Cloning, Expression and Characterization of a Novel Fructosyltransferase from Aspergillus oryzae ZZ-01 for the Synthesis of Sucrose 6-Acetate
Abstract
:1. Introduction
2. Results and Discussion
2.1. Cloning and Sequence Analysis of the aoft Gene
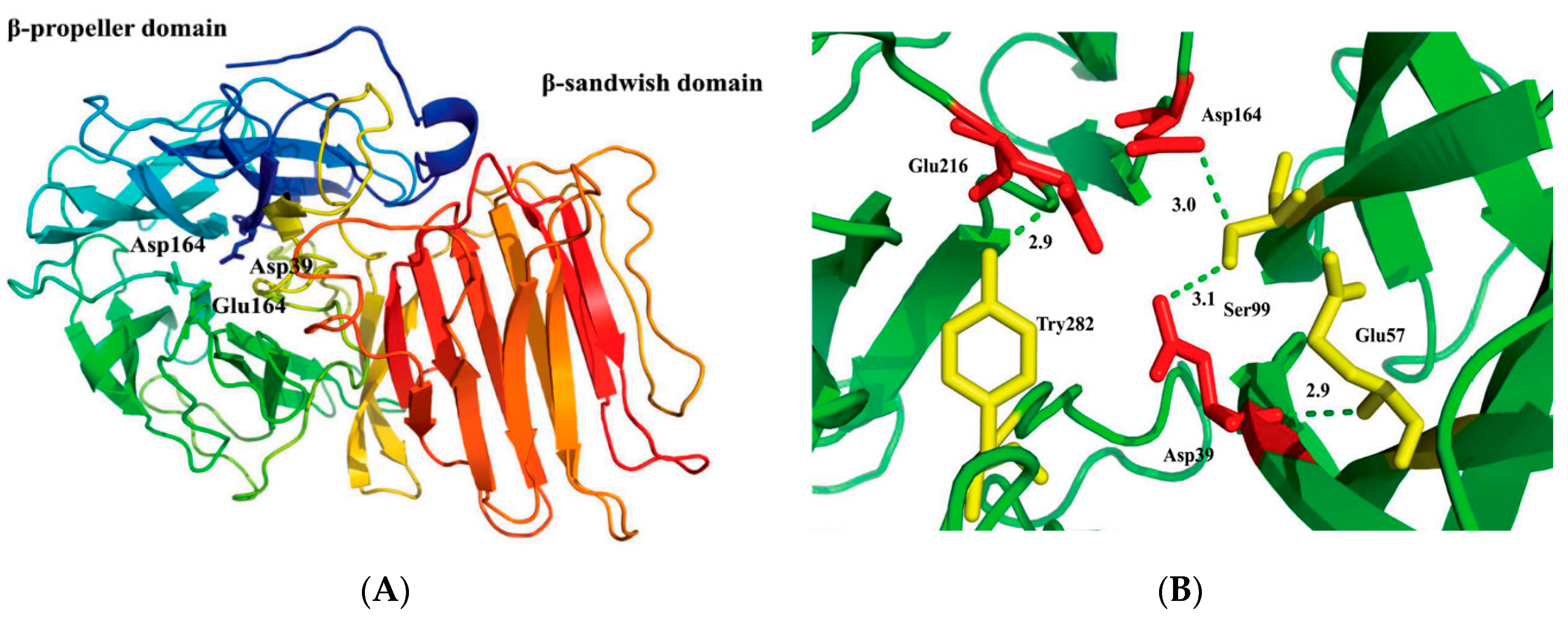
2.2. Structural Model of r-AoFT
2.3. Expression and Purification of r-AoFT
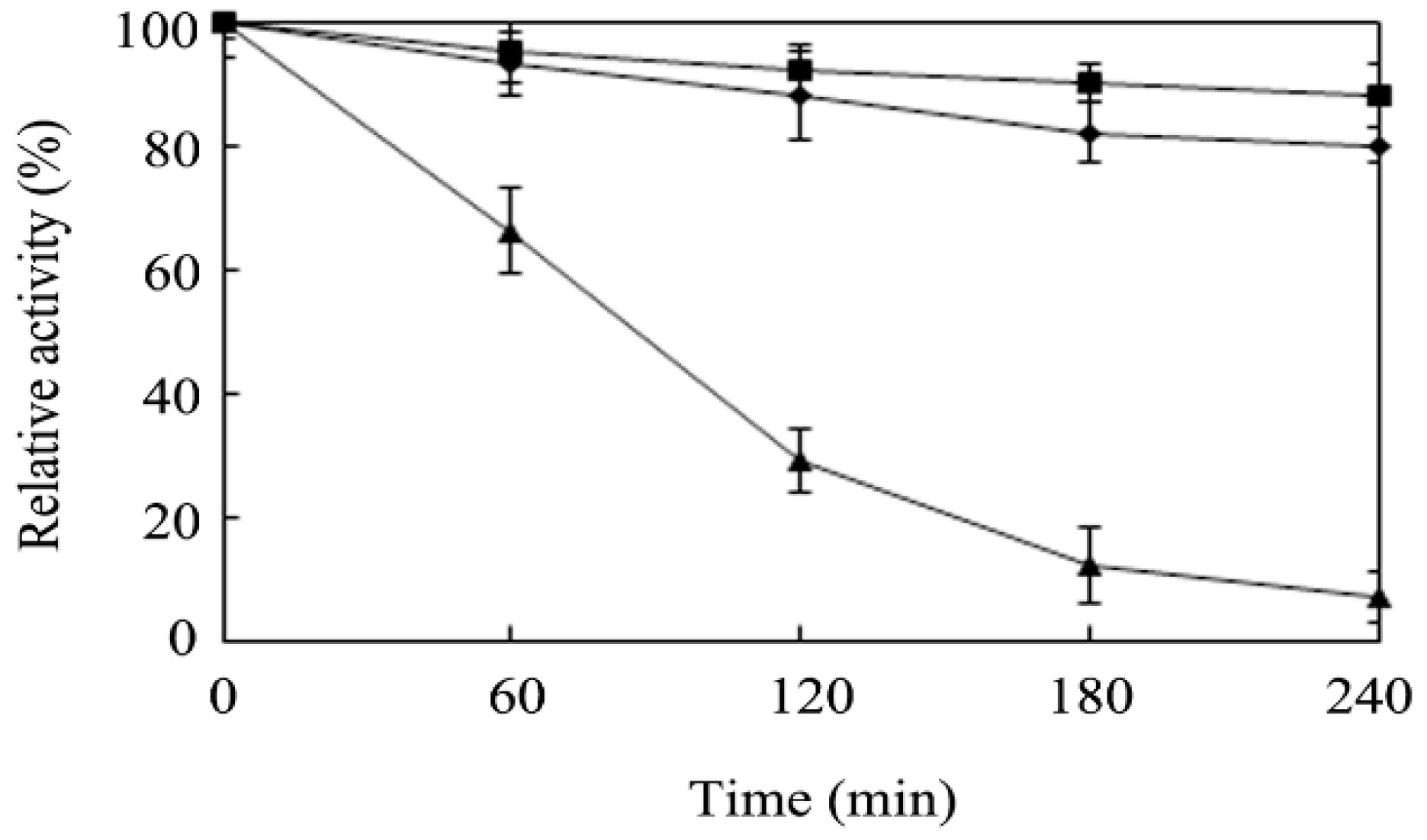
2.4. Effects of Temperature and pH on Enzyme Activity
2.5. Effects of Different Metal Ions, Organic Solvents and Detergents on the Enzymatic Activity of r-AoFT
2.6. Identification of Crucial Amino Acid Residue for r-AoFT Activity
3. Experimental Section
3.1. Chemicals, Strains and Plasmids
3.2. Cloning of the aoft Gene
3.3. Sequence Analysis
3.4. Expression and Purification of Recombinant Fructosyltransferase r-AoFT
3.5. Enzymatic Activity Assay
3.6. Determination of the Enzymatic Properties of r-AoFT
4. Conclusions
Supplementary Materials
Acknowledgments
Author contributions
Conflicts of Interest
References
- Alméciga-Díaz, C.J.; Gutierrez, A.M.; Bahamon, I.; Rodríguez, A.; Rodríguez, M.A.; Sánchez, O.F. Computational analysis of the fructosyltransferase enzymes in plants, fungi and bacteria. Gene 2011, 484, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, M.; Soliveri, J.; Plou, F.J. Synthesis of sugar esters in solvent mixtures by lipase from Thermomyces lanuginosus and Candida Antarctica B, and their antimicrobial properties. Enzym. Microb. Technol. 2005, 36, 391–398. [Google Scholar] [CrossRef]
- Jones, J.D.; Hacking, A.J.; Cheetham, P.S. Biological method for protection of 6-position of sucrose and its use in synthesis of disaccharide high-intensity sweetener. Biotechnol. Bioeng. 1992, 39, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Sabeder, S.; Habulin, M.; Knez, Z. Lipase-catalyzed synthesis of fatty acid fructose esters. J. Food. Eng. 2006, 77, 880–886. [Google Scholar] [CrossRef]
- Han, Y.W.; Liu, G.M.; Huang, D.Y.; Qiao, B.J.; Chen, L.P.; Guan, L.H.; Mao, D.B. Study on the synthesis of sucrose 6-acetate catalyzed by fructosyltransferase from Aspergillus aryzae. Nat. Biotechnol. 2011, 28, 14–18. [Google Scholar]
