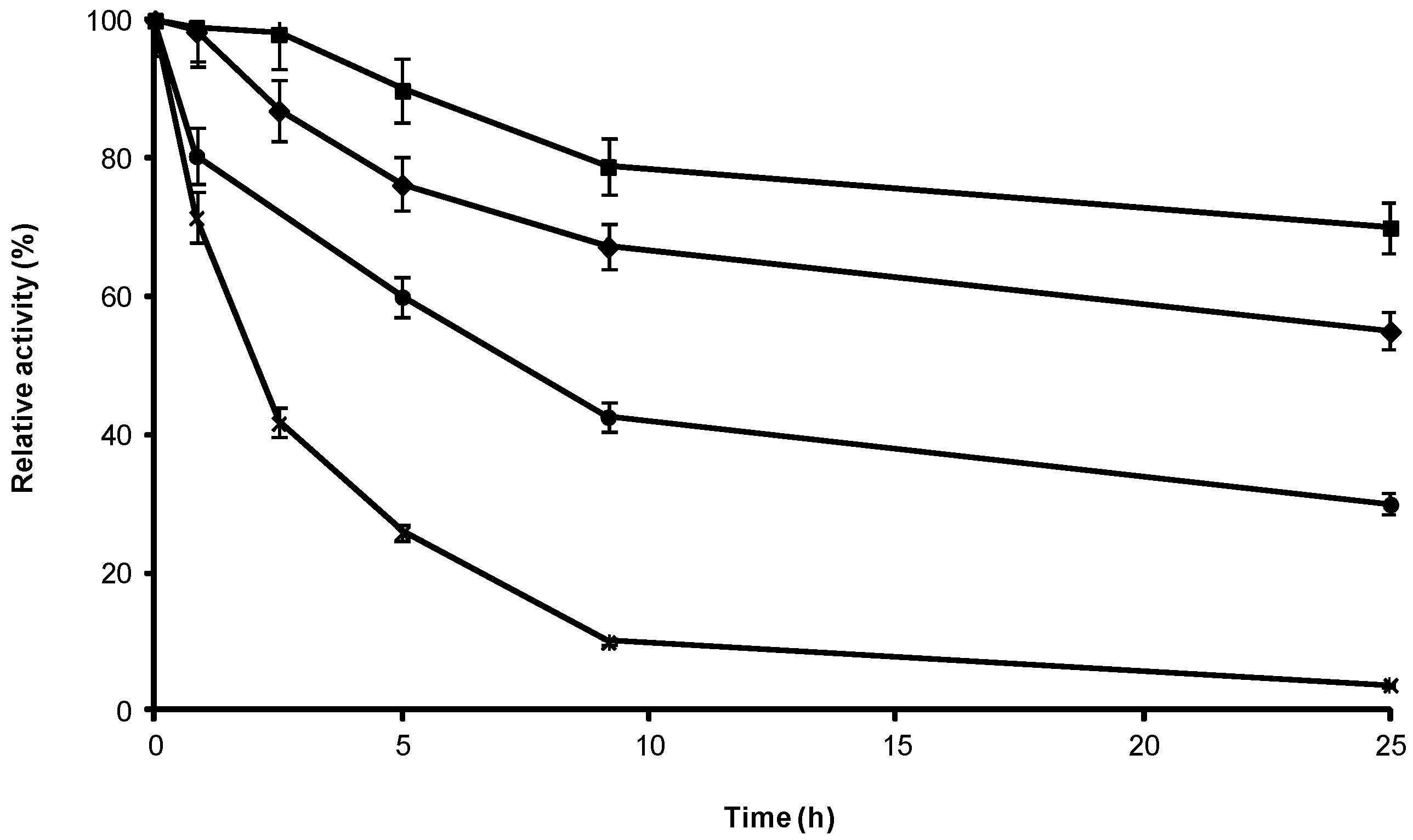
2.3. Stability of Different Covalent Immobilized Preparations of CRL
2.4. Regioselective Deprotection of Per-O-acetylated Thymidine by Immobilized CRL Biocatalysts
3. Experimental Section
3.1. Materials
3.2. Lipase Activity Assay
3.3. C. rugosa Lipase Purification
3.4. Preparation of EDA-Aldehyde–Activated Sepharose Support (Ald-EDA)
3.5. Multipoint Covalent Immobilization of CRL on Different Aldehyde-Activated Sepharose Supports (Ald)
3.5.1. Immobilization on Aldehyde-Activated Sepharose (Ald) or Aldehyde-Activated Lewatit-105 (Ald-Lew105) at Alkaline pH
3.5.2. Immobilization on Aldehyde-Activated EDA-Sepharose (Ald-EDA) at pH 8 and Incubation at pH 10
3.6. Inactivation of CRL Immobilized Preparations against T and Co-Solvent
3.7. Enzymatic Hydrolysis of 3′,5′-Di-O-Acetylthymidine (1)
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Reetz, M.T. Biocatalysis in Organic Chemistry and Biotechnology: Past, Present, and Future. J. Am. Chem. Soc. 2013, 135, 12480–12496. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Wu, C.-Y.; Branford-White, C.J.; Ning, X.; Nie, H.-L.; Zhu, L.-M. Chemical modification of stem bromelain with anhydride groups to enhance its stability and catalytic activity. J. Mol. Catal. B Enzym. 2010, 63, 188–193. [Google Scholar] [CrossRef]
- Srirangsan, P.; Kawai, K.; Hamada-Sato, N.; Watanabe, M.; Suzuki, T. Stabilizing effects of sucrose-polymer formulations on the activities of freeze-dried enzyme mixtures of alkaline phosphatase, nucleoside phosphorylase and xanthine oxidase. Food Chem. 2011, 125, 1188–1193. [Google Scholar] [CrossRef]
- Santagapita, P.R.; Brizuela, L.G.; Mazzobre, M.F.; Ramírez, H.L.; Corti, H.R.; Santana, R.V.; Buera, M.P. β-cyclodextrin modifications as related to enzyme stability in dehydrated systems: Supramolecular transitions and molecular interactions. Carbohydr. Polym. 2011, 83, 203–209. [Google Scholar] [CrossRef]
- Nascimento, C.; Leandro, J.; Lino, P.R.; Ramos, L.; Almeida, A.J.; De Almeida, I.T.; Leandro, P. Polyol additives modulate the in vitro stability and activity of recombinant human phenylalanine hydroxylase. Appl. Biochem. Biotechnol. 2010, 162, 192–207. [Google Scholar] [CrossRef] [PubMed]
- Iyer, P.V.; Ananthanarayan, L. Enzyme stability and stabilization-Aqueous and non-aqueous environment. Process Biochem. 2008, 43, 1019–1032. [Google Scholar] [CrossRef]
- Guisán, J.M. Immobilization of Enzymes and Cells, Methods in Molecular Biology, 3rd ed.; Humana Press: New York, NY, USA, 2013; pp. 1–377. [Google Scholar]
- Mateo, C.; Grazu, V.; Palomo, J.M.; Lopez-Gallego, F.; Fernandez-Lafuente, R.; Guisán, J.M. Immobilization of Enzymes on Heterofunctional Epoxy Supports. Nat. Protoc. 2007, 2, 1022–1033. [Google Scholar] [CrossRef] [PubMed]
