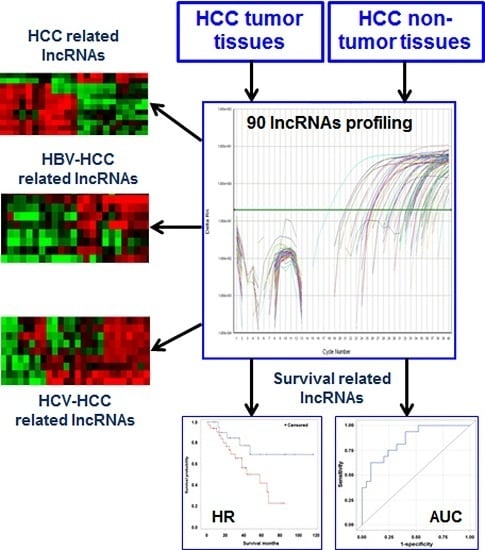
Exploration of Deregulated Long Non-Coding RNAs in Association with Hepatocarcinogenesis and Survival
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Tissue Samples
2.2. RNA Extraction and lncRNA Measurement
2.3. Statistical Analysis
3. Results
3.1. Clinical and Pathological Characteristics
| Variables | No. of Cases (%) |
|---|---|
| Age at diagnosis (yrs), Mean ± SD | 59.5 ± 14.7 |
| Age group | |
| <60 years | 28 (43) |
| ≥60 years | 37 (57) |
| Gender | |
| Male | 49 (75) |
| Female | 16 (25) |
| Ethnicity | |
| Caucasian | 33 (51) |
| African-American | 6 (9) |
| Hispanic | 6 (9) |
| Asian | 14 (22) |
| Unknown/Other | 6 (9) |
| Viral infection | |
| HBV (−), HCV (−) | 18 (28) |
| HBV (−), HCV (+) | 19 (29) |
| HBV (+), HCV (−) | 13 (20) |
| HBV (+), HCV (+) | 4 (6) |
| Missing | 11 (17) |
| Cigarette smoking | |
| No | 26 (40) |
| Yes | 35 (54) |
| Missing | 4 (6) |
| Alcohol drinking | |
| No | 27 (42) |
| Yes | 35 (54) |
| Missing | 3 (4) |
| Cirrhosis | |
| No | 17 (26) |
| Yes | 47 (72) |
| Missing | 1 (2) |
| Tumor size (cm), Mean ± SD | 6.1 (4.8) |
| Tumor grade * | |
| I–II | 23 (35) |
| III | 21 (32) |
| IV | 18 (28) |
| Missing | 3 (5) |
3.2. Differentially Expressed lncRNAs in HCC Tissues
| lncRNAs | Tumor | Non-Tumor | Fold-Change | Unadjusted p-Value | FDR |
|---|---|---|---|---|---|
| lincRNA-VLDLR | −4.6 | −3.4 | −2.4 | 0.005 | 0.188 |
| MEG9 | −5.6 | −4.2 | −2.8 | 0.010 | 0.188 |
| H19 antisense | −1.3 | 0.2 | −2.7 | 0.010 | 0.188 |
| ncR-uPAR | −1.3 | −0.03 | −2.6 | 0.018 | 0.188 |
| NEAT1 (family) | −2.4 | −1.3 | −2.2 | 0.021 | 0.188 |
| LUST | −0.5 | 0.6 | −2.1 | 0.037 | 0.287 |
| UM9-5 | −8.7 | −7.6 | −2.5 | 0.042 | 0.287 |
| HOTAIR | −8.8 | −7.1 | −2.6 | 0.048 | 0.287 |
3.3. Deregulated lncRNAs in HBV- or HCV-Related HCC
| lncRNAs | Tumor | Non-Tumor | Fold-Change | Unadjusted p-value | FDR |
|---|---|---|---|---|---|
| Kcnq1ot1 | −8.9 | −5.2 | −9.1 | 0.005 | 0.215 |
| TncRNA | −5.5 | −4.0 | −2.9 | 0.009 | 0.215 |
| lincRNA-VLDLR | −3.8 | −1.8 | −4.2 | 0.014 | 0.215 |
| Zfhx2as | −2.0 | 0.8 | −6.7 | 0.019 | 0.215 |
| NRON | −6.1 | −3.7 | −5.3 | 0.020 | 0.215 |
