2.1. MUC1-TRIM46-KRTCAP2 Is a Recurrent Chimeric RNA in HGSC
The Cancer Genome Atlas (TCGA) has performed extensive high-throughput transcriptome sequencing on HGSC patient samples [
17]. For our analysis, we utilized the sequencing reads from 130 HGSC patient samples, a sample size that is sufficient to identify highly recurrent chimeric RNAs. Our strategy for identifying chimeric transcripts was to search for paired “chimeric” reads with each read mapping to a different gene either in the genome or transcriptome using our previously established pipeline [
18]. This strategy led to the identification of nearly 1383 chimeric RNA candidates from the 130 cancer samples. We selected 33 out of 1383 chimeric RNA candidates for experimental validation based on the criteria that the chimeric RNA must be present in at least five cancer samples. To experimentally validate these candidates, we designed specific primers with each primer targeting one parental gene, therefore specifically amplifying only the chimeric RNA but not the parental gene transcripts. For 9 of the 33 chimeric RNA candidates, we were able to obtain RT-PCR (reverse transcription PCR) products from in-house tumor samples. The RT-PCR bands were excised and sequenced by Sanger sequencing. This led to the confirmation of the presence of nine chimeric RNAs, and the identification of their exact RNA junction (
Table S1). Since TCGA transcriptome sequencing collection does not contain organ specific controls, we examined these nine chimeric RNAs using our in-house non-cancerous ovary samples to answer the question of whether these candidates are also present in non-cancerous tissue in addition to the cancer samples. The frequency of occurrence for the nine chimeric RNAs in non-cancerous ovary samples is listed in
Table S2. Among them, MUC1-TRIM46 is the only chimeric RNA that was highly recurrent in HGSC tumor samples but absent in non-cancer samples (see below).
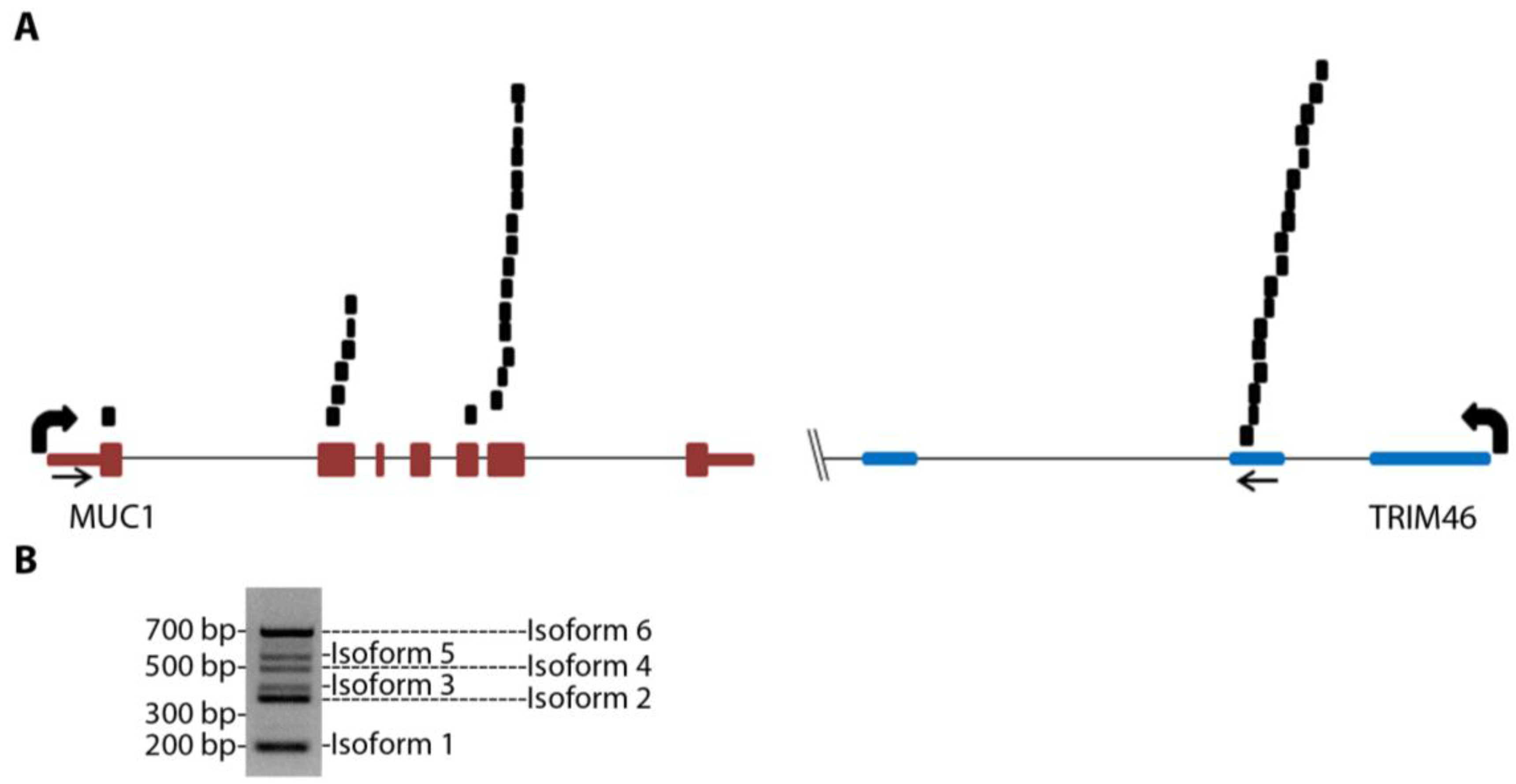
MUC1-TRIM46 chimeric RNA is novel and has not been previously reported. The presence of this fusion transcript in TCGA cancer samples is supported by 25 paired “chimeric” reads with one read mapping to MUC1 and the other to TRIM46 (
Figure 1A and
Table S3, using GRCh37/hg19). RT-PCR analysis using several in-house cancer samples (see
Table S4 for primers) resulted in six bands of different sizes (an example is shown in