- Katchalski-Katzir, E. Immobilized enzymes—Learning from past successes and failures. Trends Biotechnol. 1993, 11, 471–478. [Google Scholar] [CrossRef]
- Cao, L. Immobilised enzymes: Science or art? Curr. Opin. Chem. Biol. 2005, 9, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Mateo, C.; Palomo, J.M.; Fernandez-Lorente, G.; Guisan, J.M.; Fernandez-Lafuente, R. Improvement of enzyme activity, stability and selectivity via immobilization techniques. Enzyme Microb. Technol. 2007, 40, 1451–1463. [Google Scholar] [CrossRef]
- Pedroche, J.; Yust, M.M.; Mateo, C.; Fernández-Lafuente, R.; Girón-Calle, J.; Alaiz, M.; Vioque, J.; Guisan, J.M.; Millan, F. Effect of the support and experimental conditions in the intensity of the multipoint covalent attachment of proteins on aldehyde activated-Sepharose supports: Correlation between enzyme–support linkages and thermal stability. Enzyme Microb. Technol. 2007, 40, 1160–1167. [Google Scholar] [CrossRef]
- Bolivar, J.M.; Rocha-Martin, J.; Mateo, C.; Cava, F.; Berenguer, J.; Vega, D.; Fernandez-Lafuente, R.; Guisan, J.M. Purification and stabilization of a glutamate dehygrogenase from Thermus thermophilus via oriented multisubunit plus multipoint covalent immobilization. J. Mol. Catal. B Enzym. 2009, 58, 158–163. [Google Scholar] [CrossRef]
- Klibanov, A.M. Stabilization of Enzymes against Thermal Inactivation. Adv. Appl. Microbiol. 1983, 29, 1–28. [Google Scholar] [PubMed]
- Mateo, C.; Abian, O.; Bernedo, M.; Cuenca, E.; Fuentes, M.; Fernandez-Lorente, G.; Palomo, J.M.; Grazu, V.; Pessela, B.C.C.; Giacomini, C.; et al. Some special features of aldehyde activated supports to immobilize proteins. Enzyme Microb. Technol. 2005, 37, 456–462. [Google Scholar] [CrossRef]
- Blanco, R.M.; Calvete, J.J.; Guisan, J.M. Immobilization-stabilization of enzymes; variables that control the intensity of the trypsin (amine)-Sepharose (aldehyde) multipoint attachment. Enzyme Microb. Technol. 1989, 11, 353–359. [Google Scholar] [CrossRef]
- Kolodiazhnyi, O.I. Recent developments in the asymmetric synthesis of -chiral phosphorus compounds. Tetrahedron Asymmetry 2012, 23, 1–46. [Google Scholar]
- Filice, M.; Guisan, J.M.; Terreni, M.; Palomo, J.M. Regioselective monodeprotection of peracetylated carbohydrates. Nat. Protoc. 2012, 7, 1783–1796. [Google Scholar] [CrossRef] [PubMed]
- Domínguez de María, P.; Alcántara, A.R.; Carballeira, J.D.; de la Casa, R.M.; García-Burgos, C.A.; Hernáiz, M.J.; Sánchez-Montero, J.M.; Sinisterra, J.V. Candida rugosa Lipase: A traditional and complex biocatalyst. Curr. Org. Chem. 2006, 10, 1053–1066. [Google Scholar] [CrossRef]
