Development of Deep Learning with RDA U-Net Network for Bladder Cancer Segmentation
Abstract
:Simple Summary
Abstract
1. Introduction
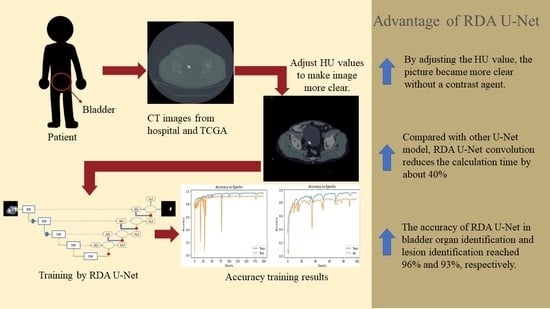
- Improve the speed and accuracy of identifying organs and lesions without contrast agent injection.
- Compared with Attention U-Net and Attention Dense-U-Net, RDA U-Net convolution reduces the calculation time by about 40%.
- With a mixture of hospital and open-source image data, RDA U-Net can identify various abdominal organs accurately.
2. Method
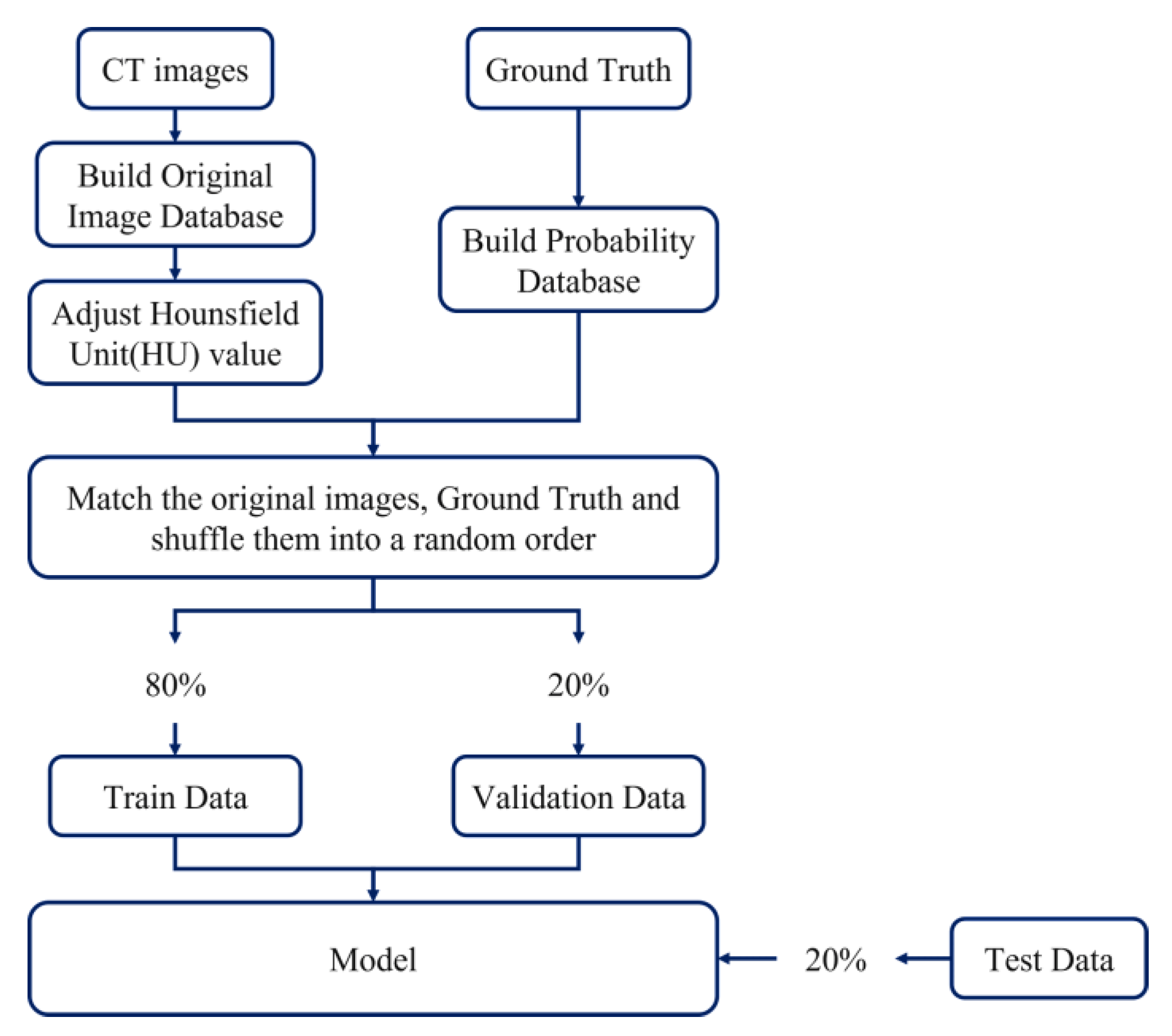
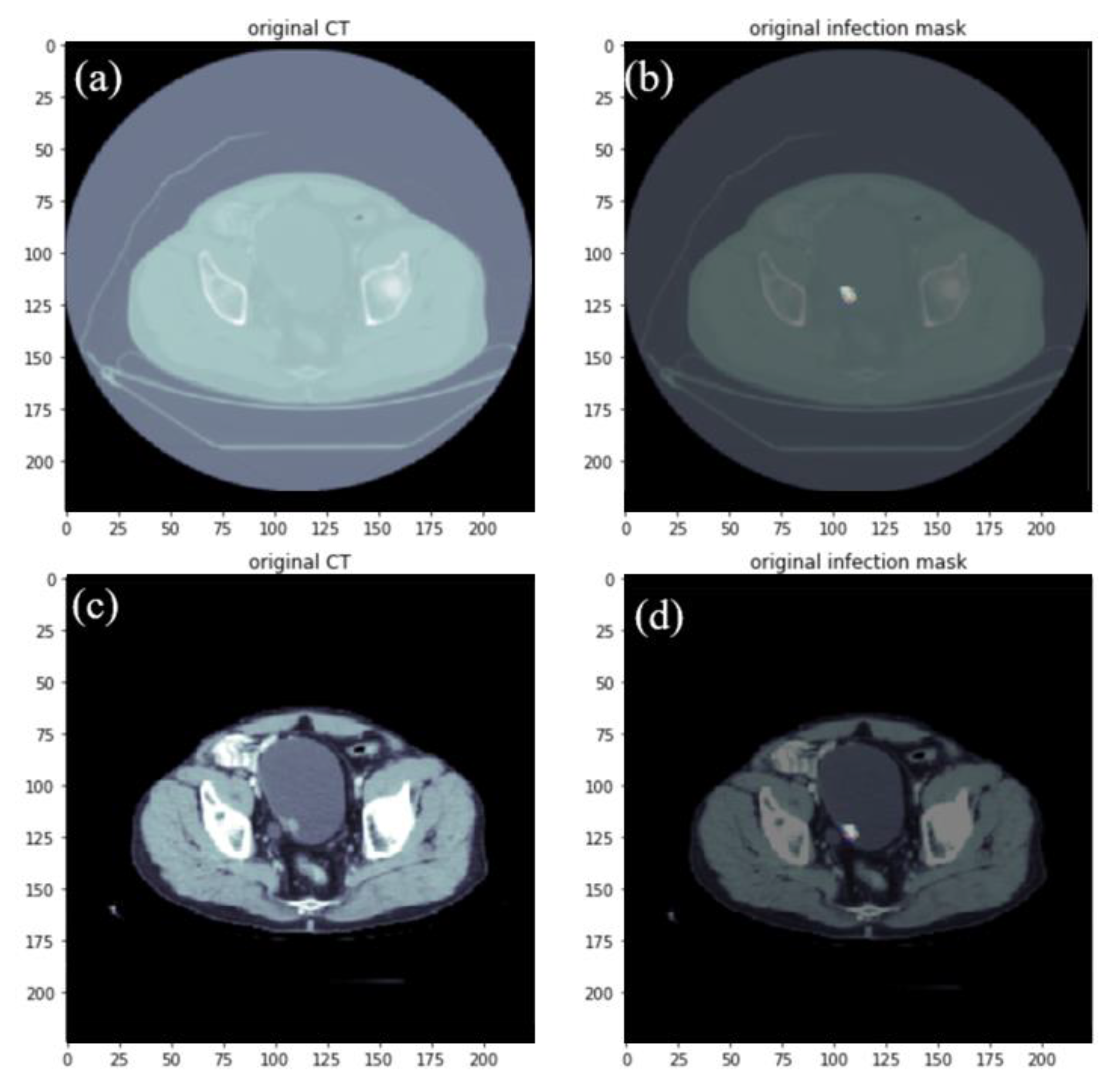
2.1. Data Set-Up and Image Pre-Processing
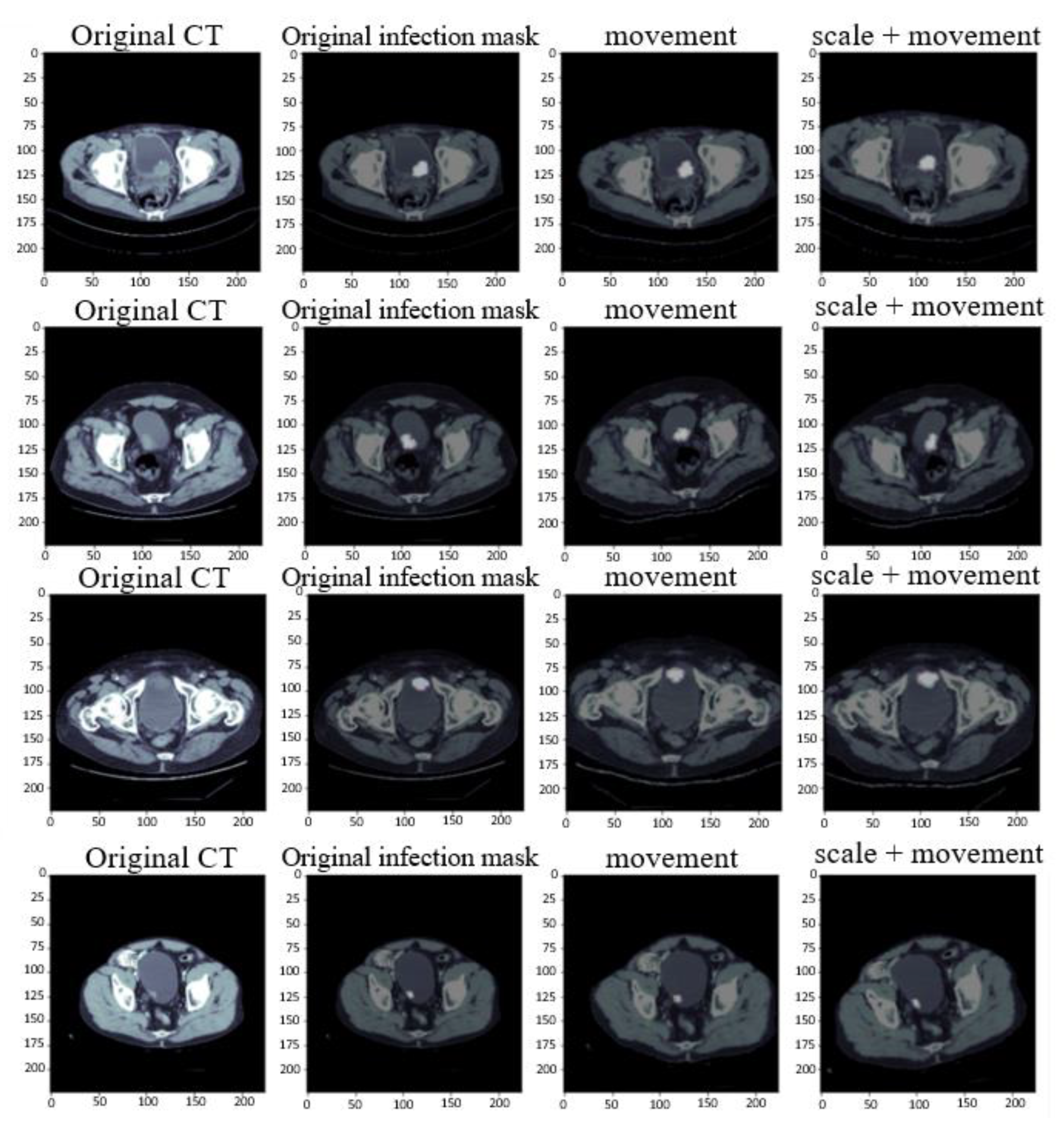
2.2. Data Augmentation
- Size scaling: For zooming in and out of the image, this research uses the original size of the image and zooms to 120% of the original size for training.
- Rotation: The image is randomly translated and rotated at the specified angle. This research uses a rotation translation of plus or minus 15 degrees.