- Yang, X.; Zheng, P.; Ni, Y.; Sun, Z.H. Highly efficient biosynthesis of sucrose 6-acetate with cross-linked aggregates of Lipozyme TL 100L. J. Biotechnol. 2012, 161, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Qian, J.Q.; Guo, H.; Hu, Y.Y.; Liu, M. Biosynthesis of sucrose-6-acetate catalyzed by surfactant-coated Candida rugosa lipase immobilized on sol–gel supports. Bioprocess. Biosyst. Eng. 2014, 37, 813–818. [Google Scholar] [CrossRef] [PubMed]
- Heyer, A.G.; Wendenburg, R. Gene cloning and functional characterization by heterologous expression of the fructosyltransferase of Aspergillus sydowi IAM 2544. Appl. Environ. Microbiol. 2001, 67, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Yu, X.; Wang, Y.Y.; Zhu, Y.H.; Du, C.C.; Jia, C.X.; Mao, D.B. Purification and evaluation of the enzymatic properties of a novel fructosyltransferase from Aspergillus oryzae: A potential biocatalyst for the synthesis of sucrose 6-acetate. Biotechnol. Lett. 2014, 36, 1015–1020. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Jia, W.W.; Yu, X.; Mao, D.B. Enhancement of enzymatic synthesis of sucrose 6-acetate with Aspergillus oryzae fructosyltransferase using ionic liquid as a cosolvent. J. Mol. Catal. B Enzym. 2016, 123, 100–106. [Google Scholar] [CrossRef]
- Gallagher, J.; Cairns, A.; Pollock, C. Cloning and characterization of a putative fructosyltransferase and two putative invertase genes from the temperate grass Lolium temulentum L. J. Exp. Bot. 2004, 55, 557–569. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.W.; Chen, L.P.; Mao, D.B.; Tang, L.J.; Guan, L.H. Expression and activity analysis of sucrose: Sucrose 1-fructosyltransferase from onion. Nat. Biotechnol. 2010, 27, 324–329. [Google Scholar]
- Ueno, K.; Onodera, S.; Kawakami, A.; Yoshida, M.; Shiomi, N. Molecular characterization and expression of a cDNA encoding fructan: Fructan 6G-fructosyltransferase from asparagus (Asparagus officinalis). New Phytol. 2005, 165, 813–824. [Google Scholar] [CrossRef] [PubMed]