- Palomo, J.M.; Fernández-Lorente, G.; Mateo, C.; Ortiz, C.; Fernandez-Lafuente, R.; Guisan, J.M. Modulation of the enantioselectivity of lipases via controlled immobilization and medium engineering Hydrolytic resolution of Mandelic acid esters. Enzyme Microb. Technol. 2002, 31, 775–783. [Google Scholar] [CrossRef]
- Knezevic, Z.; Milosavic, N.; Bezbradica, D.; Jakovljevic, Z.; Prodanovic, R. Immobilization of lipase from Candida rugosa on Eupergit® C supports by covalent attachment. Biochem. Eng. J. 2006, 30, 269–278. [Google Scholar] [CrossRef]
- Noriko, M.; Miok, K.; Tadao, K. Stabilization of l-Ascorbic acid by superoxide dismutase and catalase. Biosci. Biotechnol. Biochem. 1999, 63, 54–57. [Google Scholar]
- Lozano, P.; Combes, D.; Iborra, J.L. Effects of polyols on chymotrypsin thermostability a mechanistic analysis of the enzyme stabilization. J. Biotechnol. 1994, 35, 9–18. [Google Scholar] [CrossRef]
- Castro, G.R. Properties of soluble α-chymotrypsin in neat glycerol and water. Enzyme Microb. Technol. 2000, 27, 143–150. [Google Scholar] [CrossRef]
- Combes, D.; Yoodikhya, T.; Girbal, E.; Willemot, R.M.; Monsan, P. Mechanism of Enzyme Stabilization. Ann. N. Y. Acad. Sci. 1987, 501, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Salahas, G.; Peslis, B.; Georgiou, C.D.; Gavalas, N.A. Trehalose, an extreme temperature protector of phosphoenolpyruvate carboxylase from the C4-plant Cynodon dactylon. Phytochemistry 1997, 46, 1331–1334. [Google Scholar] [CrossRef]
- Breccia, J.D.; Moran, A.C.; Castro, G.R.; Siñeriz, F. Thermal stabilization by polyols of β-xylanase from Bacillus amyloliquefaciens. J. Chem. Technol. Biotechnol. 1998, 71, 241–245. [Google Scholar] [CrossRef]
- Jordheim, L.P.; Durantel, D.; Zoulim, F.; Dumontet, C. Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases. Nat. Rev. Drug Discov. 2013, 12, 447–464. [Google Scholar] [CrossRef] [PubMed]
- Rawat, S.; Raman Suri, C.; Sahoo, D.K. Molecular mechanism of polyethylene glycol mediated stabilization of protein. Biochem. Biophys. Res. Commun. 2010, 392, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Lorente, G.; Lopez-Gallego, F.; Bolivar, J.M.; Rocha-Martin, J.; Moreno-Perez, S.; Guisan, J.M. Immobilization of proteins on highly activated glyoxyl supports: Dramatic increase of the enzyme stability via multipoint immobilization on pre-existing carriers. Curr. Org. Chem. 2015, 19, 1719–1731. [Google Scholar] [CrossRef]
- Palomo, J.M. Lipases Enantioselectivity Alteration by Immobilization Techniques. Curr. Biol. Compet. 2008, 4, 126–138. [Google Scholar] [CrossRef]
- Guajardo, N.; Bernal, C.; Wilson, L.; Cabrera, Z. Selectivity of R-α-monobenzoate glycerol synthesis catalyzed by Candida antarctica lipase B immobilized on heterofunctional supports. Process Biochem. 2015, 50, 1870–1877. [Google Scholar] [CrossRef]