Figure 1B). These bands were gel purified, cloned, subjected to Sanger sequencing, and confirmed to harbor different exons of MUC1 joined to the same genomic sequence of TRIM46 by annotated splice sites (
Figure 2A). This indicates that the RNA junctions of these chimeric RNAs are the result of splicing.
Figure 1.
MUC1 chimeric RNAs identified in TCGA database of HGSC patient samples. (A) Schematic showing the position of 25 paired chimeric reads aligning to both MUC1 and TRIM46 genes identified from nine patients in the 130 TCGA cohort. Arrows indicate PCR primer targeting locations. (B) An example of RT-PCR validation for MUC1-TRIM46 chimeric RNA using one of the in-house HGSC patient samples. The bands corresponding to the six isoforms are labeled as shown.
Figure 1.
MUC1 chimeric RNAs identified in TCGA database of HGSC patient samples. (A) Schematic showing the position of 25 paired chimeric reads aligning to both MUC1 and TRIM46 genes identified from nine patients in the 130 TCGA cohort. Arrows indicate PCR primer targeting locations. (B) An example of RT-PCR validation for MUC1-TRIM46 chimeric RNA using one of the in-house HGSC patient samples. The bands corresponding to the six isoforms are labeled as shown.
To annotate the ends of the MUC1-TRIM46 chimeric RNAs, we analyzed available EST collections. The analysis revealed two ESTs (CD366871.1 and BF870262.1) that contain a sequence from either MUC1 or TRIM46 extending into a third gene KRTCAP2. To test if the chimeric RNAs we identified indeed extend into KRTCAP2, we designed a primer pair to amplify from the beginning of the 5’ UTR of MUC1 to the very end of the 3’ UTR of KRTCAP2. RT-PCR performed on cancer samples yielded six full-length bands. Sequencing of cloned bands revealed that they correspond to the six isoforms described in
Figure 1B, but now with confirmed mRNA sequences extending from the 5’ end of MUC1 till the 3’ end of KRTCAP2 with one exon in between originating from the TRIM46 sequence (
Figure 2A,
Table S5-sequences). The six isoforms differ in the MUC1 region where different exons are recruited but they all have an identical TRIM46 and KRTCAP2 sequence (
Figure 2A). Two unique RNA fusion junctions were identified among the six isoforms (
Table S5). One is unique to isoform 1 and the other is common among isoforms 2–6. Henceforth, these chimeric RNAs are referred to as MUC1-TRIM46-KRTCAP2 instead of MUC1-TRIM46.