| HOTTIP | −9.8 | −7.8 | −4.2 | 0.025 | 0.231 |
| lincRNA-RoR | −9.5 | −7.7 | −3.4 | 0.046 | 0.273 |
| LncRNAs | Viral Negative (N = 18) | HBV Positive (N = 13) | Fold-Change | Unadjusted p-Value | FDR |
|---|---|---|---|---|---|
| Kcnq1ot1 | −8.9 | −5.2 | 12.6 | 0.004 | 0.193 |
| NRON | −6.2 | −3.7 | 5.4 | 0.042 | 0.947 |
3.4. Aberrant Expression of lncRNAs in Tumor Tissue and Prediction for HCC Survival
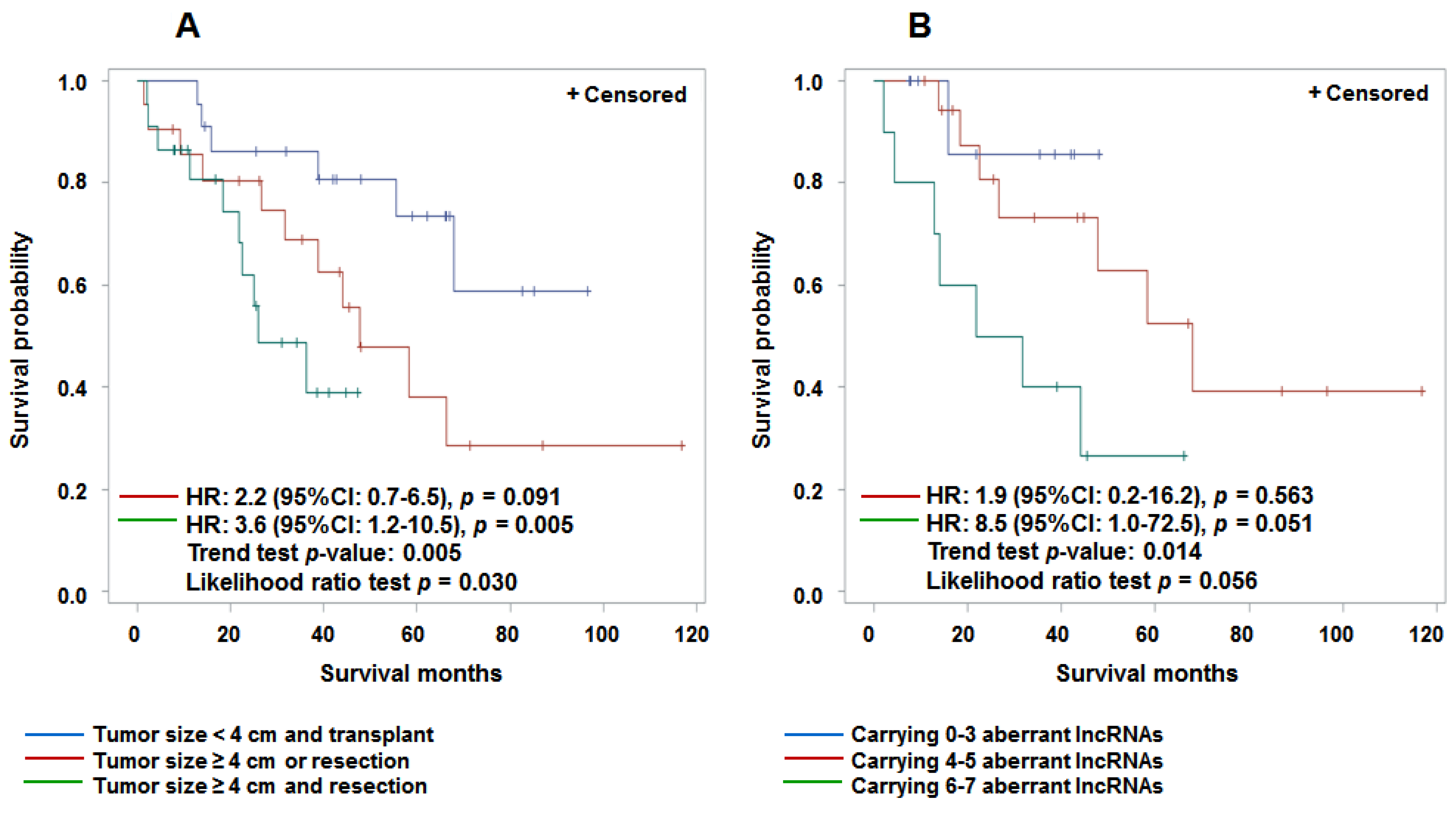
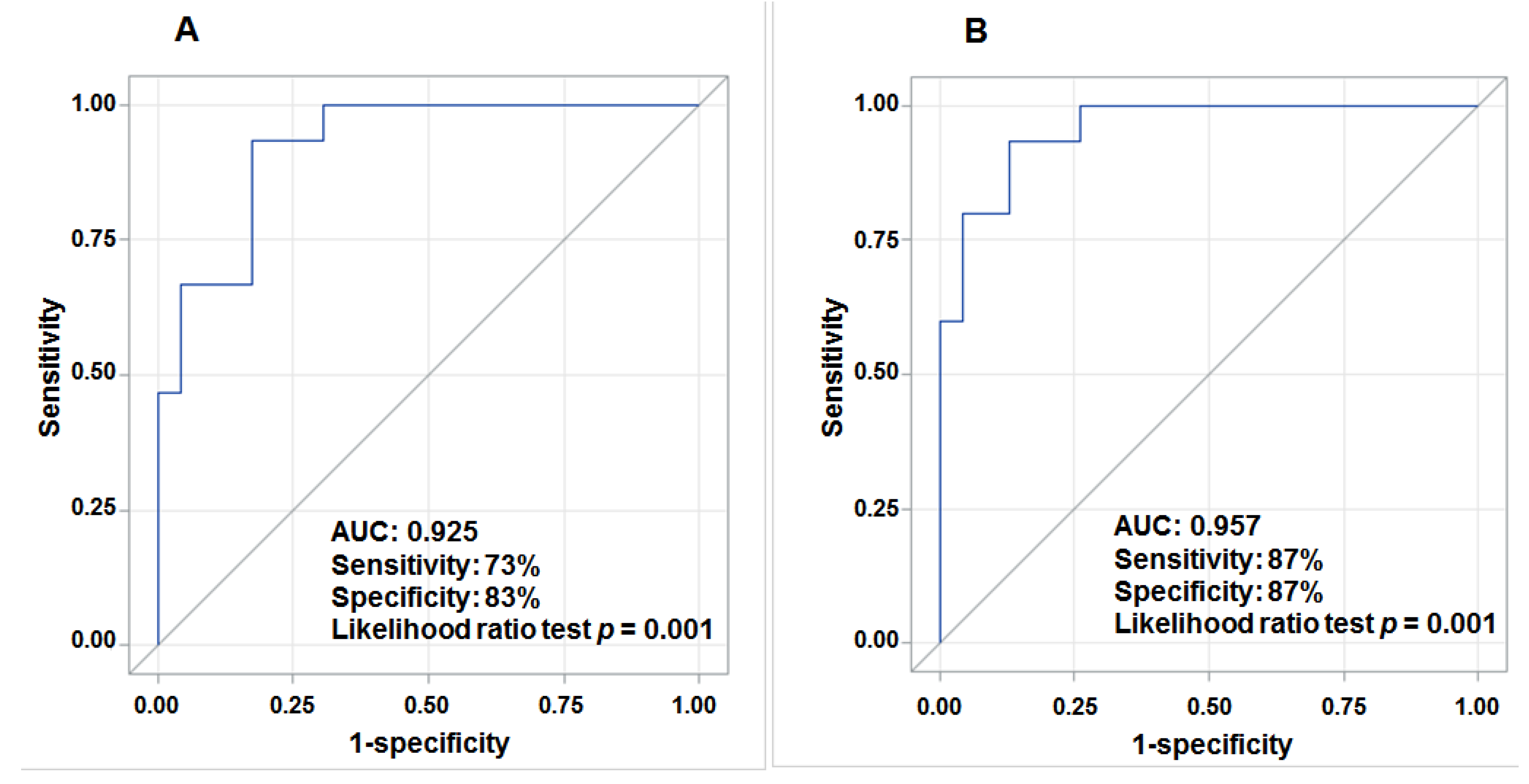
4. Discussion
5. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- El Serag, H.B.; Rudolph, K.L. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology 2007, 132, 2557–2576. [Google Scholar] [CrossRef] [PubMed]
- Jemal, A.; Siegel, R.; Ward, E.; Hao, Y.; Xu, J.; Thun, M.J. Cancer statistics, 2009. CA Cancer J. Clin. 2009, 59, 225–249. [Google Scholar] [CrossRef] [PubMed]
- Altekruse, S.F.; McGlynn, K.A.; Reichman, M.E. Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J. Clin. Oncol. 2009, 27, 1485–1491. [Google Scholar] [CrossRef] [PubMed]
- El Serag, H.B. Hepatocellular carcinoma. N. Engl. J. Med. 2011, 365, 1118–1127. [Google Scholar] [CrossRef] [PubMed]
- Birney, E.; Stamatoyannopoulos, J.A.; Dutta, A.; Guigo, R.; Gingeras, T.R.; Margulies, E.H.; Weng, Z.; Snyder, M.; Dermitzakis, E.T.; Thurman, R.E.; et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007, 447, 799–816. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S. The genetic signatures of noncoding RNAs. PLoS Genet. 2009, 5, e1000459. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S. The functional genomics of noncoding RNA. Science 2005, 309, 1527–1528. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Makunin, I.V. Non-coding RNA. Hum. Mol. Genet. 2006, 15, R17–R29. [Google Scholar] [CrossRef] [PubMed]
- Reis, E.M.; Verjovski-Almeida, S. Perspectives of long non-coding RNAs in cancer diagnostics. Front. Genet. 2012, 3, 32. [Google Scholar] [CrossRef] [PubMed]
- Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.C.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; Wells, C.; et al. The transcriptional landscape of the mammalian genome. Science 2005, 309, 1559–1563. [Google Scholar] [PubMed]