- Arand, M.; Golubev, A.; Neto, J.; Polikarpov, I.; Wattiez, R.; Korneeva, O.; Eneyskaya, E.; Kulminskaya, A.; Shabalin, K.; Shishliannikov, S.; et al. Purification, characterization, gene cloning and preliminary X-ray data of the exo-inulinase from Aspergillus awamori. Biochem. J. 2002, 362, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, S.; Tanaka, H.; Uwataki, M.; Muguruma, M.; Ohta, K. Molecular cloning and characterization of an exoinulinase gene from Aspergillus niger strain 12 and its expression in Pichia pastoris. J. Biosci. Bioeng. 2003, 96, 324–331. [Google Scholar] [CrossRef]
- Isono, N.; Tochihara, T.; Kusnadi, Y.; Win, T.; Watanabe, K.; Obae, K.; Ito, H.; Matsui, H. Cloning and heterologous expression of a β-fructofuranosidase gene from Arthrobacter globiformis IFO 3062, and site-directed mutagenesis of the essential aspartic acid and glutamic acid of the active site. J. Biosci. Bioeng. 2004, 97, 244–249. [Google Scholar] [CrossRef]
- Janer, C.; Rohr, L.; Pelaez, C.; Laloi, M.; Cleusix, V.; Requena, T.; Meile, L. Hydrolysis of oligofructoses by the recombinant β-fructofuranosidase from Bifidobacterium lactis. Syst. Appl. Microbiol. 2004, 27, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Ryan, S.; Fitzgerald, G.; van Sinderen, D. Transcriptional regulation and characterization of a novel β-fructofuranosidase-encoding gene from Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 2005, 71, 3475–3482. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.L.; Goosen, C.; Kools, H.; van der Maarel, M.; van den Hondel, C.; Dijkhuizen, L.; Ram, A.F.J. Database mining and transcriptional analysis of genes encoding inulin-modifying enzymes of Aspergillus niger. Microbiology 2006, 152, 3061–3073. [Google Scholar] [CrossRef] [PubMed]
- Goosen, C.; Yuan, X.L.; van Munster, J.; Ram, A.; van der Maarel, M.; Dijkhuizen, L. Molecular and biochemical characterization of a novel intracellular invertase from Aspergillus niger with transfructosylating activity. Eukaryot. Cell 2007, 6, 674–681. [Google Scholar] [CrossRef] [PubMed]
- Yanai, K.; Nakane, A.; Kawate, A.; Hirayama, M. Molecular cloning and characterization of the fructooligosaccharide-producing β-fructofuranosidase gene from Aspergillus niger ATCC20611. Biosci. Biotechnol. Biochem. 2001, 65, 766–773. [Google Scholar] [CrossRef] [PubMed]