Figure 2.
MUC1-TRIM46-KRTCAP2 chimeric RNA isoforms and predicted protein consequences. (A) Schematic of the parental MUC1 and the six isoforms of MUC1-TRIM46-KRTCAP2 chimeric RNAs are shown with MUC1 (red), TRIM46 (blue) and KRTCAP2 (green) regions. The indicated coding and non-coding regions of MUC1 are based on the annotation of specific transcripts in the UCSC genome browser. (B) The expected protein products of these chimeric RNAs are shown with the domains indicated.
Figure 2.
MUC1-TRIM46-KRTCAP2 chimeric RNA isoforms and predicted protein consequences. (A) Schematic of the parental MUC1 and the six isoforms of MUC1-TRIM46-KRTCAP2 chimeric RNAs are shown with MUC1 (red), TRIM46 (blue) and KRTCAP2 (green) regions. The indicated coding and non-coding regions of MUC1 are based on the annotation of specific transcripts in the UCSC genome browser. (B) The expected protein products of these chimeric RNAs are shown with the domains indicated.
To test whether the transcription of MUC1-TRIM46-KRTCAP2 chimeric RNA is the result of a genomic segment deletion between MUC1 and TRIM46 gene, we employed the same primer pair used in
Figure 1A to detect the presence of genomic deletion from HGSC patients’ genomic DNA. However, genomic DNA PCR showed the same size bands as compared to reference human genomic DNA (data not shown), suggesting that the chimeric RNAs are not the result of genomic rearrangement, but are generated at the transcriptional level either by trans-splicing or read-through splicing.
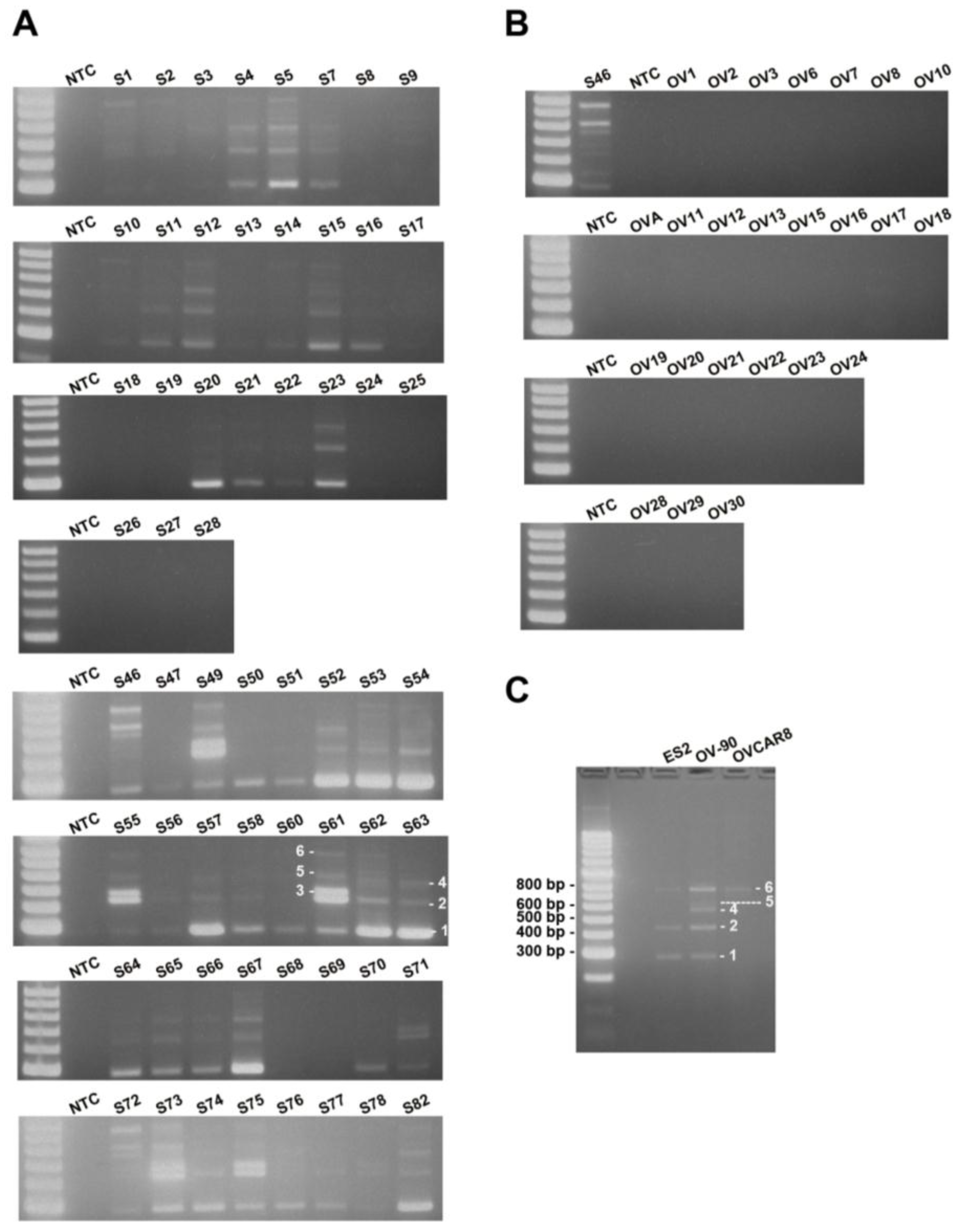
To estimate the frequency of occurrence of MUC1-TRIM46-KRTCAP2 chimeric RNAs, we performed RT-PCR on a cohort of 59 HGSC patient samples. The results showed that MUC1-TRIM46-KRTCAP2 chimeric RNAs are highly recurrent with 75% of the cancer samples containing at least one isoform (