- Shifting: The pixel moves the image horizontally and vertically. This research moves the image on the x and y axes by plus or minus 10.
- Shearing: The random amount of the image is clipped according to the set angle range. The set value in this research is between plus and minus 5 degrees.
- Horizontal flip: The image is flipped based on the horizontal direction. In this paper, 50% of the images are converted.
2.3. Model Structure
2.4. Encoder
2.5. Decoder
2.6. Training Data
3. Evaluation Metrics
4. Result and Discussion
4.1. Parameter Setting of Experimental Learning Rate
4.2. Comparison of Bladder Training Time and Convolution Parameters
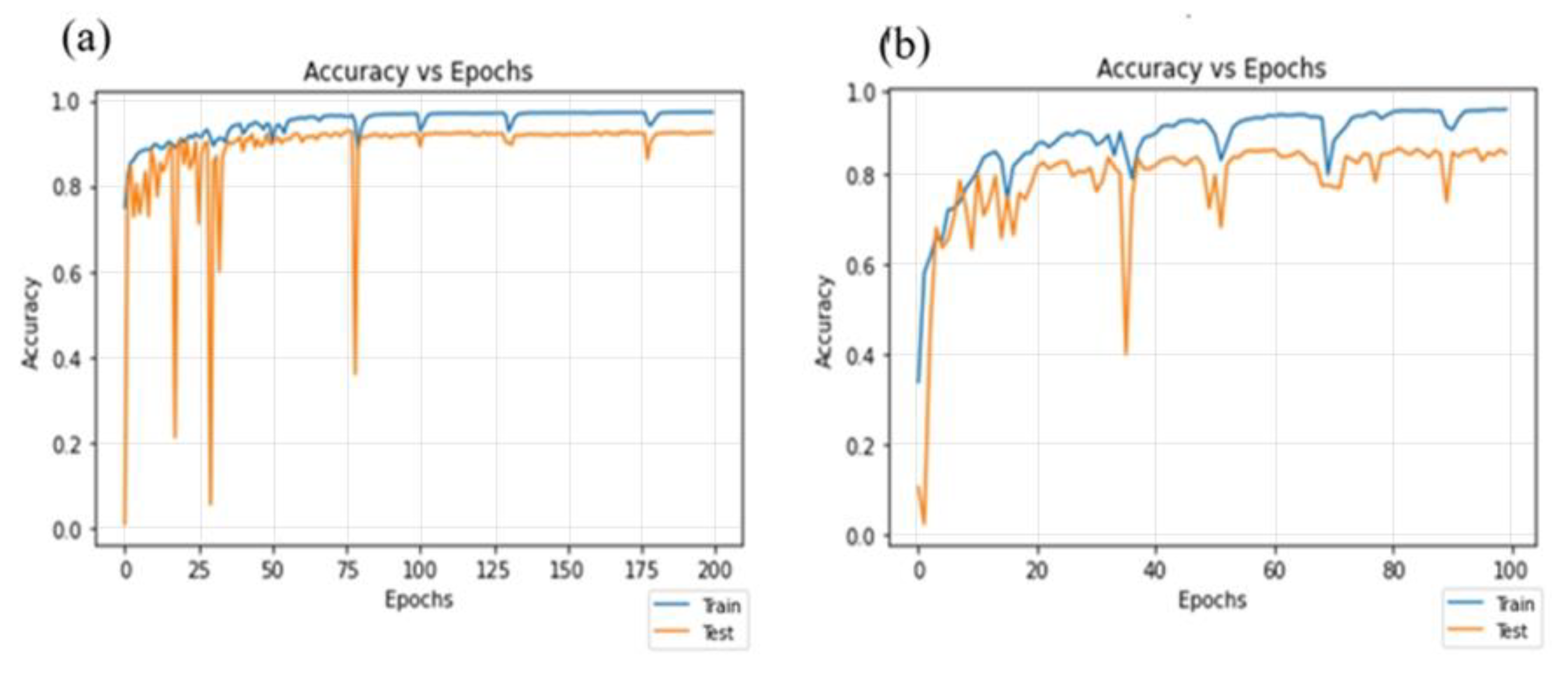
4.3. Residual-Dense Attention U-Net Model Convergence Curve
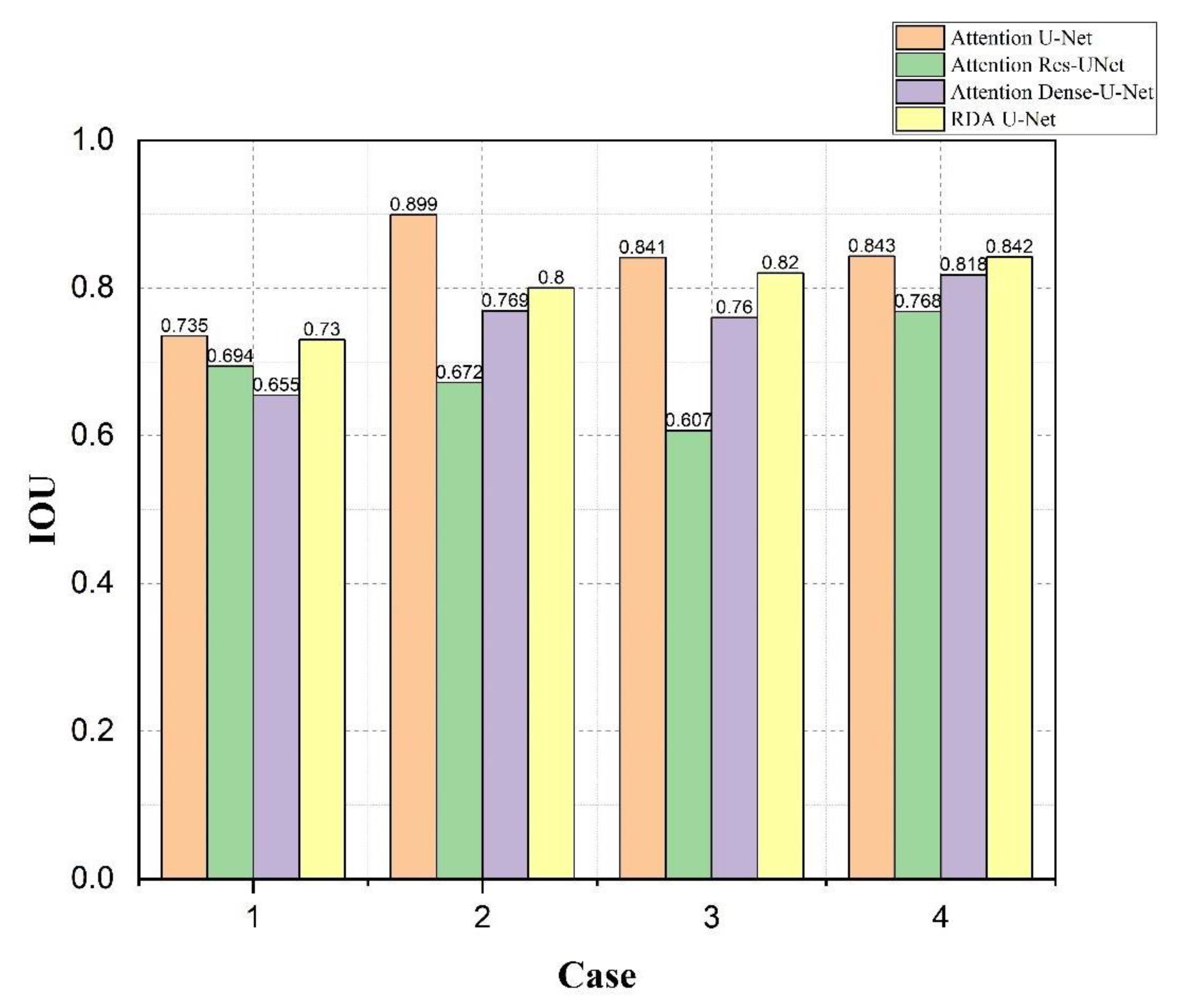
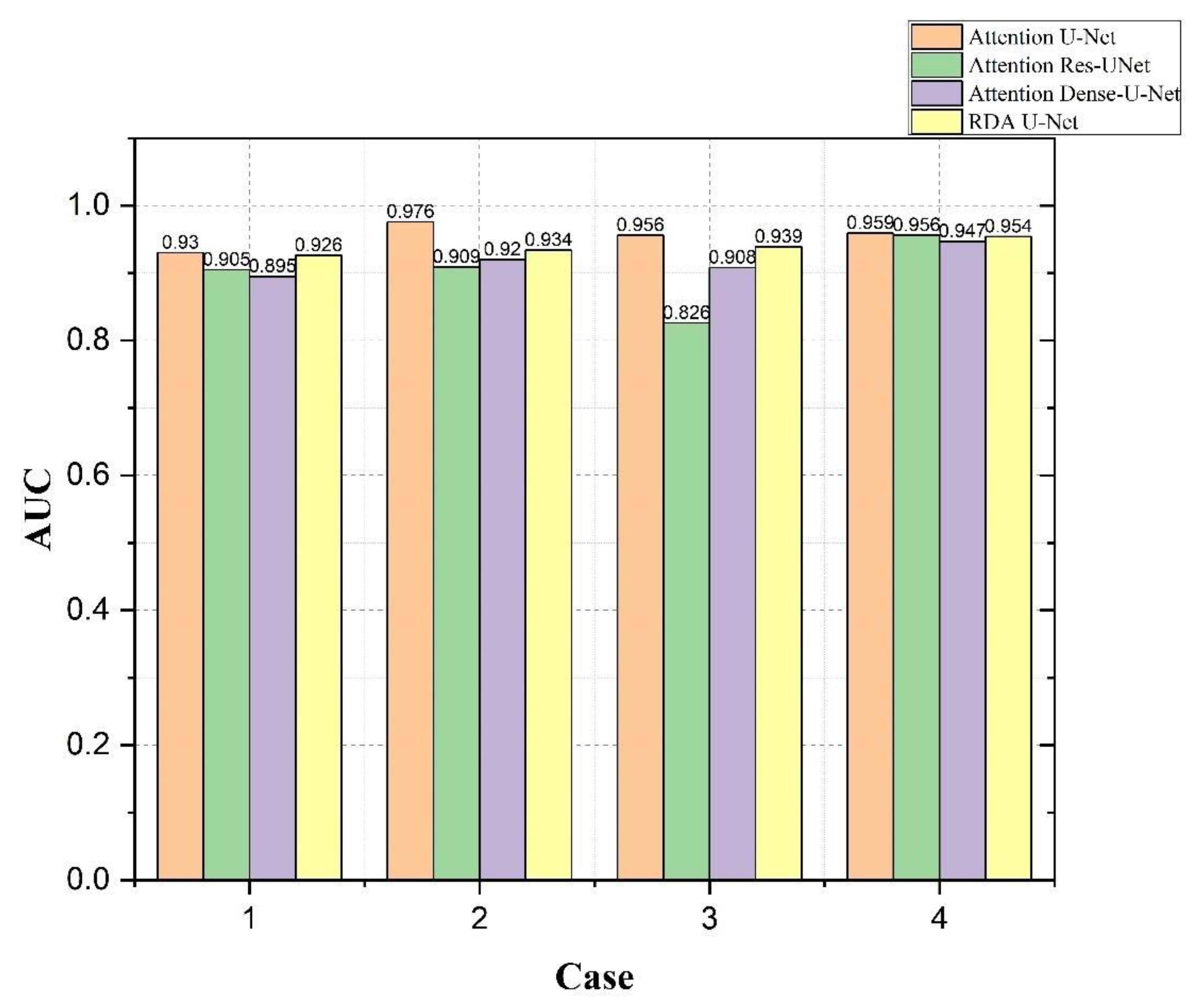
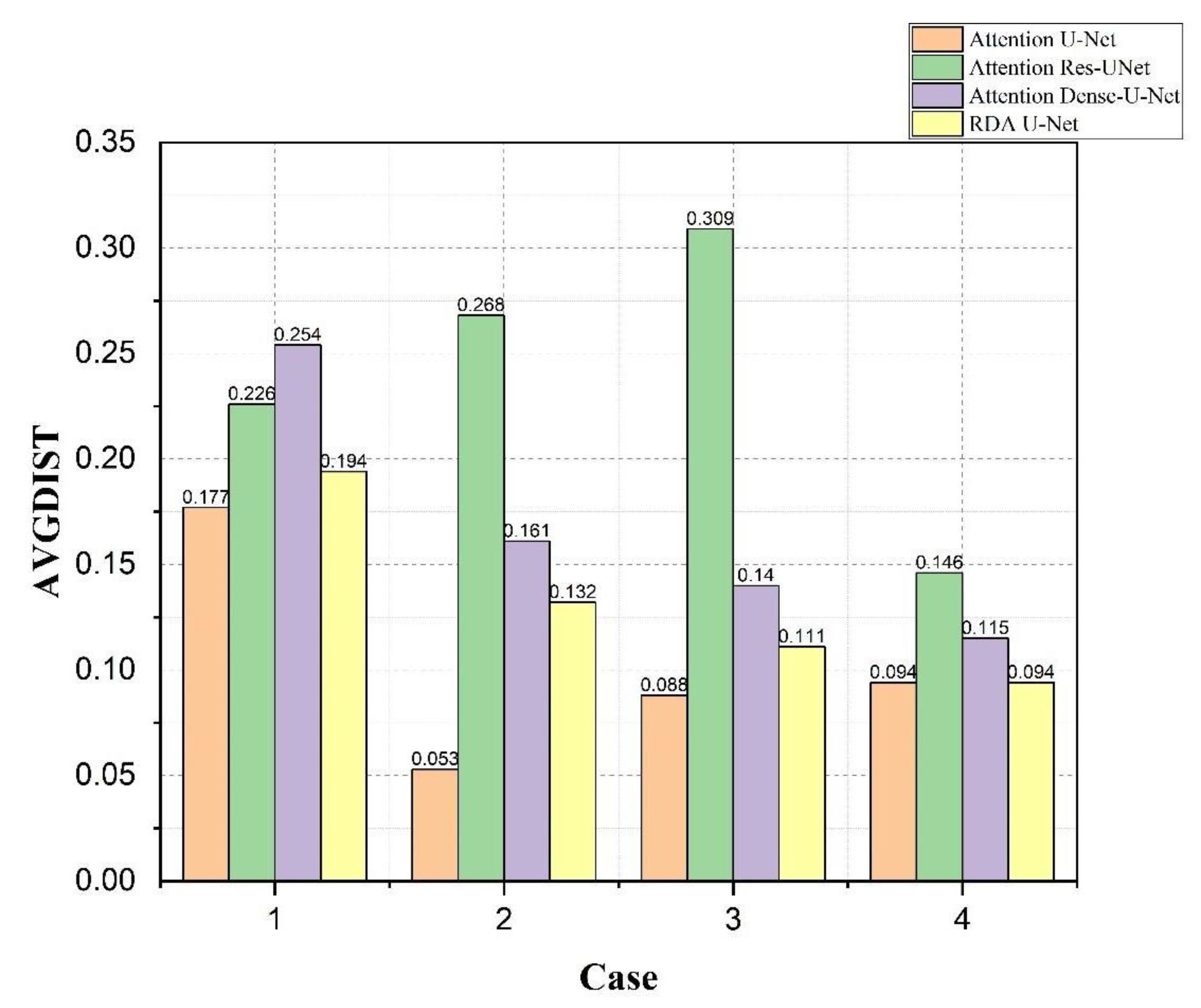
4.4. Bladder and Lesion Segmentation Results
4.5. Analysis of the Results of Bladder Cancer
5. Conclusions
- By changing the HU value, we made the picture clearer without a contrast agent. Through experiments, we confirmed that the training speed of our model was much lower than that of other models.
- Through experiments, it was confirmed that RDA U-NET reduced the computing time by about 40% compared with the proposed strategy.
- With the data from Kaohsiung Medical University and Open-Source Imaging (TCGA), the accuracy of RDA U-Net in bladder organ identification and lesion identification reached 96% and 93%, respectively.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lenis, A.T.; Patrick, M.; Lec, M.P.; Chamie, K. Bladder cancer: A review. JAMA 2020, 324, 1980–1991. Available online: https://jamanetwork.com/journals/jama/articlepdf/2772966/jama_lenis_2020_rv_200011_1605024530.47615.pdf (accessed on 11 October 2022). [CrossRef]
- KBcommonhealth “Bladder Cancer”. Available online: https://kb.commonhealth.com.tw/library/273.html?from=search#data-1-collapse (accessed on 13 September 2022).