- Liebl, W.; Brem, D.; Gotschlich, A. Analysis of the gene for β-fructosidase (invertase, inulinase) of the hyperthermophilic bacterium Thermotoga maritima, and characterisation of the enzyme expressed in Escherichia coli. Appl. Microbiol. Biotechnol. 1998, 50, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Olivares-Illana, V.; Lopez-Munguıa, A.; Olvera, C. Molecular characterization of inulosucrase from Leuconostoc citreum: a fructosyltransferase within a glucosyltransferase. J. Bacteriol. 2003, 185, 3606–3612. [Google Scholar] [CrossRef] [PubMed]
- Fu, R.; Wang, Y.; Sung, H. Cloning, characterization and functional expression of a new β-d-fructofuranosidase (Os beta fruct2) cDNA from Oryza sativa. Biotechnol. Lett. 2003, 25, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, A.; Yoshida, M. Molecular characterization of sucrose: Sucrose 1-fructosyltransferase and sucrose:fructan 6-fructosyltransferase associated with fructan accumulation in winter wheat during cold hardening. Biosci. Biotechnol. Biochem. 2002, 66, 2297–2305. [Google Scholar] [CrossRef] [PubMed]
- Lammens, W.; Le Roy, K.; Schroeven, L.; van Laere, A.; Rabijns, A.; van den Ende, W. Structural insights into glycoside hydrolase family 32 and 68 enzymes: functional implications. J. Exp. Bot. 2009, 60, 727–740. [Google Scholar] [CrossRef] [PubMed]
- Velazquez-Hernandez, M.L.; Baizabal-Aguirre, V.M.; Bravo-Patino, A.; Cajero-Juarez, M.; Chavez-Moctezuma, M.P.; Valdez-Alarcon, J.J. Microbial fructosyltransferases and the role of fructans. J. Appl. Microbiol. 2009, 106, 1763–1778. [Google Scholar] [CrossRef] [PubMed]
- Dordick, J.S.; Hacking, A.J.; Khan, R.A. Selective Acrylation of Sugars. U.S. Patent 5,128,248, 7 July 1992. [Google Scholar]
- Fernandez-Lafuente, R. Lipase from Thermomyces lanuginosus: Uses and prospects as an industrial biocatalyst. J. Mol. Catal. B Enzym. 2010, 62, 197–212. [Google Scholar] [CrossRef]
- Ratnam, R.; Aurora, S.; Subramaniyam, P. Method of Producing Sucrose-6-Acetate by Whole-Cell Biocatalysis. W.O. Patent 2007/054972 A2, 18 May 2007. [Google Scholar]
- Rodriguez, M.A.; Sanchez, O.A.; Almeciga-Diaz, C.J. Gene cloning and enzyme structure modeling of the Aspergillus oryzae N74 fructosyltransferase. Mol. Biol. Rep. 2011, 38, 1151–1161. [Google Scholar] [CrossRef] [PubMed]