Figure 3A). MUC1-TRIM46-KRTCAP2 isoform 1 is the most common chimeric RNA and is observed in 64% of the cancer samples, while isoform 3 is the least common chimeric RNA and is expressed in 17% of the cohort (
Table S6). In contrast, none of the isoforms are detected in the 24 non-cancerous ovary samples (
Figure 3B). Thus, the results suggest that MUC1-TRIM46-KRTCAP2 chimeric RNAs are highly cancer-enriched.
Since MUC1-TRIM46-KRTCAP2 family has a high frequency of occurrence among cancer samples, we speculated that this chimeric RNA might also be present in established HGSC cell lines. This indeed is the case. By RT-PCR screening of three serous type cancer cell lines (ES2, OV-90 and OVCAR8), we found that isoforms of MUC1-TRIM46-KRTCAP2 are expressed in all three cell lines (
Figure 3C). However, different isoforms are selectively expressed in each cell line, a pattern also found in patient samples. The presence of these chimeric RNAs in established serous type cancer cell lines further support the potential significance of the MUC1-TRIM46-KRTCAP2 in HGSC.
Figure 3.
MUC1-TRIM46-KRTCAP2 is a highly recurrent chimeric RNA in HGSC patient tumor samples and cell lines. (A) The results of RT-PCR for MUC1-TRIM46-KRTCAP2 in 59 HGSC tumor samples (denoted by “S”). (B) The results of 24 non-cancerous ovary samples (“OV”) are shown. NTC refers to “no cDNA control”. The different isoforms are indicated on samples S61 and S63. (C) The results of RT-PCR for MUC1-TRIM46-KRTCAP2 in three HGSC cell lines (ES2, OV90 and OVCAR8) are shown.
Figure 3.
MUC1-TRIM46-KRTCAP2 is a highly recurrent chimeric RNA in HGSC patient tumor samples and cell lines. (A) The results of RT-PCR for MUC1-TRIM46-KRTCAP2 in 59 HGSC tumor samples (denoted by “S”). (B) The results of 24 non-cancerous ovary samples (“OV”) are shown. NTC refers to “no cDNA control”. The different isoforms are indicated on samples S61 and S63. (C) The results of RT-PCR for MUC1-TRIM46-KRTCAP2 in three HGSC cell lines (ES2, OV90 and OVCAR8) are shown.