- Lerner, S.P.; Schoenberg, M.; Sternberg, C. Textbook of Bladder Cancer, 1st ed.; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar]
- Lerner, S.P.; Schoenberg, M.; Sternberg, C.N. Bladder Cancer: Diagnosis and Clinical Management, 1st ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2015. [Google Scholar]
- Gassenmaier, S.; Warm, V.; Nickel, D.; Weiland, E.; Herrmann, J.; Almansour, H.; Wessling, D.; Afat, S. Thin-Slice Prostate MRI Enabled by Deep Learning Image Reconstruction. Cancers 2023, 15, 578. [Google Scholar] [CrossRef]
- Parikh, A.R.; Leshchiner, I.; Elagina, L.; Goyal, L.; Levovitz, C.; Siravegna, G.; Livitz, D.; Rhrissorrakrai, K.; Martin, E.E.; Seventer, E.E.V.; et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat. Med. 2019, 25, 1415–1421. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, J.; Nett, B.; Yu, Z.; Sauer, K.; Thibault, J.B.; Bouman, C.A. Recent Advances in CT Image Reconstruction. Curr. Radiol. Rep. 2013, 1, 39–51. [Google Scholar] [CrossRef] [Green Version]
- Yasaka, K.; Akai, H.; Abe, O.; Kiryu, S. Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast-enhanced CT: A preliminary study. Radiology 2018, 286, 887–896. Available online: https://pubs.rsna.org/doi/pdf/10.1148/radiol.2017170706 (accessed on 29 December 2022). [CrossRef] [PubMed] [Green Version]
- Tanabe, K.; Ikeda, M.; Hayashi, M.; Matsuo, K.; Yasaka, M.; Machida, H.; Shida, M.; Katahira, T.; Imanishi, T.; Hirasawa, T.; et al. Comprehensive Serum Glycopeptide Spectra Analysis Combined with Artificial Intelligence (CSGSA-AI) to Diagnose Early-Stage Ovarian Cancer. Cancers 2020, 12, 2373. [Google Scholar] [CrossRef] [PubMed]
- Janiesch, C.; Zschech, P.; Heinrich, K. Machine learning and deep learning. Electron. Mark. 2021, 31, 685–695. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. nature 2015, 521, 436–444. Available online: https://www.nature.com/articles/nature14539.pdf (accessed on 13 September 2022). [CrossRef]
- Esteva, A.; Robicquet, A.; Ramsundar, B.; Kuleshov, V.; DePristo, M.; Chou, K.; Cui, C.; Corrado, G.; Thrun, S.; Dean, J. A guide to deep learning in healthcare. Nat. Med. 2019, 25, 24–29. [Google Scholar] [CrossRef]
- Erickson, B.J.; Korfiatis, P.; Akkus, Z.; Kline, T.L. Machine learning for medical imaging. Radiographics 2017, 37, 505. Available online: https://pubs.rsna.org/doi/pdf/10.1148/rg.2017160130 (accessed on 20 December 2022). [CrossRef] [Green Version]
- Giger, M.L. Machine learning in medical imaging. J. Am. Coll. Radiol. 2018, 15, 512–520. [Google Scholar] [CrossRef] [PubMed]
- Pratondo, A.; Chui, C.H.; Ong, S.H. Integrating machine learning with region-based active contour models in medical image segmentation. J. Vis. Commun. Image Represent. 2017, 43, 1–9. [Google Scholar] [CrossRef]
- Robinson, K.R. Machine Learning on Medical Imaging for Breast Cancer Risk Assessment; The University of Chicago: Chicago, IL, USA, 2019. [Google Scholar]
- Wernick, M.N.; Yang, Y.; Brankov, J.G.; Yourganov, G.; Strother, S.C. Machine learning in medical imaging. IEEE Signal Process. Mag. 2010, 27, 25–38. [Google Scholar] [CrossRef] [Green Version]
- Obayya, M.; Maashi, M.S.; Nemri, N.; Mohsen, H.; Motwakel, A.; Osman, A.E.; Alneil, A.A.; Alsaid, M.I. Hyperparameter Optimizer with Deep Learning-Based Decision-Support Systems for Histopathological Breast Cancer Diagnosis. Cancers 2023, 15, 885. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Jiang, W.; Qiu, X. Deep learning for COVID-19 detection based on CT images. Sci. Rep. 2021, 11, 14353. [Google Scholar] [CrossRef] [PubMed]
- Zováthi, B.H.; Mohácsi, R.; Szász, A.M.; Cserey, G. Breast Tumor Tissue Segmentation with Area-Based Annotation Using Convolutional Neural Network. Diagnostics 2022, 12, 2161. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi-Zadeh, S.A.; Amini, A.; Soltanian-Zadeh, H. Brain Tumor Segmentation Using U-net and U-net++ Networks. In Proceedings of the 2022 30th International Conference on Electrical Engineering (ICEE), Tehran, Iran, 17–19 May 2022; pp. 841–845. [Google Scholar]
- Baressi Šegota, S.; Lorencin, I.; Smolić, K.; Anđelić, N.; Markić, D.; Mrzljak, V.; Štifanić, D.; Musulin, J.; Španjol, J.; Car, Z. Semantic segmentation of urinary bladder cancer masses from ct images: A transfer learning approach. Biology 2021, 10, 1134. [Google Scholar] [CrossRef]
- Hu, H.; Zheng, Y.; Zhou, Q.; Xiao, J.; Chen, S.; Guan, Q. MC-Unet: Multi-scale convolution unet for bladder cancer cell segmentation in phase-contrast microscopy images. In Proceedings of the 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), San Diego, CA, USA, 18–21 November 2019; pp. 1197–1199. [Google Scholar]
- Yu, J.; Cai, L.; Chen, C.; Fu, X.; Wang, L.; Yuan, B.; Yang, X.; Lu, Q. Cascade Path Augmentation Unet for bladder cancer segmentation in MRI. Med. Phys. 2022, 49, 4622–4631. [Google Scholar] [CrossRef]
- Shan, H.; Padole, A.; Homayounieh, F.; Kruger, U.; Khera, R.D.; Nitiwarangkul, C.; Kalra, M.K.; Wang, G. Can deep learning outperform modern commercial CT image reconstruction methods? arXiv 2018, arXiv:1811.03691. [Google Scholar]
- National Cancer Institute. The Cancer Genome Atlas (TCGA). National Cancer Institute at the National Institutes of Health. Available online: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga (accessed on 2 September 2022).