- Mateo, C.; Palomo, J.M.; Fuentes, M.; Betancor, L.; Grazu, V.; Lopez-Gallego, F.; Pessela, B.C.C.; Hidalgo, A.; Fernandez-lorente, G.; Fernandez-lafuente, R.; et al. Glyoxyl agarose: A fully inert and hydrophilic support for immobilization and high stabilization of proteins. Enzyme Microb. Technol. 2006, 39, 274–280. [Google Scholar] [CrossRef]
- Romero, O.; Filice, M.; de Las Rivas, B.; Carrasco-Lopez, C.; Klett, J.; Morreale, A.; Hermoso, J.A.; Guisan, J.M.; Abian, O.; Palomo, J.M. Semisynthetic peptide-lipase conjugates for improved biotransformations. Chem. Commun. 2012, 48, 9053–9055. [Google Scholar] [CrossRef] [PubMed]
- Bastida, A.; Sabuquillo, P.; Armisen, P.; Fernandez-Lafuente, R.; Huguet, J.; Guisan, J.M. A single step purification, immobilization, and hyperactivation of lipases via interfacial adsorption on strongly hydrophobic supports. Biotechnol. Bioeng. 1998, 58, 486–493. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgramquantities of protein utilizing the principle of protein dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]



| Support | Immobilization yield (%) a | Retained Activity (%) |
|---|---|---|
| Ald-Sepharose | 89 | 53 |
| Ald-Lew105 | 69 | 39 |
| Ald-EDA-Sepharose | 95 | 37 |
| Biocatalyst | pH | Initial Rate a | Reaction Time (h) | Yield b (%) | 2 (%) | 3 (%) | Thymidine |
|---|---|---|---|---|---|---|---|
| free CRL | 5.0 | 0.08 | 104 | 99 | 81 | 9 | 10 |
| Ald-CRL | 5.0 | 0.24 | 71 | 100 | 91 | 3 | 6 |
| Ald-EDA-CRL | 5.0 | 0.09 | 144 | 100 | 70 | 10 | 20 |
| Lew105-CRL | 5.0 | 0.06 | 144 | 100 | 17 | 15 | 67 |
| free CRL | 7.0 | 0.08 | 104 | 99 | 90 | 8 | 2 |
| Ald-CRL | 7.0 | 0.26 | 51 | 100 | 90 | 4 | 6 |
| Ald-EDA | 7.0 | 0.09 | 152 | 100 | 88 | 10 | 3 |
| Lew105 | 7.0 | 0.04 | 150 | 62 | 28 | 32 | 2 |
| free CRL | 8.0 | 0.08 | 120 | 100 | 75 | 11 | 13 |
| Ald-CRL | 8.0 | 0.11 | 73 | 100 | 88 | 8 | 4 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rivero, C.W.; Palomo, J.M. Covalent Immobilization of Candida rugosa Lipase at Alkaline pH and Their Application in the Regioselective Deprotection of Per-O-acetylated Thymidine. Catalysts 2016, 6, 115. https://doi.org/10.3390/catal6080115
Rivero CW, Palomo JM. Covalent Immobilization of Candida rugosa Lipase at Alkaline pH and Their Application in the Regioselective Deprotection of Per-O-acetylated Thymidine. Catalysts. 2016; 6(8):115. https://doi.org/10.3390/catal6080115
Chicago/Turabian StyleRivero, Cintia W., and Jose M. Palomo. 2016. "Covalent Immobilization of Candida rugosa Lipase at Alkaline pH and Their Application in the Regioselective Deprotection of Per-O-acetylated Thymidine" Catalysts 6, no. 8: 115. https://doi.org/10.3390/catal6080115