- Chuankhayan, P.; Hsieh, C.Y.; Huang, Y.C.; Hsieh, Y.Y.; Guan, H.H.; Hsieh, Y.C.; Tien, Y.C.; Chen, C.D.; Chiang, C.M.; Chen, C.J. Crystal structures of Aspergillus japonicas fructosyltransferase complex with donor/acceptor substrates reveal complete subsites in the active site for catalysis. J. Biol. Chem. 2010, 285, 23251–23264. [Google Scholar] [CrossRef] [PubMed]
- Bujacz, A.; Jedrzejczak-Krzepkowska, M.; Bielecki, S.; Redzynia, I.; Bujacz, G. Crystal structures of the apo form of β-fructofuranosidase from Bifidobacterium longum and its complex with fructose. FEBS J. 2011, 278, 1728–1744. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda, R.H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Van den Ende, W.; Lammens, W.; van Laere, A.; Schroeven, L.; Le Roy, K. Donor and acceptor substrate selectivity among plant glycoside hydrolase family 32 enzymes. FEBS J. 2009, 276, 5788–5798. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Le Roy, K.; Venken, T.; Lammens, W.; van den Ende, W.; De Maeyer, M. pKa modulation of the acid/base catalyst within GH32 and GH68: A role insubstrate/inhibitor specificity? PLoS ONE 2012, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Aharoni, A.; Gaidukov, L.; Yagur, S.; Toker, L.; Silman, I.; Tawfik, D.S. Directed evolution of mammalian paraoxonases PON1 and PON3 for bacterial expression and catalytic specialization. Proc. Natl. Acad. Sci. USA 2004, 101, 482–487. [Google Scholar] [CrossRef] [PubMed]
- Harel, M.; Aharoni, A.; Gaidukov, L.; Brumshtein, B.; Khersonsky, O.; Meged, R.; Dvir, H.; Ravelli, R.B.; McCarthy, A.; Toker, L.; et al. Structure and evolution of the serum paraoxonase family of detoxifying and anti-atherosclerotic enzymes. Nat. Struct. Mol. Biol. 2004, 11, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Kadowaki, H.; Kadowaki, T.; Wondisford, F.E.; Taylor, S.I. Use of polymerase chain reaction catalyzed by Taq DNA polymerase for site-specific mutagenesis. Gene 1989, 76, 161–166. [Google Scholar] [CrossRef]
- Kelley, L.; Sternberg, M. Protein structure prediction on the web: A case study using the Phyre server. Nat. Protoc. 2009, 4, 363–371. [Google Scholar] [CrossRef] [PubMed]


| Step | Total Protein (mg) | Total Activity (U) | Specific Activity (U/mg) | Purification Fold | Yield (%) |