2.2. MUC1-TRIM46-KRTCAP2 Isoforms Are Translated into Intracellular Fusion Proteins
The complete cDNA sequence of MUC1-TRIM46-KRTCAP2 obtained from RT-PCR enabled us to predict the protein consequences of these highly recurrent chimeric RNAs. Isoform 4 is predicted to yield no protein since the annotated start codon is immediately followed by a stop codon. The remaining five isoforms are predicted to translate different fusion proteins with the MUC1 C-terminal domain replaced by a common sequence composed of 53 amino acids from TRIM46 and 54 amino acids from KRTCAP2 (
Figure 2B and
Table S7). However, the TRIM46 sequence in the chimeric RNAs is in the antisense direction and the KRTCAP2 sequence is out of frame. Therefore, these newly added common amino acids are unrelated to parental TRIM46 or KRTCAP2 proteins. These predicted protein isoforms lack the VNTR region, which is the site of glycosylation in parental MUC1 (
Figure 2B). The signal peptide is present in isoforms 1, 2, 3 and 5, while the transmembrane domain is present only in isoforms 3 and 6. The MUC1 cytoplasmic tail is maintained in all isoforms except for isoform 1. The differences in the domains retained in the isoforms suggest that they could result in varied cellular localization.
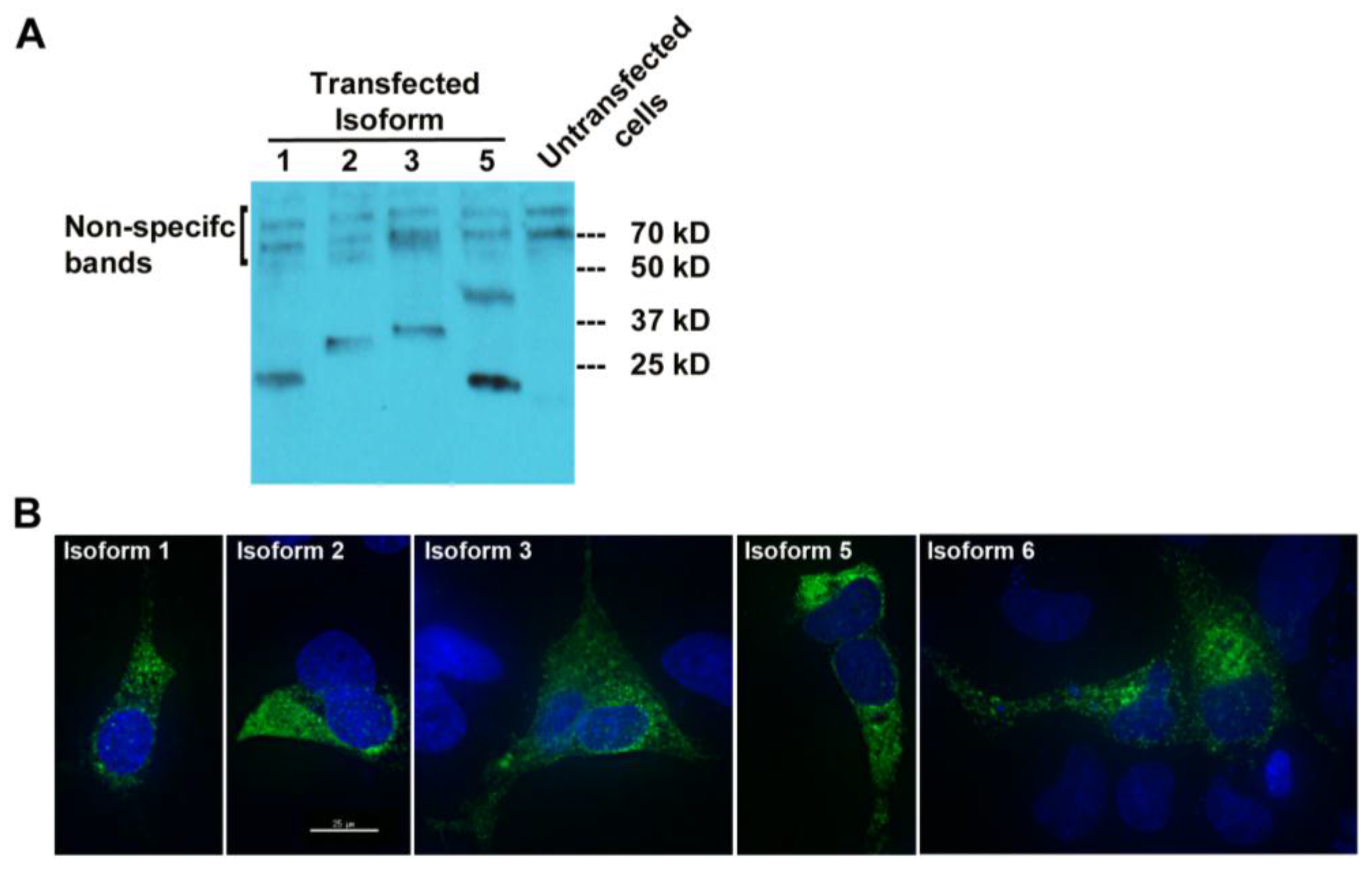
To check whether MUC1-TRIM46-KRTCAP2 chimeric RNAs are translated into fusion proteins, we tagged the full-length cDNA of five isoforms (1, 2, 3, 5 and 6) with a FLAG at the C-terminus. The predicted sizes for the isoforms are between 23 and 30 kD (
Figure 2A). Transfection in OVCAR8 cell line and the subsequent Western blot analysis revealed that all isoforms are translated with the exception of isoform 6 (
Figure 4A). The results confirmed that most of the MUC1-TRIM46-KRTCAP2 fusion proteins are translated as predicted. However, we are not able to confirm the presence of endogenous MUC1-TRIM46-KRTCAP2 isoforms expressed in tumor tissue and in cancer cell lines because (1) most of the commercially available antibodies target the VNTR domain that is lacking in our fusion protein isoforms and (2) the sizes of these fusion protein isoforms are very similar to the various protein isoforms of parental MUC1, making it difficult to conclusively distinguish between the two groups.