- Dinger, M.E.; Pang, K.C.; Mercer, T.R.; Mattick, J.S. Differentiating protein-coding and noncoding RNA: Challenges and ambiguities. PLoS Comput. Biol. 2008, 4, e1000176. [Google Scholar] [CrossRef] [PubMed]
- Frith, M.C.; Bailey, T.L.; Kasukawa, T.; Mignone, F.; Kummerfeld, S.K.; Madera, M.; Sunkara, S.; Furuno, M.; Bult, C.J.; Quackenbush, J.; et al. Discrimination of non-protein-coding transcripts from protein-coding mRNA. RNA Biol. 2006, 3, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Gibb, E.A.; Brown, C.J.; Lam, W.L. The functional role of long non-coding RNA in human carcinomas. Mol. Cancer 2011, 10, 38. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Huarte, M.; Rinn, J.L. Large non-coding RNAs: Missing links in cancer? Hum. Mol. Genet. 2010, 19, R152–R161. [Google Scholar] [CrossRef] [PubMed]
- Prensner, J.R.; Iyer, M.K.; Balbin, O.A.; Dhanasekaran, S.M.; Cao, Q.; Brenner, J.C.; Laxman, B.; Asangani, I.A.; Grasso, C.S.; Kominsky, H.D.; et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat. Biotechnol. 2011, 29, 742–749. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Cheetham, S.W.; Gruhl, F.; Mattick, J.S.; Dinger, M.E. Long noncoding RNAs and the genetics of cancer. Br. J. Cancer 2013, 108, 2419–2425. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Kong, G.; You, X.; Zhang, S.; Zhang, T.; Gao, Y.; Ye, L.; Zhang, X. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J. Biol. Chem. 2012, 287, 26302–26311. [Google Scholar] [CrossRef] [PubMed]
- Gabory, A.; Jammes, H.; Dandolo, L. The H19 locus: Role of an imprinted non-coding RNA in growth and development. BioEssays 2010, 32, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.J.; Xie, S.L.; Li, Q.; Ma, J.; Wang, G.Y. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J. Int. Med. Res. 2011, 39, 2119–2128. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.C.; Yang, Z.; Zhou, L.; Zhu, Q.Q.; Xie, H.Y.; Zhang, F.; Wu, L.M.; Chen, L.M.; Zheng, S.S. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med. Oncol. 2012, 29, 1810–1816. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Maeda, S.; Liu, C.; Karin, M.; Edgington, T.S. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene 2007, 26, 851–858. [Google Scholar] [CrossRef] [PubMed]