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of theInternational Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Cham, Switzerland; pp. 234–241. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Razi, T.; Niknami, M.; Ghazani, F.A. Relationship between Hounsfield unit in CT scan and gray scale in CBCT. J. Dent.Res. Dent.Clin. Dent. Prospect. 2014, 8, 107. [Google Scholar]
- Varma, V.; Mehta, N.; Kumaran, V.; Nundy, S. Indications and contraindications for liver transplantation. Int. J. Hepatol. 2011, 2011, 121862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Dumoulin, V.; Visin, F. A guide to convolution arithmetic for deep learning. arXiv 2016, arXiv:1603.07285. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Radke, K.L.; Kors, M.; Müller-Lutz, A.; Frenken, M.; Wilms, L.M.; Baraliakos, X.; Wittsack, H.-J.; Distler, J.H.W.; Abrar, D.B.; Antoch, G.; et al. Adaptive IoU Thresholdingfor Improving Small Object Detection:A Proof-of-Concept Study of HandErosions Classification of Patientswith Rheumatic Arthritis on X-ray Images. Diagnostics 2023, 13, 104. [Google Scholar] [CrossRef]
- Aydin, O.U.; Taha, A.A.; Hilbert, A.; Khalil, A.A.; Galinovic, I.; Fiebach, J.B.; Frey, D.; Madai, V.I. On the usage of average Hausdorff distance for segmentation performance assessment: Hidden error when used for ranking. Eur. Radiol. Exp. 2021, 5, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.-F.; Ou, H.-Y.; Lin, H.-Y.; Wei, C.-P.; Liao, C.-C.; Cheng, Y.-F.; Pan, C.-T. Development of Novel Residual-Dense-Attention (RDA) U-Net Network Architecture for Hepatocellular Carcinoma Segmentation. Diagnostics 2022, 12, 1916. [Google Scholar] [CrossRef]
- Li, S.; Dong, M.; Du, G.; Mu, X. Attention dense-u-net for automatic breast mass segmentation in digital mammogram. IEEE Access 2019, 7, 59037–59047. [Google Scholar] [CrossRef]
- Maji, D.; Sigedar, P.; Singh, M. Attention Res-U-Net with Guided Decoder for semantic segmentation of brain tumors. Biomed. Signal Process. Control. 2022, 71, 103077. [Google Scholar] [CrossRef]
- Namdar, K.; Haider, M.A.; Khalvati, F.A. Modified AUC for Training Convolutional Neural Networks: Taking Confidence Into Account. Front. Artif. Intell. 2021, 4, 582928. [Google Scholar] [CrossRef] [PubMed]






| Method | Parameter | Bladder Tumor-Training Time (s) | Bladder-Training Time (s) |
|---|---|---|---|
| RDA U-Net | 13,053,861 | 41 | 42 |
| Attention U-Net [32] | 35,238,293 | 56 | 57 |
| Attention Dense-U-Net [38] | 14,374,021 | 59 | 60 |
| Attention Res-U Net [39] | 12,981,573 | 40 | 40 |
| Method | ACC | DSC | IoU | AVGDIST |
|---|---|---|---|---|
| RDA U-Net | 0.9656 | 0.9745 | 0.9505 | 0.0269 |
| Attention U-Net [32] | 0.9610 | 0.9771 | 0.9553 | 0.0245 |
| Attention Dense-U-Net [38] | 0.9721 | 0.9811 | 0.9631 | 0.0187 |
| Attention Res-U-Net [39] | 0.9614 | 0.9717 | 0.9452 | 0.0291 |
| Method | ACC | DSC | IoU | AVGDIST |
|---|---|---|---|---|
| RDA U-Net | 0.9394 | 0.8895 | 0.8024 | 0.1279 |
| Attention U-Net [32] | 0.9330 | 0.8993 | 0.8184 | 0.1141 |
| Attention Dense-U-Net [38] | 0.9258 | 0.8797 | 0.7869 | 0.1406 |
| Attention Res-U-Net [39] | 0.8928 | 0.8322 | 0.7150 | 0.2080 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, M.-C.; Wang, S.-Y.; Pan, C.-T.; Chien, M.-Y.; Li, W.-M.; Xu, J.-H.; Luo, C.-H.; Shiue, Y.-L. Development of Deep Learning with RDA U-Net Network for Bladder Cancer Segmentation. Cancers 2023, 15, 1343. https://doi.org/10.3390/cancers15041343
Lee M-C, Wang S-Y, Pan C-T, Chien M-Y, Li W-M, Xu J-H, Luo C-H, Shiue Y-L. Development of Deep Learning with RDA U-Net Network for Bladder Cancer Segmentation. Cancers. 2023; 15(4):1343. https://doi.org/10.3390/cancers15041343
Chicago/Turabian StyleLee, Ming-Chan, Shao-Yu Wang, Cheng-Tang Pan, Ming-Yi Chien, Wei-Ming Li, Jin-Hao Xu, Chi-Hung Luo, and Yow-Ling Shiue. 2023. "Development of Deep Learning with RDA U-Net Network for Bladder Cancer Segmentation" Cancers 15, no. 4: 1343. https://doi.org/10.3390/cancers15041343
APA StyleLee, M.-C., Wang, S.-Y., Pan, C.-T., Chien, M.-Y., Li, W.-M., Xu, J.-H., Luo, C.-H., & Shiue, Y.-L. (2023). Development of Deep Learning with RDA U-Net Network for Bladder Cancer Segmentation. Cancers, 15(4), 1343. https://doi.org/10.3390/cancers15041343