To test whether these translated protein isoforms are secreted in the media, we performed Western blotting analyses using the culture media collected from transfected cells. As shown in
Figure S1, none of the MUC1-TRIM46-KRTCAP2 isoforms were detected in the culture media, indicating they are not secreted. Immunocytochemical imaging using anti-FLAG antibody showed that the MUC1-TRIM46-KRTCAP2 fusion proteins are mainly located in the cytoplasm as opposed to the cell membrane of transfected OVCAR8 cells (
Figure 4B). Surprisingly, isoform 6, which is not detected by Western blotting, is present in the cytoplasm by immunocytochemistry. This suggests that isoform 6 is also translated but the FLAG epitope can only be detected when the protein is in the native conformation. Alternatively, the FLAG epitope may be cleaved off from isoform 6, resulting in a fragment too small to be detected on the Western blot. The cytoplasmic localization of these isoforms is in contrast to parental MUC1, which is a transmembrane protein, indicating that MUC1-TRIM46-KRTCAP2 fusion proteins may have protein functions that are very different from parental MUC1 protein functions.
Figure 4.
MUC1-TRIM46-KRTCAP2 chimeric RNAs give rise to fusion proteins. (A) MUC1-TRIM46-KRTCAP2 isoforms were cloned with a C-terminal FLAG tag and expressed in OVCAR8 cells. Western blot of protein extracts using FLAG antibody shows that most of the isoforms are translated with the expected sizes lacking glycosylation. Isoform 5 appears to form a homodimer. (B) Immunocytochemistry of OVCAR8 cells transfected with different MUC1-TRIM46-KRTCAP2-FLAG expression constructs. The fusion protein isoforms are seen mainly in the cytoplasm as visualized by FLAG antibody. Images were taken using deconvolution microscopy.
Figure 4.
MUC1-TRIM46-KRTCAP2 chimeric RNAs give rise to fusion proteins. (A) MUC1-TRIM46-KRTCAP2 isoforms were cloned with a C-terminal FLAG tag and expressed in OVCAR8 cells. Western blot of protein extracts using FLAG antibody shows that most of the isoforms are translated with the expected sizes lacking glycosylation. Isoform 5 appears to form a homodimer. (B) Immunocytochemistry of OVCAR8 cells transfected with different MUC1-TRIM46-KRTCAP2-FLAG expression constructs. The fusion protein isoforms are seen mainly in the cytoplasm as visualized by FLAG antibody. Images were taken using deconvolution microscopy.