- Matouk, I.J.; Degroot, N.; Mezan, S.; Ayesh, S.; Abu-Lail, R.; Hochberg, A.; Galun, E. The H19 non-coding RNA is essential for human tumor growth. PLoS ONE 2007, 2, e845. [Google Scholar] [CrossRef] [PubMed]
- Panzitt, K.; Tschernatsch, M.M.; Guelly, C.; Moustafa, T.; Stradner, M.; Strohmaier, H.M.; Buck, C.R.; Denk, H.; Schroeder, R.; Trauner, M.; et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology 2007, 132, 330–342. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, X.; Wu, H.; Ni, P.; Gu, Z.; Qiao, Y.; Chen, N.; Sun, F.; Fan, Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010, 38, 5366–5383. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhou, L.; Wu, L.M.; Lai, M.C.; Xie, H.Y.; Zhang, F.; Zheng, S.S. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann. Surg. Oncol. 2011, 18, 1243–1250. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Liu, S.; Ye, F.; Shen, Y.; Tie, Y.; Zhu, J.; Jin, Y.; Zheng, X.; Wu, Y.; Fu, H. The long noncoding RNA expression profile of hepatocellular carcinoma identified by microarray analysis. PLoS ONE 2014, 9, e101707. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Ying, H.Q.; He, B.S.; Pan, Y.Q.; Deng, Q.W.; Sun, H.L.; Chen, J.; Liu, X.; Wang, S.K. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget 2015, 6, 7899–7917. [Google Scholar] [PubMed]
- Quagliata, L.; Matter, M.S.; Piscuoglio, S.; Arabi, L.; Ruiz, C.; Procino, A.; Kovac, M.; Moretti, F.; Makowska, Z.; Boldanova, T.; et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology 2014, 59, 911–923. [Google Scholar] [CrossRef] [PubMed]
- Tu, Z.Q.; Li, R.J.; Mei, J.Z.; Li, X.H. Down-regulation of long non-coding RNA GAS5 is associated with the prognosis of hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 2014, 7, 4303–4309. [Google Scholar] [PubMed]
- Yang, F.; Zhang, L.; Huo, X.S.; Yuan, J.H.; Xu, D.; Yuan, S.X.; Zhu, N.; Zhou, W.P.; Yang, G.S.; Wang, Y.Z.; et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology 2011, 54, 1679–1689. [Google Scholar] [CrossRef] [PubMed]