|---|---|---|---|---|---|
| Crude cell extract | 48.2 | 528.9 | 11.0 | 1 | 100 |
| Heat treatment | 30.1 | 396.7 | 13.2 | 1.2 | 75.0 |
| Ni-NTA affinity | 13.5 | 265.3 | 19.7 | 1.8 | 50.0 |
| Superdex-200 gel filtration | 4.8 | 182.5 | 38.0 | 3.5 | 34.5 |
| Metals or EDTA | Concentration | Relative Activity (%) |
|---|---|---|
| None | - | 100 |
| Ca2+ | 5 mM | 108 ± 3 |
| Mn2+ | 5 mM | 103 ± 1 |
| Zn2+ | 5 mM | 30 ± 5 |
| Mg2+ | 5 mM | 135 ± 3 |
| Cu2+ | 5 mM | 13 ± 2 |
| Fe2+ | 5 mM | 85 ± 1 |
| Ni2+ | 5 mM | 8 ± 2 |
| Co2+ | 5 mM | 84 ± 2 |
| EDTA | 5 mM | 92 ± 1 |
| Organic Solvents or Detergents | Concentration (%) | Relative Activity (%) |
|---|---|---|
| None | - | 100 |
| Methanol | 50 (v/v) | 92 ± 5 |
| Ethanol | 50 (v/v) | 76 ± 1 |
| Acetone | 50 (v/v) | 83 ± 2 |
| Chloroform | 50 (v/v) | 103 ± 1 |
| Toluene | 50 (v/v) | 112 ± 3 |
| DMF | 50 (v/v) | 89 ± 2 |
| DMSO | 50 (v/v) | 96 ± 3 |
| DTT | 5 mM | 94 ± 2 |
| Urea | 4 M | 22 ± 3 |
| SDS | 1% (w/v) | 25 ± 5 |
| Tween 20 | 1% (w/v) | 118 ± 2 |
| Triton X-100 | 1% (w/v) | 116 ± 3 |
| Mutants | Specific Activities (μmol/min/mg) | Km (μM) | Vmax (μmol/min/mg) |
|---|---|---|---|
| r-AoFT | 38 | 21 | 75 |
| N38L | 42.1 | 24.3 | 89.2 |
| S99A | 40.1 | 22.2 | 81.2 |
| Y282A | 55.2 | 42.1 | 98.2 |
| D39A | 0 | ND | ND |
| D164A | 0 | ND | ND |
| E216A | 0 | ND | ND |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, T.; Huang, S.; Zang, J.; Jia, C.; Mao, D. Cloning, Expression and Characterization of a Novel Fructosyltransferase from Aspergillus oryzae ZZ-01 for the Synthesis of Sucrose 6-Acetate. Catalysts 2016, 6, 67. https://doi.org/10.3390/catal6050067
Wei T, Huang S, Zang J, Jia C, Mao D. Cloning, Expression and Characterization of a Novel Fructosyltransferase from Aspergillus oryzae ZZ-01 for the Synthesis of Sucrose 6-Acetate. Catalysts. 2016; 6(5):67. https://doi.org/10.3390/catal6050067
Chicago/Turabian StyleWei, Tao, Shen Huang, Jie Zang, Chunxiao Jia, and Duobin Mao. 2016. "Cloning, Expression and Characterization of a Novel Fructosyltransferase from Aspergillus oryzae ZZ-01 for the Synthesis of Sucrose 6-Acetate" Catalysts 6, no. 5: 67. https://doi.org/10.3390/catal6050067