- Edge, S.B.; Compton, C.C. The American Joint Committee on Cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann. Surg. Oncol. 2010, 17, 1471–1474. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Zweig, M.H.; Campbell, G. Receiver-operating characteristic (ROC) plots: A fundamental evaluation tool in clinical medicine. Clin. Chem. 1993, 39, 561–577. [Google Scholar] [PubMed]
- Pandey, R.R.; Mondal, T.; Mohammad, F.; Enroth, S.; Redrup, L.; Komorowski, J.; Nagano, T.; Mancini-Dinardo, D.; Kanduri, C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell 2008, 32, 232–246. [Google Scholar] [CrossRef] [PubMed]
- Willingham, A.T.; Orth, A.P.; Batalov, S.; Peters, E.C.; Wen, B.G.; Aza-Blanc, P.; Hogenesch, J.B.; Schultz, P.G. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 2005, 309, 1570–1573. [Google Scholar] [CrossRef] [PubMed]
- Shou, J.; Jing, J.; Xie, J.; You, L.; Jing, Z.; Yao, J.; Han, W.; Pan, H. Nuclear factor of activated T cells in cancer development and treatment. Cancer Lett. 2015, 361, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Liu, Y.J.; Xia, L.M.; Yan, W.; Zhu, Q.; Tian, D.A. Very low density lipoprotein receptor subtype II silencing by RNA interference inhibits cell proliferation in hepatoma cell lines. Hepatogastroenterology 2010, 57, 882–890. [Google Scholar] [PubMed]
- Zhou, H.; Guo, W.; Zhao, Y.; Wang, Y.; Zha, R.; Ding, J.; Liang, L.; Yang, G.; Chen, Z.; Ma, B.; Yin, B. MicroRNA-135a acts as a putative tumor suppressor by directly targeting very low density lipoprotein receptor in human gallbladder cancer. Cancer Sci. 2014, 105, 956–965. [Google Scholar] [CrossRef] [PubMed]
- Takada, H.; Imoto, I.; Tsuda, H.; Nakanishi, Y.; Sakakura, C.; Mitsufuji, S.; Hirohashi, S.; Inazawa, J. Genomic loss and epigenetic silencing of very-low-density lipoprotein receptor involved in gastric carcinogenesis. Oncogene 2006, 25, 6554–6562. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, L.C.; Wang, K.; Robinson, A.G. RBM5 as a putative tumor suppressor gene for lung cancer. J. Thorac. Oncol. 2010, 5, 294–298. [Google Scholar] [CrossRef] [PubMed]
- Rintala-Maki, N.D.; Sutherland, L.C. Identification and characterisation of a novel antisense non-coding RNA from the RBM5 gene locus. Gene 2009, 445, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Nakano, S.; Murakami, K.; Meguro, M.; Soejima, H.; Higashimoto, K.; Urano, T.; Kugoh, H.; Mukai, T.; Ikeguchi, M.; Oshimura, M. Expression profile of LIT1/KCNQ1OT1 and epigenetic status at the KvDMR1 in colorectal cancers. Cancer Sci. 2006, 97, 1147–1154. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.J.; Bikle, D.D. LncRNA profiling reveals new mechanism for VDR protection against skin cancer formation. J. Steroid Biochem. Mol. Biol. 2014, 144, 87–90. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, F.; Mondal, T.; Guseva, N.; Pandey, G.K.; Kanduri, C. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development 2010, 137, 2493–2499. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Huang, M.; Zhao, H.; Wang, C.; Zhao, X.; Jiang, X.; Bian, S.; He, Y.; Gao, Y. A novel tetranucleotide repeat polymorphism within KCNQ1OT1 confers risk for hepatocellular carcinoma. DNA Cell Biol. 2013, 32, 628–634. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, B.A.; Weng, Y.I.; Liu, T.M.; Zuo, T.; Hsu, P.Y.; Lin, C.H.; Cheng, A.L.; Cui, H.; Yan, P.S.; Huang, T.H. Estrogen-mediated epigenetic repression of the imprinted gene cyclin-dependent kinase inhibitor 1C in breast cancer cells. Carcinogenesis 2011, 32, 812–821. [Google Scholar] [CrossRef] [PubMed]
- Imam, H.; Bano, A.S.; Patel, P.; Holla, P.; Jameel, S. The lncRNA NRON modulates HIV-1 replication in a NFAT-dependent manner and is differentially regulated by early and late viral proteins. Sci. Rep. 2015, 5, 8639. [Google Scholar] [CrossRef] [PubMed]
- Kornienko, A.E.; Guenzl, P.M.; Barlow, D.P.; Pauler, F.M. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013, 11, 59. [Google Scholar] [CrossRef] [PubMed]
- Da Sacco, L.; Baldassarre, A.; Masotti, A. Bioinformatics Tools and Novel Challenges in Long Non-Coding RNAs (lncRNAs) Functional Analysis. Int. J. Mol. Sci. 2012, 13, 97–114. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Shen, B.; Tan, M.; Mu, X.; Qin, Y.; Zhang, F.; Liu, Y. Long non-coding RNA UCA1 increases chemoresistance of bladder cancer cells by regulating Wnt signaling. FEBS J. 2014, 281, 1750–1758. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Li, X.; Wang, Y.; Zhao, L.; Chen, W. Long non-coding RNA UCA1 regulated cell cycle distribution via CREB through PI3-K dependent pathway in bladder carcinoma cells. Gene 2012, 496, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Glover, A.R.; Zhao, J.T.; Ip, J.C.; Lee, J.C.; Robinson, B.G.; Gill, A.J.; Soon, P.S.; Sidhu, S.B. Long noncoding RNA profiles of adrenocortical cancer can be used to predict recurrence. Endocr. Relat. Cancer 2015, 22, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Szegedi, K.; Sonkoly, E.; Nagy, N.; Nemeth, I.B.; Bata-Csorgo, Z.; Kemeny, L.; Dobozy, A.; Szell, M. The anti-apoptotic protein G1P3 is overexpressed in psoriasis and regulated by the non-coding RNA, PRINS. Exp. Dermatol. 2010, 19, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Gao, T.; Sun, Y.; Zheng, X.; Wang, J.; Ma, J.; Hu, X.; Li, J.; Hu, M. LncRNA expression profile reveals the potential role of lncRNAs in gastric carcinogenesis. Cancer Biomark. 2015. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Kim, C.; Ku, J.L.; Kim, W.; Yoon, S.K.; Kuh, H.J.; Lee, J.H.; Nam, S.W.; Lee, E.K. A long non-coding RNA snaR contributes to 5-fluorouracil resistance in human colon cancer cells. Mol. Cells 2014, 37, 540–546. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, J.; Siegel, A.B.; Remotti, H.; Wang, Q.; Shen, Y.; Santella, R.M. Exploration of Deregulated Long Non-Coding RNAs in Association with Hepatocarcinogenesis and Survival. Cancers 2015, 7, 1847-1862. https://doi.org/10.3390/cancers7030865
Shen J, Siegel AB, Remotti H, Wang Q, Shen Y, Santella RM. Exploration of Deregulated Long Non-Coding RNAs in Association with Hepatocarcinogenesis and Survival. Cancers. 2015; 7(3):1847-1862. https://doi.org/10.3390/cancers7030865
Chicago/Turabian StyleShen, Jing, Abby B. Siegel, Helen Remotti, Qiao Wang, Yueyue Shen, and Regina M. Santella. 2015. "Exploration of Deregulated Long Non-Coding RNAs in Association with Hepatocarcinogenesis and Survival" Cancers 7, no. 3: 1847-1862. https://doi.org/10.3390/cancers7030865